Abstract
Although importin α (Imp α) has been shown to act as the receptor for basic nuclear localization signals (NLSs) and to mediate their recruitment to the importin β nuclear import factor, little is known about the functional domains present in Imp α, with the exception that importin β binding is known to map close to the Imp α NH2 terminus. Here, we demonstrate that sequences essential for binding to the CAS nuclear export factor are located near the Imp α COOH terminus and include a critical acidic motif. Although point mutations introduced into this acidic motif inactivated both CAS binding and Imp α nuclear export, a putative leucine-rich nuclear export signal proved to be neither necessary nor sufficient for Imp α nuclear export. Analysis of sequences within Imp α that bind to the SV-40 T antigen NLS or to the similar LEF-1 NLS revealed that both NLSs interact with a subset of the eight degenerate armadillo (Arm) repeats that form the central part of Imp α. However, these two NLS-binding sites showed only minimal overlap, thus suggesting that the degeneracy of the Arm repeat region of Imp α may serve to facilitate binding to similar but nonidentical basic NLSs. Importantly, the SV-40 T NLS proved able to specifically inhibit the interaction of Imp α with CAS in vitro, thus explaining why the SV-40 T NLS is unable to also function as a nuclear export signal.
Keywords: importin α, nuclear export, nuclear import, nuclear localization signal
Nuclear import of proteins bearing a nuclear localization signal (NLS)1 of the basic type first defined in SV-40 T antigen and nucleoplasmin is dependent on two cellular factors termed importin α (Imp α) or karyopherin α and importin β (Imp β) or karyopherin β1 (for review see Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Weis, 1998). Imp α functions as the NLS receptor (Görlich et al., 1994; Weis et al., 1995) and also directly binds to Imp β via a specific domain, located between residues 10 and 55, termed the Imp β binding (IBB) domain (Görlich et al., 1996a ; Weis et al., 1996). The resultant heterotrimeric complex is _targeted to the nuclear pore complex due to the direct interaction of Imp β with specific nucleoporins (Görlich et al., 1995; Moroianu et al., 1995). The heterotrimer is then translocated into the nucleus via a poorly understood, energy-dependent process that also requires the activity of the Ran GTPase and a Ran cofactor termed NTF2 or p10 (Moore and Blobel, 1993; Nehrbass and Blobel, 1996; Paschal et al., 1996). Once the heterotrimer reaches the nucleus, Imp β directly interacts with GTP-bound Ran, a form of Ran that is largely confined to the nucleus, resulting in the release of Imp α and the NLS protein cargo (Rexach and Blobel, 1995; Görlich et al., 1996b ; Izaurralde et al., 1997). Both Imp α and Imp β are then separately recycled back to the cytoplasm, where they can once again support nuclear import. Although Imp α is a critical participant in basic NLS-dependent protein import, it actually serves only as an adapter to recruit NLS-containing proteins to Imp β, which is the true mediator of nuclear transport. This is demonstrated by the finding that the Imp α IBB domain is itself a potent NLS when attached to a carrier protein (Görlich et al., 1996a ; Weis et al., 1996) and also by the identification of at least one other NLS that is Imp β dependent but Imp α independent for nuclear import (Truant et al., 1998a ).
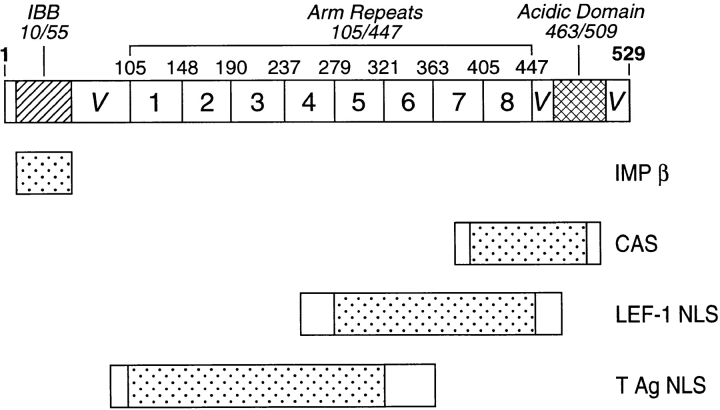
Although Imp β is a member of a family of related proteins, including the distinct nuclear import factor transportin and the nuclear export factors Crm1 and CAS, only Imp β is believed to participate in Imp α–dependent nuclear import processes (Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Weis, 1998). On the other hand, there are at least five different human forms of Imp α that display significantly different tissue expression patterns (Cortes et al., 1994; Cuomo et al., 1994; Görlich et al., 1994; O'Neill and Palese, 1995; Weis et al., 1995; Köhler et al., 1997; Tsuji et al., 1997; Nachury et al., 1998) and that, in some cases, have been shown to interact differentially with specific basic NLS sequences (Miyamoto et al., 1997; Nadler et al., 1997; Sekimoto et al., 1997). As suggested by Köhler et al. (1997), these Imp α family members are here termed Imp α1 (also termed hSRP1 or NPI-1), Imp α2 (also termed Rch1, hSRP1α, or pendulin), Imp α3 (also termed Qip1), Imp α4 (also termed hSRP1γ), and Imp α6. These five variants, which vary in size between 521 and 538 amino acids (aa) in length, can be grouped into three Imp α subfamilies consisting, respectively, of α2, of α1 and α6, and finally of α3 and α4, where proteins within one grouping are ∼80% identical and proteins in different groups are ∼50% identical (Köhler et al., 1997; Tsuji et al., 1997). Alignment of these five proteins has permitted a rough structural domain organization of Imp α to be proposed (see Fig. 1). In brief, the core of all five human Imp α proteins consists of eight degenerate repeats of an ∼42 aa, relatively hydrophobic sequence termed the armadillo (Arm) motif. Two additional conserved domains can also be identified. The first of these coincides with the IBB domain whereas the second conserved sequence, which is rich in acidic residues, extends from approximately residue 463– 509 (Köhler et al., 1997). Other regions of Imp α, i.e., the residues flanking the IBB and the acidic motif, tend to be poorly conserved in the different Imp α variants and have therefore been termed variable regions (Tsuji et al., 1997).
Figure 1.
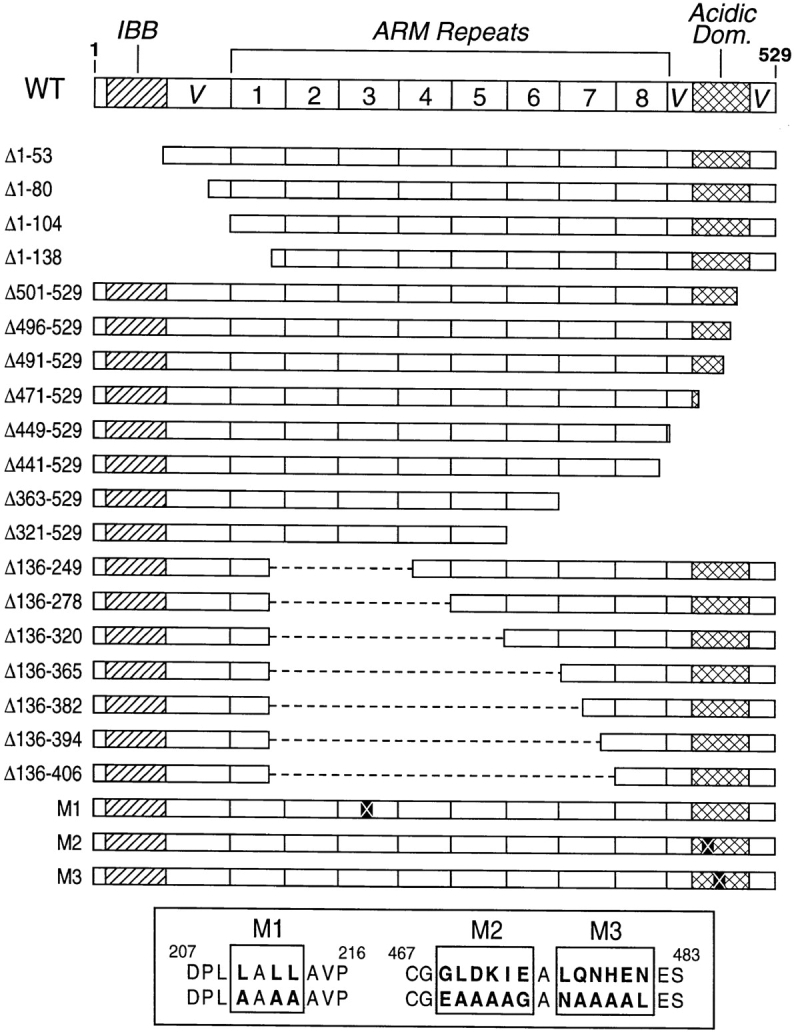
Structure of Imp α2 mutants. The conserved structural domains present in all Imp α family members are shown at the top of the figure whereas the structure of the Imp α2 deletion and missense mutants used in this work are given below. The names of the deletion mutants indicate which NH2 residues are missing in the particular mutant protein. The location of the M1, M2, and M3 missense mutations within Imp α2, and the identity of the introduced mutation, are given at the bottom.
In addition to sequences that interact with Imp β, Imp α is predicted to contain a binding site(s) for basic NLS sequences and also a nuclear export signal (NES) to permit its rapid return to the cytoplasm. Only in one case has the binding site for a basic NLS been mapped, i.e., for the RAG 1 NLS on Imp α1, where binding was shown to require the Arm motifs four through seven or five through eight (Cortes et al., 1994). Based on this result, and also on the finding that a form of Imp α1 lacking the first 77 aa and the last 63 aa retained the ability to bind the SV-40 T antigen NLS (T NLS), it has been suggested that basic NLS binding maps to the Arm repeat region of Imp α (Sekimoto et al., 1997). The issue of the Imp α NES is more complex, in that Imp α has, on one hand, been proposed to exit the nucleus via a leucine-rich NES located between residues 207 and 217 in Arm repeat 3 (Boche and Fanning, 1997), that would be predicted to be dependent on the Crm1 nuclear export factor (Fornerod et al., 1997; Stade et al., 1997), whereas on the other hand Imp α has also been proposed to exit the nucleus via an unmapped NES that is dependent on the distinct nuclear export factor CAS (Kutay et al., 1997a ).
In this article, we have attempted to more fully define the domain organization of the α2 form of Imp α and, in particular, to map Imp α2 sequences involved in NLS binding and nuclear export. We demonstrate that Imp α2 binds to CAS, but not to Crm1, in vivo and show that CAS binding is critically dependent on the conserved acidic domain located near the Imp α COOH terminus. Mutation of this acidic domain is shown to block the ability of Imp α2 to exit the nucleus whereas mutation of the putative Imp α2 leucine-rich NES has no effect. We also demonstrate that NLS binding by Imp α2 involves not only the Arm repeats but also the flanking variable regions and show that different NLS sequences bind to distinct regions within Imp α2. Finally, we demonstrate that Imp α is unable to simultaneously bind a basic NLS and the CAS nuclear export factor, thus providing an explanation for why such basic NLS sequences are unable to also function as an NES.
Materials and Methods
Construction of Molecular Clones
The human CAS cDNA sequence was amplified by PCR using primers containing unique 5′-BamHI and 3′-SalI restriction sites using pCAD/ CAS as a template (Brinkmann et al., 1995) and inserted into the polylinker sites BamHI and SalI present in pGBT9 (Clontech Laboratories, Palo Alto, CA). The resultant pGAL4/CAS plasmid is predicted to express a fusion protein consisting of the DNA-binding domain of GAL4 (aa 1–147) fused to the NH2 terminus of full-length CAS. pGAL4/Imp β was made by amplifying a full-length importin Imp β1 cDNA, using primers that introduce BamHI sites on both ends, with plasmid pGST/Imp β as a template (Truant et al., 1998a ). The PCR product was then cut with BamHI and ligated into pGBT9 digested with BamHI. The truncated form of Imp β was made similarly, using a 5′ primer that starts at aa 262. pGAL4/TNLS was made by inserting annealed oligos encoding the amino acids NH2-GGTPPKKKRKVEDP-COOH (boldfaced letters represent the T NLS) into the restriction sites EcoRI and SalI present in pGBT9. Plasmid pGAL4/LEF-1 encodes the GAL4 DNA-binding domain fused to aa 297–399 of LEF-1, which includes the LEF-1 NLS, and has been described (Prieve et al., 1996).
A cDNA encoding full-length human Imp α2 (Rch1) was isolated from a human T cell cDNA library by PCR amplification with primers introducing unique EcoRI (5′) and XhoI (3′) restriction sites. This cDNA was found to encode a predicted protein identical to the published Imp α2 (Rch1) sequence (Cuomo et al., 1994) except that residue 455 was lysine instead of glutamic acid. The Imp α2 cDNA was attached 3′ to, and in frame with, sequences encoding the VP16 transcription activation domain by insertion into the EcoRI and XhoI restriction sites present in pVP16 (Bogerd et al., 1995). Deletion mutants of Imp α2 were generated similarly by inserting PCR products (template pVP16/Imp α2) encoding the indicated amino acids (see Fig. 1) into the EcoRI and XhoI restriction sites of pVP16. The sequences encoding VP16/α2M1, M2, and M3 were generated using recombinant PCR with overlapping primers that introduced the missense mutations indicated in Fig. 1. The outside primers used in the second round of amplification were designed to allow efficient in-frame insertion into pVP16. The integrity of the resultant Imp α2 clones was confirmed by sequencing, using an Abi Prism 377 DNA Sequencer (Perkin Elmer Corp., Foster City, CA).
The pVP16/Imp α4 expression plasmid contains a cDNA encoding full-length human Imp α4 (hSRP1γ) (Köhler et al., 1997) isolated from a human HeLa cDNA library (Clontech Laboratories) by PCR amplification using primers that introduced flanking 5′ EcoRI and 3′ BglII restriction sites. The resulting PCR product was inserted into pVP16 digested with EcoRI and BamHI. The pVP16/Imp α1 expression plasmid contains a cDNA encoding full-length Imp α1 (hSRP1) (Weis et al., 1995) obtained by PCR amplification using the same HeLa cDNA library. The primers used introduced unique 5′ EcoRI and 3′ BamHI restriction sites that were then used to insert the open reading frame into the EcoRI and BamHI restriction sites present in the pVP16 polylinker.
The plasmid pSG424 (Sadowski and Ptashne, 1989) was used for construction of plasmids that allow expression of GAL4(1–147) fusion proteins in mammalian cells. To generate pGAL4/CAS, a cDNA sequence encoding full-length CAS was obtained by digesting the yeast expression plasmid pGAL4/CAS with BamHI and SalI and inserting the resultant fragment into pSG424 cut with the same restriction enzymes. Plasmid pGAL4/ImpβΔ1/261 was made by inserting a BamHI fragment derived from the corresponding yeast expression plasmid into the BamHI restriction site present in pSG424.
The VP16 fusion proteins used in the mammalian system were generated by inserting PCR products encoding Imp α2 or its mutant forms into pBC12/cytomegalovirus (CMV)/VP16 (Bogerd et al., 1995) using the restriction sites NcoI and BglII. Templates for amplification were the corresponding cDNA sequences present in the yeast expression plasmids. The resulting plasmids allow the expression of Imp α2 fusion proteins that contain a COOH-terminal VP16 domain. The mammalian indicator plasmid pG6(−31)HIVLTRΔTARCAT, referred to here as pG6/CAT, and the internal control plasmid pBC12/CMV/β-Gal have been described (Southgate and Green, 1991; Fridell et al., 1997), as have the mammalian two-hybrid expression constructs pGAL4/Crm1, pRev/VP16, and pRevM10/VP16 (Bogerd et al., 1995).
The pGST/α2/HIS expression plasmid, encoding wild-type Imp α2 fused to glutathione-S-transferase (GST) (NH2 terminal) and a 6× His tag (COOH terminal), was made by insertion of cDNA sequences encoding wild-type Imp α2 into pGEX 4T-1 (Pharmacia Biotech, Piscataway, NJ) using the restriction sites EcoRI and XhoI. A plasmid encoding a GST/ α2M1/HIS fusion protein was constructed using the identical approach. In both cases, the COOH-terminal 6× His tag was added by cutting the plasmid with the restriction enzymes PpuMI and XhoI. This digestion removes a fragment encoding the last 5 aa of Imp α2 including the stop codon. Then annealed oligos encoding the missing 5 aa, and a 6× His tag followed by a stop codon were inserted. The inserted sequence also contained an EcoRI and a SalI restriction site between the last aa of Imp α2 and the His tag. The pGST/α2 M2/HIS plasmid, containing the M2 missense mutation of Imp α2, was generated by inserting a NgoM1/PpuM1 fragment from the yeast expression plasmid pVP16/M2 into the same restriction sites present in pGST/α2/HIS. Plasmid pGST/α2Δ491/529/HIS was made by PCR amplification of a cDNA sequence encoding Imp α2 aa 364–490, using primers that contained a 5′ NgoMI restriction site as well as 3′ EcoRI and SalI restriction sites followed by a 6× HIS tag, a stop codon, and an XhoI site. This PCR product was then inserted into GST/ α2/HIS digested with NgoMI and XhoI.
Plasmid pHT-CAS was constructed by BamHI and SalI digesting plasmid pGAL4/CAS and ligating the resulting fragment into BamHI-SalI– digested pQE32 vector (QIAGEN, Santa Clarita, CA). The resultant construct encodes the CAS protein with an NH2-terminal 6-His tag. Plasmids pGSTα2 and pGSTα2M2 were constructed similarly by digesting plasmids pVP16/Impα2 and pVP16/Impα2M2 with EcoRI and XhoI and ligating the resulting fragments into EcoRI-XhoI–digested pGEX 4T-1 (Pharmacia Biotech). The resulting plasmids encode the Impα2wt or Impα2M2 protein fused to GST separated by a thrombin protease cleavage site. Plasmid pGSTRanQ69L has been described (Truant et al., 1998a ).
Yeast Two-hybrid Analysis
The interaction between Impα1, Impα4, and wild-type and mutant forms of Imp α2, on the one hand and CAS, wild-type and Δ1/261-deleted Imp β, the LEF-1 NLS and the T NLS on the other, was assayed using the yeast two-hybrid protein interaction system (Fields and Song, 1989). Plasmids encoding the appropriate GAL4(1–147) DNA-binding domain and VP16 fusion proteins were transformed into the yeast indicator strain Y190 (Harper et al., 1993) by standard techniques. After 3 d of growth at 30°C on selective culture plates, double transformants were transferred to selective medium. The following day, cultures were assayed for β-galactosidase (β-gal) activity (Bogerd et al., 1995). However, due to the slower growth of yeast cells containing pGAL4/LEF-1(297–399), these transformants were incubated for 5 d on plates and liquid cultures were incubated for ∼36 h at 30°C before β-gal activity was measured.
Mammalian Two-hybrid Assay
Human 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum, gentamycin, fungizone, and glutamine. On the day before transfection, cells were seeded in 6-well dishes at a density of 3 × 105 cells per 35-mm well. The cells were transfected with 0.5 μg of the reporter plasmid pG6/CAT, 2 μg of the plasmids encoding the appropriate VP16 and GAL4 fusion proteins, and 100 ng of pBC12/CMV/β-gal using the calcium phosphate method. At ∼48 h posttransfection, cell lysates were prepared and CAT and β-gal levels quantified as described (Bogerd et al., 1995).
Recombinant Protein Expression
GST fusion proteins were expressed in the Escherichia coli strains BL21 (Novagen, Madison, WI) (GST/α2/HIS, GST/α2 M2/HIS) or TOPP1 (Stratagene, La Jolla, CA) (GST/α2 M1/HIS, GST/α2Δ491/529/HIS). Overnight cultures were diluted 1:5 and then grown for 4.5 h without induction. Cells were collected by centrifugation, resuspended in GST buffer (20 mM Hepes, pH 7.4, 0.5 M NaCl, 10% glycerol) and lysed by sonication. The lysate was centrifuged and the GST fusion protein present in the supernatant absorbed to glutathione–Sepharose 4B (Pharmacia Biotech.), washed with GST buffer, and then eluted with elution buffer (1× PBS, 10 mM glutathione, pH 7.4). The proteins were then concentrated in a centricon-10 concentrator (Amicon, Beverly, MA).
For use in affinity chromatography assays, wild-type and M2 mutant forms of Impα2, as well as the Q69L mutant of Ran, which does not support hydrolysis of bound GTP (Bischoff et al., 1994), were expressed, purified, and cleaved as described (Truant et al., 1998a ). CAS protein was expressed in the E. coli DH5α strain. After 2 h of growth of a 1:10 dilution from an overnight culture in 2XYT medium at 37°C, the culture was induced with 0.5 mM isopropylthio-β-d-galactoside (IPTG) for 4 h at 37°C. After harvesting by centrifugation, the cells were lysed by sonication, centrifuged, and then the supernatant was saved. The pellet was detergent extracted (B-Per; Pierce Chemical Co., Rockford, IL), recentrifuged, and the second supernatant was incubated along with the original supernatant with nickel-agarose beads (QIAGEN). The His-tagged CAS protein was eluted from the nickel-agarose beads on an imidazole gradient and then further purified on a Q–Sepharose ion exchange column (Pharmacia Biotech). SV-40 T NLS peptides were made synthetically. The peptide sequences used were: T NLS wild type, 125-YPKKKRKVEDP-135 and T NLS mutant(K128T), 125-YPKtKRKVEDP-135.
Protein Affinity Chromatography
Purified wild-type and M2 mutant Imp α2 proteins were covalently coupled at a 6 mg/ml concentration to active ester-agarose beads (Affi-10; Bio-Rad Laboratories, Hercules, CA). The protein-coupled beads were then used to make 10-μl columns in 100-μl borosilicate glass micropipets. Approximately 4 μg of CAS protein was preincubated on ice with 6 μg RanQ69L and 1 mM GTP in ACB buffer (10 mM Hepes, pH 7.4, 10% glycerol, 50 mM NaCl, 1 mM MgCl2) in a 75-μl final volume. This entire mix was then loaded onto microcolumns containing either wild-type or M2 mutant Imp α2. The columns were then washed with 6-column vol of 100 mM NaCl ACB buffer and finally eluted with 5-column vol 500 mM MgCl2. The entire bound fraction was analyzed by 10% SDS-PAGE (Ready-gels; Bio-Rad Laboratories) and visualized by Coomassie blue staining (R-250, Life Technologies, Inc.). For SV-40 T NLS peptide competition experiments, 100 μg (∼100 fold molar excess) of SV-40 T NLS wild-type or mutant peptides were mixed with the CAS/RanQ69L/GTP mixture before loading onto the Impα2 columns.
Mammalian Cell Microinjection
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum, gentamycin, fungizone, and glutamine. 2 d before microinjection, HeLa cells were seeded onto CELLocate microgrid coverslips (Eppendorf Scientific, Hamburg, Germany) at a density of 2 × 105 cells per 35-mm dish. To increase the percentage of multinucleated cells, cultures were serum starved overnight. The next day, cells were replenished with serum containing media 4–6 h before microinjection. The proteins (final concentration in PBS ∼2 μg/μl) were coinjected with a rhodamine-conjugated goat IgG tracer (final concentration 1.5 μg/μl; Cappel Laboratories, Malvern, PA) to verify the site of injection. After injection, cells were incubated at 37°C for 35 min and then fixed with 3% paraformaldehyde in PBS (Fridell et al., 1997). The GST fusion protein was visualized by indirect immunofluorescence using a polyclonal affinity-purified rabbit anti–GST antibody and fluorescein-conjugated donkey anti–rabbit antiserum. Cellular localization of the injected proteins was determined using a Leica DMRB fluorescence microscope (Leica USA, Deerfield, IL) at a 100× magnification.
Results
As noted above, Imp α is predicted to interact with Imp β, with a range of different basic NLS sequences and with at least one nuclear export factor. To test whether this prediction is correct, we asked whether the α2 (RchI) form of Imp α would be able to interact with Imp β, with CAS, and with two different NLS sequences, derived from the T NLS (Kalderon et al., 1984) or the transcription factor LEF-1 (Prieve et al., 1996), in the yeast two-hybrid protein–protein interaction assay.
As shown in Table I, full-length Imp α2 proved able to specifically interact with full-length human CAS and with both the T NLS and the LEF-1 NLS, as shown by the readily detectable activation of β-gal expression in cells expressing a VP16/Imp α2 fusion protein and the appropriate GAL4 fusions. However, Imp α2 failed to interact with the GAL4 DNA-binding domain itself or with full-length human Imp β. This latter result is expected, as the interaction of Imp β with Ran-GTP in the nucleus is predicted to block Imp α binding (Rexach and Blobel, 1995; Görlich et al., 1996b ). We therefore removed most of the Ran-GTP–binding domain of Imp β, which is known to be located at the Imp β NH2 terminus (Chi and Adam, 1997; Kutay et al., 1997b ), by deletion of Imp β residues 1–261. The resultant Imp βΔ1/261 protein proved able to effectively interact with Imp α2 in the two-hybrid assay (Table I).
Table I.
Interaction of Imp α2 with Various Cellular _targets Measured by Yeast Two-hybrid Analysis
| DNA-binding domain hybrid | β-gal activity (mOD/ml)* | |||
|---|---|---|---|---|
| VP16 | VP16/Imp α2 | |||
| GAL4 | 5 | 7 | ||
| GAL4/Imp β | 3 | 8 | ||
| GAL4/Imp βΔ1/261 | 4 | 1,198 | ||
| GAL4/CAS | 3 | 434 | ||
| GAL4/T-NLS | 4 | 1,211 | ||
| GAL4/LEF-1 (297–399) | 4 | 130 | ||
Data are given as milli optical density units per ml of yeast extract observed in yeast transformants expressing the indicated GAL4 and VP16 fusion proteins. Induced β-gal activities were determined as described previously (Bogerd et al., 1995) and are representative of values obtained in three independent experiments.
As noted above, there are five known forms of Imp α that can be subdivided into three families consisting of α1 and α6, α3 and α4, and finally α2. To test whether the ability of α2 to interact with CAS and Imp β was a general property of these family members, we asked whether α1 and α4 would also be able to interact with these two proteins in the yeast two-hybrid assay. As shown in Table II, both Imp α1 and Imp α4 interacted strongly with the GAL4/Imp–βΔ1/261 fusion protein and less strongly, but still detectably, with CAS. We therefore conclude that both Imp β and CAS binding are conserved properties of Imp α family members and would therefore be predicted to map to sequences conserved among Imp α family members.
Table II.
Both CAS and Imp β Binding Are Conserved among Different Imp α Family Members
| DNA-binding domain hybrid | β-gal activity (mOD/ml) | |||||
|---|---|---|---|---|---|---|
| VP16 | VP16/Imp α1 | VP16/Imp α4 | ||||
| GAL4 | 5 | 4 | 3 | |||
| GAL4/Imp β | 3 | 4 | 1 | |||
| GAL4/Imp βΔ1/261 | 4 | 2,350 | 1,850 | |||
| GAL4/CAS | 3 | 101 | 174 | |||
Protein interactions were analyzed in yeast cells using the two hybrid assay, as described in Table I. Data shown are representative of three independent experiments.
Protein Interaction Domains of Imp α2
We next constructed a series of deletion mutants of Imp α2 in the context of the Vp16/Imp α2 fusion protein and determined whether these would retain the ability to bind to ImpβΔ1/261, CAS, and to the T NLS and LEF-1 NLS. It should be noted that the T NLS (NH2-PKKKRKVE-COOH) and the LEF-1 NLS (NH2-KKKKRKREK-COOH) are both lysine-rich NLS sequences that have been previously shown to specifically interact with full-length Imp α (Prieve et al., 1996; Sekimoto et al., 1997).
The structure of the 19 deletion mutants of Imp α2 used in these experiments is shown in Fig. 1. These consist of four mutants progressively deleted from the NH2 terminus, eight mutants deleted progressively from the COOH terminus, and seven mutants deleted internally, starting from residue 137. It is important to note that every one of these mutants gave rise to a readily detectable interaction with at least one of the four protein _targets described above (Table III), thus demonstrating that each mutant fusion protein was expressed at functional levels in the yeast cell nucleus.
Table III.
Protein Interaction Profiles of a Panel of Imp α2 Mutants
| Activation domain hybrid | Relative β-gal activity | |||||||
|---|---|---|---|---|---|---|---|---|
| GAL4/CAS | GAL4/Imp βΔ1/261 | GAL4/LEF-1 | GAL4/T NLS | |||||
| VP16/α2Δ1/53 | + | − | ++ | + | ||||
| VP16/α2Δ1/80 | + | − | ++ | + | ||||
| VP16/α2Δ1/104 | + | − | ++ | − | ||||
| VP16/α2Δ1/138 | + | − | ++ | − | ||||
| VP16/α2Δ501/529 | + | ++ | ++ | ++ | ||||
| VP16/α2Δ496/529 | − | ++ | ++ | ++ | ||||
| VP16/α2Δ491/529 | − | + | ++ | + | ||||
| VP16/α2Δ471/529 | − | ++ | + | + | ||||
| VP16/α2Δ449/529 | − | + | − | ++ | ||||
| VP16/α2Δ441/529 | − | + | − | + | ||||
| VP16/α2Δ363/529 | − | ++ | − | ++ | ||||
| VP16/α2Δ321/529 | − | + | − | − | ||||
| VP16/α2Δ136/249 | + | ++ | ++ | − | ||||
| VP16/α2Δ136/278 | ++ | ++ | − | − | ||||
| VP16/α2Δ136/320 | ++ | ++ | − | − | ||||
| VP16/α2Δ136/365 | + | ++ | − | − | ||||
| VP16/α2Δ136/382 | +/− | ++ | − | − | ||||
| VP16/α2Δ136/394 | − | ++ | − | − | ||||
| VP16/α2Δ136/406 | − | ++ | − | − | ||||
| VP16/α2M1 | ++ | ++ | ++ | ++ | ||||
| VP16/α2M2 | − | ++ | ++ | ++ | ||||
| VP16/α2M3 | +/− | ++ | ++ | ++ | ||||
Protein interaction data were determined by yeast two-hybrid analysis, as described in Table I, and are categorized relative to the level seen with the wild-type VP16/Imp α2 fusion protein as follows: ++, >40% of wild-type α2; +, 15–40% of wild-type; +/−, 2–5% of wild-type; −, <2% of wild-type.
As expected, deletion of the first 53 or 80 aa of Imp α2, which includes the IBB domain (Görlich et al., 1996a ; Weis et al., 1996), blocked binding to Imp β but had at most a modest effect on binding to CAS, T NLS, and LEF-1 NLS (Table III). Surprisingly, further NH2-terminal deletion, to either residue 104 or residue 138, also entirely blocked T NLS binding by Imp α2, although LEF-1 NLS and CAS binding remained readily detectable. Therefore, these data demonstrate that the LEF-1 and T NLSs bind to distinguishable domains in Imp α2 and further demonstrate that the sequences essential for T NLS binding extend NH2 terminal to the first Arm motif of Imp α2.
Progressive deletion from the COOH terminus of Imp α2 demonstrated that residues located between 496 and 501, i.e., within the conserved acidic domain, were critical for CAS binding but dispensable for binding to Imp β and to both the LEF-1 and T NLS (Table III). Further deletion of Imp α2 residues between positions 449 and 471 resulted in the additional loss of LEF-1 NLS binding but did not affect binding to Imp β or the T NLS. In fact, deletion of additional residues up to position 363, i.e., deletion of Arm repeats 7 and 8, failed to inhibit either T NLS or Imp β binding, although both LEF-1 NLS binding and CAS binding remained undetectable. However, deletion of residues 321–363, which form Arm repeat 6, did block T NLS, but not Imp β, binding by Imp α2 (Table III). These data therefore further confirm the distinction between the Imp α2 sequences required to bind the T NLS and LEF-1 NLS and demonstrate that the sequences required for LEF-1 binding extend COOH terminal to the eighth and last Arm motif.
All internal deletion mutations, starting with VP16/ α2Δ136/249, were negative for T NLS binding (Table III). However, these mutants did permit the NH2-terminal border of the CAS-binding domain of Imp α2 to be mapped to between 382 and 394 for weak CAS binding or between 365 and 382 for strong binding. Importantly, this mapping therefore excludes any role for the putative leucine-rich NES sequence, located between residues 207 and 217 in Imp α2, in mediating binding to CAS. These data also map the NH2 terminus of the LEF-1 NLS binding sequence in Imp α2 to between residues 249 and 278, i.e., to at least 145 residues COOH terminal to the border of the sequences required for binding the functionally equivalent T NLS (Table III).
The data derived from the deletion mutants presented in Fig. 1 demonstrate that CAS binding is dependent on Imp α2 sequences located in the conserved COOH-terminal acidic domain but independent of the putative leucine-rich NES located between 207 and 216 (Boche and Fanning, 1997). To more fully confirm this hypothesis, we mutated the leucine-rich sequence present in Imp α2 by substitution of alanine residues for leucines to generate mutant M1 (Fig. 1). Comparable mutations have previously been shown to entirely block NES function for several leucine-rich NESs (Malim et al., 1991; Wen et al., 1995). Similarly, we also substituted alanine in place of conserved residues located in the Imp α2 acidic domain to generate Imp α2 mutants M2 (residues 469–474) and M3 (residues 476 –481) (Fig. 1). These missense mutants were then tested for binding to Imp β, CAS, the T NLS and the LEF-1 NLS by two-hybrid analysis. As shown in Table III, the M1 mutation had no effect on Imp α2 binding to any of these four protein _targets. In contrast, the M2 mutation eliminated, and the M3 mutation strongly inhibited, CAS binding without affecting binding to Imp β, T NLS, or LEF-1 NLS. These data therefore confirm that the conserved Imp α2 acidic domain is critical for specific CAS binding in vivo.
Imp α2 Binds to CAS but Not to Crm1
We next asked whether Imp α2 would be able to bind to CAS, or to the leucine-rich NES-specific nuclear export factor Crm1 (Fornerod et al., 1997; Stade et al., 1997), in mammalian cells using a previously described mammalian version of the two-hybrid assay (Bogerd et al., 1995). In this assay, fusions of the GAL4 DNA-binding domain to full-length human Crm1 or CAS, or to the Δ1/261 deletion mutant of Impβ, were coexpressed in the mammalian cell nucleus together with fusion proteins consisting of the VP16 transcription activation domain linked to wild-type or M1 mutant forms of Impα2 or to wild-type or M10 mutant forms of the human immunodeficiency virus type 1 Rev protein. The Rev protein is known to contain an active leucine-rich NES whereas the M10 Rev mutant encodes a defective form of this NES (Malim et al., 1989; Fischer et al., 1995; Wen et al., 1995).
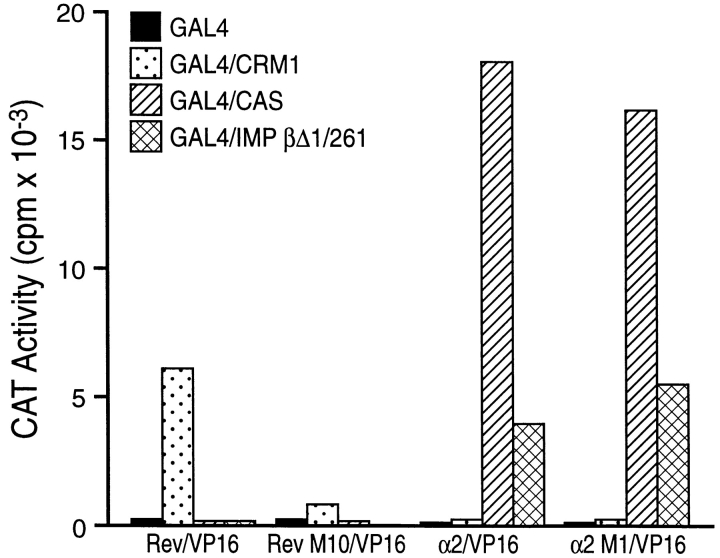
As shown in Fig. 2, we observed a readily detectable interaction between wild-type Rev and Crm1 that was blocked by the M10 NES mutation, but not between Rev and either CAS or Imp β. In contrast, both the wild-type and M1 mutant form of Imp α2 interacted strongly with both GAL4/CAS and the GAL4 ImpβΔ1/261 fusion protein but failed to interact detectably with Crm1. These data therefore suggest that Imp α2 contains a CAS-dependent, but not a Crm1-dependent, NES sequence.
Figure 2.
Interaction of Imp α2 and HIV-1 Rev with human nucleocytoplasmic transport factors measured by mammalian two-hybrid assay. Human 293T cells were transfected with one of the indicated GAL4 DNA-binding domain fusion protein expression plasmids, one of the indicated VP16 transcription activation domain fusion protein expression plasmids, the two-hybrid interaction indicator construct pG6/CAT and finally the internal control plasmid pBC12/CMV/β-gal. Induced CAT activities were measured at 48 h after transfection, as previously described (Bogerd et al., 1995), and were corrected for small differences in transfection efficiency revealed by analysis of β-gal expression levels.
In addition to the mammalian two-hybrid data presented in Fig. 2, we also assessed the ability of a subset of the α2 mutations described in Fig. 1 to interact with either CAS or ImpβΔ1/261 in the mammalian nucleus. In brief, these data (Table IV) confirmed the yeast data (Table III) showing that the NH2-terminal border of the CAS-binding domain in Imp α2 was located near to residue 365 and also located the COOH-terminal border of this domain between residues 491 and 501. In addition, these data (Table IV) revealed that both the M2 and the M3 mutant of Imp α2 are severely inhibited for CAS, but not for Imp β, binding in the mammalian cell nucleus.
Table IV.
Interaction of Imp a2 Mutants with CAS and Imp β Measured by Mammalian Two-hybrid analysis
| VP16 activation domain hybrid | Relative CAT activity | |||
|---|---|---|---|---|
| GAL4/CAS | GAL4/Imp βΔ1/261 | |||
| Imp α2/VP16 | ++ | ++ | ||
| α2Δ501/529/VP16 | ++ | ++ | ||
| α2Δ491/529/VP16 | − | + | ||
| α2Δ136/249/VP16 | + | ++ | ||
| α2Δ136/365/VP16 | +/− | ++ | ||
| α2Δ136/382/VP16 | − | ++ | ||
| α2Δ136/406/VP16 | − | ++ | ||
| α2M1/VP16 | ++ | ++ | ||
| α2M2/VP16 | − | + | ||
| α2M3/VP16 | − | ++ | ||
Protein interaction data were determined by mammalian two-hybrid analysis in transfected human 293T cells, as described in Fig. 3. Activity is given relative to the level seen with the wild-type Imp α2 fusion protein as follows: ++, >40% of wild-type; +, 14–40% of wild-type; +/−, 1–5% of wild-type; −, <0.2% of wild-type.
CAS-binding Domain of Imp α2 Mediates Nuclear Export
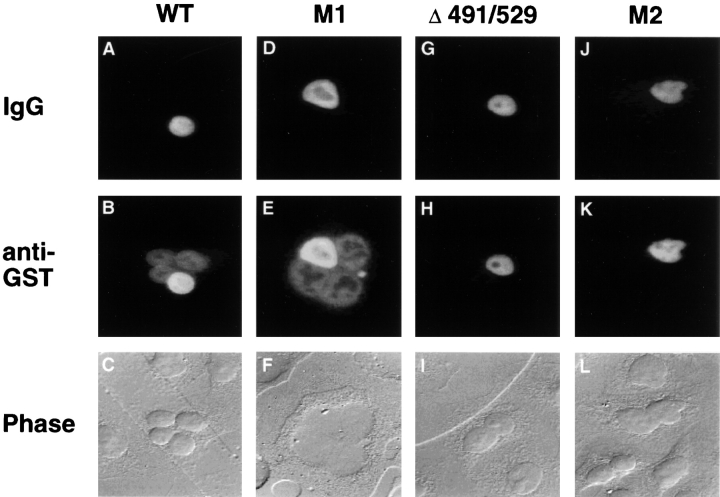
We next wished to ask whether the ability to interact with CAS was indeed critical for the nuclear export of Imp α2 from the mammalian cell nucleus. As described in more detail elsewhere (Truant et al., 1998b ), we have observed that recombinant Imp α2 accumulates in the nucleus of microinjected human cells. To demonstrate nuclear export, we therefore mixed recombinant wild-type full-length Imp α2 fused to GST with a rabbit IgG tracer protein and microinjected it into a single nucleus located in multinucleate cells. If Imp α2 contains a functional NES, to go with the NLS activity known to reside in the IBB motif, it should export from the injected nucleus and then import into the uninjected nuclei present in the same cell. This is indeed precisely the activity that was seen, in that the injected GST/Imp α2 fusion protein is redistributed to all the nuclei present in an injected cell (Fig. 3 B) whereas the IgG tracer remains in the single injected nucleus (Fig. 3 A). The same result was obtained upon injection of the M1 mutant form of full-length GST/Imp α2 into one nucleus in a multinucleate cell (Fig. 3, D and E), thus demonstrating that a functional form of the putative leucine-rich NES present between residues 207 and 217 in Impα2 is not required for nuclear export. In contrast, GST–Imp α2 fusion proteins containing either the Δ491/529 deletion mutation or the M2 missense mutation that knocks out CAS binding (Tables III and IV failed to demonstrate nucleocytoplasmic shuttling (Fig. 3, H and K) even though both proteins are efficiently imported into the nucleus when injected into the cell cytoplasm (data not shown). We therefore conclude that CAS binding is a critical step in the nuclear export of Imp α2 in human cells.
Figure 3.
CAS mediates the nuclear export of Imp α2. Recombinant fusion proteins consisting of GST linked to full-length, wild-type Imp α2 and to the M1, Δ491/529 or M2 mutants of Imp α2, were purified from bacteria, mixed with a rhodamine-conjugated rabbit IgG tracer, and then microinjected into a single nucleus in a multinucleate HeLa cell. Subcellular localization of the injected proteins was determined at 35 min after injection by double-label immunofluorescence microscopy, as previously described (Fridell et al., 1997). GST fusion proteins able to undergo nucleocytoplasmic shuttling to the uninjected nuclei present in the same cell (B and E) must contain a functional NES whereas mutants lacking nuclear export capacity (H and K), as well as the IgG tracer (A, D, G, and J), remain confined to the single injected nucleus.
Imp α Is Unable to Bind CAS and the T NLS Simultaneously
The mapping data presented in Table III, and summarized in Fig. 4, demonstrate that the CAS-binding domain in Imp α2 is located between residues 383 and 497 whereas the SV-40 T NLS binding sequences are located between residues 81 and 362. Therefore, at least in this linear representation of the Imp α2 molecule, these binding domains do not obviously overlap. However, if the T NLS and the CAS nuclear export factor could bind Imp α simultaneously, then one might predict that proteins containing the T NLS would be exported from the nucleus in a ternary complex with Imp α and CAS just as they are imported into the nucleus as a ternary complex with Imp α and Imp β.
Figure 4.
Functional domain organization of Imp α2. The figure correlates the location of the conserved structural domains of Imp α with the approximate location of sequences in Imp α2 required for binding to Imp β, to CAS, to the LEF-1 NLS and the T NLS. The speckled areas are essential for binding to each _target while the flanking clear boxes define the ambiguity as to the precise border of the critical Imp α2 sequences. IBB, Imp β binding motif; V, variable regions.
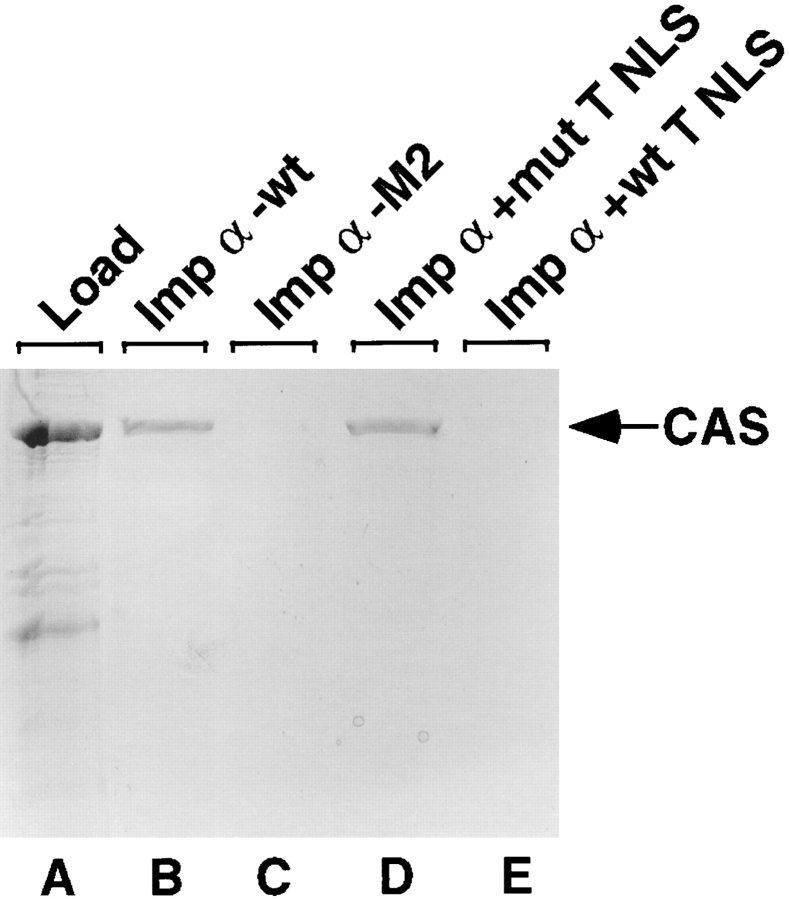
To test whether the T NLS and CAS can indeed bind Imp α2 simultaneously, we performed an in vitro protein– protein interaction assay using recombinant wild-type and M2 mutant Imp α2 and CAS as well as Ran protein (Fig. 5). As reported previously by Kutay et al. (1997a), we observed that efficient binding of Imp α by CAS only occurred in the presence of the GTP bound form of Ran (data not shown). This interaction was specific in that CAS bound the wild-type but not the M2 mutant form of Imp α2 in vitro (Fig. 5, B and C), thus also confirming the in vivo data demonstrating that the Imp α2 M2 mutant blocks CAS binding (refer to Tables III and IV. Interestingly, although the NH2 terminally HIS-tagged recombinant CAS protein preparation used in this analysis contained a significant level of breakdown products that presumably represent a nested set of COOH terminally truncated forms of CAS, only the full-length form of CAS was observed to bind to Imp α2 (Fig. 5, compare lane A with B). This further confirms the specificity of this protein–protein interaction and suggests that sequences located towards the CAS COOH terminus are critical for Imp α2 binding.
Figure 5.
Analysis of CAS binding to Imp α2 in vitro. Recombinant wild-type or M2 mutant Imp α2 proteins were coupled to agarose beads and used to construct microaffinity columns. Recombinant, NH2 terminally His-tagged CAS protein was preincubated on ice with Ran and GTP before loading onto the columns either directly or after addition of a 100-fold molar excess of a wild-type or defective mutant T NLS peptide. After extensive washing, bound proteins were eluted from the affinity columns and analyzed by SDS-PAGE.
To examine whether the T NLS would interfere with CAS binding to Imp α2, we repeated this binding assay in the presence of an ∼100-fold molar excess of synthetic peptides containing either the wild-type SV-40 T NLS or a mutant, nonfunctional form of the T NLS (K128 to T, Kalderon et al., 1984). As shown in Fig. 5, the wild-type T NLS peptide entirely blocked CAS binding by Imp α2 whereas the mutant peptide had no effect. We therefore conclude that Imp α2 is unable to simultaneously bind to both CAS and the T NLS.
Discussion
A major goal of the research described in this article was the determination of whether Crm1 (Boche and Fanning, 1997) or CAS (Kutay et al., 1997a ) was required for the nuclear export of Imp α and if the latter, the identification and delineation of the CAS-dependent NES present in Imp α. Using the two-hybrid assay for in vivo protein– protein interactions, we have demonstrated that the distinct Imp α2, Imp α1, and Imp α4 forms of Imp α are all able to specifically interact with human CAS in the yeast two-hybrid assay (refer to Tables I and II. Using a mammalian two-hybrid assay, we confirmed the specific interaction of Imp α2 with CAS and showed that Imp α2 failed to interact with Crm1 under conditions where the interaction of Crm1 with the leucine-rich NES present in HIV-1 Rev was readily detectable (refer to Fig. 2). We therefore conclude that Imp α can specifically bind to CAS but not to Crm1 in vivo. By deletion and missense mutation analysis (Tables III and IV, we have further demonstrated that a proposed leucine-rich NES sequence (Boche and Fanning, 1997), located within the third Arm repeat of Imp α2, is entirely dispensable for CAS binding, which instead was shown to largely map to a conserved acidic domain located proximal to the COOH terminus of Imp α2 (refer to Fig. 4). Finally, we demonstrate that nucleocytoplasmic shuttling of Imp α2 is blocked by two distinct mutations within the acidic domain that inhibit CAS binding but is not affected by mutation of the putative leucine-rich NES (refer to Fig. 3). We therefore conclude that the leucine-rich sequence present in Imp α2 Arm repeat 3 is neither necessary nor sufficient for the nuclear export of Imp α2 and suggest that the published identification of this sequence as the Imp α NES (Boche and Fanning, 1997) is incorrect. Rather, our data confirm the proposal by Kutay et al. (1997a) that Imp α nuclear export is dependent on the interaction of Imp α with the CAS nuclear export factor and map the Imp α sequences required for this interaction to the conserved Imp α acidic domain, together with some adjacent flanking sequences (refer to Fig. 4). Although the CAS-binding domain defined in this analysis is quite large, being minimally ∼120 aa in length, it is clearly distinct in sequence from the much smaller leucine-rich NES sequences recognized by the Crm1 nuclear export factor (Fornerod et al., 1997; Stade et al., 1997).
In addition to defining sequences in Imp α2 that mediate CAS binding, we wished to also determine which Imp α2 sequences might be required for basic NLS recognition. Surprisingly, with the exception of a manuscript that mapped RAG1 NLS binding to Arm repeats 4–8 of Imp α1 (Cortes et al., 1994), little had been reported about the identity of Imp α sequences required for this critical step in nuclear import at the time that this work was initiated. The two basic NLS sequences chosen for analysis, the T NLS and the LEF-1 NLS, are both lysine-rich NLS sequences that had been previously shown to bind to Imp α specifically (Prieve et al., 1996; Sekimoto et al., 1997).
Analysis of the sequence requirements for T NLS and LEF-1 NLS binding by Imp α2 produced two surprising findings. First, the Imp α2 sequences required for binding to these two NLS sequences were clearly distinct, with T NLS binding mapping between Imp α2 residues 81–362 whereas LEF-1 NLS binding mapped between residues 250 and 470 (refer to Table III and Fig. 4). The second unexpected result was that the Imp α2 sequences required for basic NLS binding extended at least somewhat beyond the Arm repeat sequences into flanking variable regions. Specifically, T NLS binding required Imp α2 sequences located NH2 terminal to residue 105, the first residue of the first Arm repeat, whereas LEF-1 NLS binding clearly required sequences located COOH terminal to residue 447, the last amino acid of the eighth Arm repeat (refer to Table III and Fig. 4). These observations therefore demonstrate that the identity of the Arm repeats required for any given NLS interaction is likely to be quite variable and further reveal that the Imp α Arm repeats, although critical for basic NLS binding, are not necessarily sufficient, in that flanking variable regions may also make an essential contribution. In this regard, it is interesting to note that several papers have reported that certain NLS sequences can be bound by particular members of the Imp α family of NLS receptors but not by others (Miyamoto et al., 1997; Nadler et al., 1997; Sekimoto et al., 1997). As highly variable regions in Imp α appear to make a significant contribution to NLS binding, this result is not, in retrospect, surprising.
Although the interaction of Imp β with Imp α in the cell nucleus is blocked by Ran-GTP (Rexach and Blobel, 1995; Görlich et al., 1996b ), it has remained unclear why NLS proteins are released by Imp α. Although it has been suggested that basic NLS binding to an Imp α/Imp β heterodimer is of higher affinity than binding by Imp α alone (Rexach and Blobel, 1995; Efthymiadis et al., 1998), it is nevertheless apparent that NLS binding by Imp α can be readily detected in the nucleus of cells under conditions where Imp α and Imp β do not interact (refer to Tables I and III (Cuomo et al., 1994; Cortes et al., 1995; O'Neill et al., 1995). Why then doesn't the NLS protein return to the cytoplasm along with the Imp α/CAS complex in a process analogous to its import by the Imp α/Imp β complex? One possibility is that CAS binding and basic NLS binding by Imp α is mediated by two distinct, mutually incompatible Imp α protein conformations, a mechanism which has also been invoked to explain why Imp β is unable to simultaneously bind to Imp α and Ran-GTP. Alternately, CAS binding and NLS binding might simply sterically interfere with one another. In either case, CAS binding, and hence Imp α export from the nucleus, could only occur once the NLS protein was released into the nucleoplasm. It has, in fact, previously been demonstrated that CAS binds to NLS-free Imp α preferentially, although the underlying mechanism for this effect has remained unclear (Kutay et al., 1997a ). Although the extensive overlap between the Imp α2 sequences that mediate binding to the LEF-1 NLS and to CAS, respectively (refer to Fig. 4), certainly seems consistent with the hypothesis that these protein–protein interactions might be mutually exclusive, this is less obvious in the case of the SV-40 T NLS, which binds to Imp α2 sequences that do not obviously conflict with the CAS binding site (refer to Fig. 4). Nevertheless, in vitro binding analysis reveals that the SV-40 T NLS, but not a nonfunctional mutant form thereof, can entirely block CAS binding by Imp α2 (refer to Fig. 5). These data demonstrate that Imp α2 is unable to bind to the T NLS and to CAS simultaneously and therefore provide a mechanistic explanation for the finding that the SV-40 T NLS is unable to function as an Imp α–dependent NES.
After submission of this article, a paper was published that bears on the issue of the functional domain organization of Imp α2. Specifically, Conti et al. (1998) have published the crystal structure of a truncated form of yeast Imp α bound to two molecules of an SV-40 T NLS peptide. Although yeast Imp α is somewhat different in structure to human Imp α2 (for example, it appears to contain 10 rather than 8 Arm repeats), this report is striking in that the two T NLS peptides were found to bind to Arm repeats one through four and seven through nine, respectively. The authors proposed that the larger, more NH2-terminal NLS binding site was likely to be the functionally relevant site on yeast Imp α for T NLS binding whereas the smaller, more COOH-terminal site might instead play a role in the recognition of bipartite NLSs, such as the one observed in nucleoplasmin. In contrast, the data presented in our article suggest that Imp α2 contains two distinct binding sites for monopartite NLS sequences, with the more NH2-terminal site being specific for NLSs equivalent in sequence to the SV-40 T NLS whereas the more COOH-terminal site may interact specifically with NLSs that are similar to the LEF-1 NLS. From this perspective, the weak binding observed by Conti et al. (1998) between the T NLS peptide and the more COOH-terminal binding site in yeast Imp α may therefore simply reflect the fact that the structure of the T NLS is suboptimal for binding to this latter site, rather than suggesting that this site is not an autonomous binding site for other, more optimal monopartite NLSs.
Acknowledgments
The authors thank G. Grosveld (St. Jude Children's Research Hospital, Memphis, TN) for the human Crm1 cDNA, U. Brinkmann (National Cancer Institute, Bethesda, MD) for the human CAS cDNA clone, M. Waterman (University of California, Irvine, CA) for pGAL4/LEF-1, and M. Malim (University of Pennsylvania, Philadelphia, PA) for wild-type and mutant forms of the SV-40 T NLS peptide.
Abbreviations used in this paper
- aa
amino acid(s)
- Arm
armadillo motif
- β-gal
β-galactosidase
- CMV
cytomegalovirus
- GST
glutathione-S-transferase
- IBB
importin β binding domain
- Imp α
importin α
- Imp β
importin β
- NES
nuclear export signal
- NLS
nuclear localization signal
- T NLS
SV-40 T antigen NLS
Footnotes
This research was funded by the Howard Hughes Medical Institute. A. Herold was supported by funds from the German Academic Exchange Organization (DAAD).
Address all correspondence to B.R. Cullen, Department of Genetics, Room 426 CARL Building, Research Drive, Box 3025, Duke University Medical Center, Durham, NC 27710. Tel.: (919) 684-3369. Fax: (919) 681-8979. E-mail: culle002@mc.duke.edu
References
- Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boche I, Fanning E. Nucleocytoplasmic recycling of the nuclear localization signal receptor α subunit in vivo is dependent on a nuclear export signal, energy, and RCC1. J Cell Biol. 1997;139:313–325. doi: 10.1083/jcb.139.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular co-factor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Brinkmann U, Brinkmann E, Gallo M, Pastan I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. . Proc Natl Acad Sci USA. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam SA. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell. 1997;8:945–956. doi: 10.1091/mbc.8.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Cortes P, Ye Z-S, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Kirch SA, Gyuris J, Brent R, Oettinger MA. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiadis A, Briggs LJ, Jans DA. The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J Biol Chem. 1998;273:1623–1628. doi: 10.1074/jbc.273.3.1623. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O-K. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO (Eur Mol Biol Organ) J. 1996a;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996b;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor o G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur Mol Biol Organ) J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Köhler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-α protein family. FEBS (Fed Eur Biochem Soc) Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997a;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO (Eur Mol Biol Organ) J. 1997b;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Böhnlein S, Hauber J, Cullen BR. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- Malim MH, McCarn DF, Tiley LS, Cullen BR. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: The soluble phase. Annu Rev Biochem. 1998;67:256–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1 β and α2 β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Ryder UW, Lamond AI, Weis K. Cloning and characterization of hSRP1γ, a tissue-specific nuclear transport factor. Proc Natl Acad Sci USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler SG, Tritschler D, Haffar OK, Blake J, Bruce AG, Cleaveland JS. Differential expression and sequence-specific interaction of karyopherin α with nuclear localization sequences. J Biol Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- O'Neill RE, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Delphin C, Gerace L. Nucleotide-specific interaction of Ran/TC4 with nuclear transport factors NTF2 and p97. Proc Natl Acad Sci USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieve MG, Guttridge KL, Munguia JE, Waterman ML. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J Biol Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucl Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-_targeting complex formation with NP1-1, but not Rch1. EMBO (Eur Mol Biol Organ) J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southgate CD, Green MR. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Truant R, Fridell RA, Benson RE, Bogerd H, Cullen BR. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+RNA binding protein. Mol Cell Biol. 1998a;18:1449–1458. doi: 10.1128/mcb.18.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant, R., R.A. Fridell, E.R. Benson, A. Herold, and B.R. Cullen. 1998b Nucleocytoplasmic shuttling by protein nuclear import factors. Eur. J. Cell Biol. In press. [DOI] [PubMed]
- Tsuji L, Takumi T, Imamoto N, Yoneda Y. Identification of novel homologues of mouse importin α, the α subunit of the nuclear pore-_targeting complex, and their tissue-specific expression. FEBS (Fed Eur Biochem Soc) Lett. 1997;416:30–34. doi: 10.1016/s0014-5793(97)01092-2. [DOI] [PubMed] [Google Scholar]
- Weis K. Importins and exportins: How to get in and out of the nucleus. Trends Cell Biol. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- Weis K, Mattaj IW, Lamond AI. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- Weis K, Ryder U, Lamond AI. The conserved amino-terminal domain of hSRP1α is essential for nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]