Abstract
Proteins that can interact with multiple partners play central roles in the network of protein–protein interactions. They are called hub proteins, and recently it was suggested that an abundance of intrinsically disordered regions on their surfaces facilitates their binding to multiple partners. However, in those studies, the hub proteins were identified as proteins with multiple partners, regardless of whether the interactions were transient or permanent. As a result, a certain number of hub proteins are subunits of stable multi-subunit proteins, such as supramolecules. It is well known that stable complexes and transient complexes have different structural features, and thus the statistics based on the current definition of hub proteins will hide the true nature of hub proteins. Therefore, in this paper, we first describe a new approach to identify proteins with multiple partners dynamically, using the Protein Data Bank, and then we performed statistical analyses of the structural features of these proteins. We refer to the proteins as transient hub proteins or sociable proteins, to clarify the difference with hub proteins. As a result, we found that the main difference between sociable and nonsociable proteins is not the abundance of disordered regions, in contrast to the previous studies, but rather the structural flexibility of the entire protein. We also found greater predominance of charged and polar residues in sociable proteins than previously reported.
Keywords: hub protein, protein–protein interaction, transient interaction, three-dimensional structure, intrinsic disorder
The vast abundance of protein–protein interaction data generated by large-scale experiments, such as yeast two-hybrid techniques (Uetz et al. 2000; Ito et al. 2001), is now providing keys toward understanding the comprehensive topology of protein interaction networks (Giot et al. 2003; Li et al. 2004). The construction of interaction networks facilitates the identification of the functions of novel genes or hypothetical proteins. In addition to these functional aspects of proteins, the interaction network itself is also analyzed intensively. One such analysis revealed that only a limited number of proteins can interact with many other proteins, while most proteins interact with a small number of partners. This is a typical feature of a scale-free network, and protein–protein interaction networks are proposed to be a type of the scale-free network (Jeong et al. 2000). The scale-free network is characterized by highly connected nodes called hub nodes (Jeong et al. 2000), which are known for their critical importance. If a few hub nodes are eliminated, then the biological network will change the topology of the network drastically. By analogy to the importance of hub nodes, the hub proteins are considered as the essential proteins to construct the biological networks (Jeong et al. 2001).
Because of the importance of hub proteins in biological systems, the reasons they can function as hubs or can interact with multiple partners were investigated. The authors of several recent studies (Dunker et al. 2005; Patil and Nakamura 2006) reported that an abundance of intrinsically disordered regions is found in hub proteins, which led to the proposal that disordered regions differentiate hub proteins from the others. Intrinsically disordered regions cannot have rigid and unique three-dimensional structures without any interaction partners, such as small molecules, DNA, or other proteins. Induced folding of intrinsically disordered regions first drew attention in DNA binding proteins (Spolar and Record 1994). Since then, many intrinsically disordered proteins have been found to undergo coupled folding upon binding with partners, and it is now widely accepted that they play important roles in the functions of signaling and regulatory proteins (Kriwacki et al. 1996; McEwan et al. 1996; Iakoucheva et al. 2002; Friedler et al. 2005). The transition upon binding will potentially lead to high specificity coupled with low affinity (Schulz 1979), which would be a binding characteristic of hub proteins. On the other hand, charged or polar residues on the interface are also thought to be involved in the binding of hub proteins, such as Rap1A and p53 (Sheinerman and Honig 2002; Friedler et al. 2005), which are known to bind different peptides or proteins in overlapping regions, with different affinity.
The structural features of hub proteins obtained by past studies seem to be convincing, but the protein interaction network was treated as a static one, although protein–protein interactions are known to change dynamically, according to the different cell conditions. As a result of the static treatment, the so-called hub proteins that interact with multiple partners can be the components of stable supramolecules, such as ribosomes and nucleosome complexes, as opposed to the normal concepts of hub proteins.
Therefore, we tried to carry out further analyses of such hub proteins that interact with other proteins transiently, which we refer to as transient hub proteins or sociable proteins, to clarify the difference from hub proteins. In this study, we first developed a method to identify sociable proteins, using known protein complexes in the Protein Data Bank (PDB) (Berman et al. 2000), and then carried out systematic analyses to clarify the difference between sociable and nonsociable proteins. The differences between hub proteins and sociable proteins were also analyzed and discussed. We concluded that structural flexibility, rather than the ratio of disordered regions, is the critical difference between sociable and nonsociable proteins.
Results and Discussion
Data sets
X-ray structures with 3.0 Å or better resolution were selected from the PDB, as of July 2006. From them, short peptide chains (<30 residues) were removed, to focus on protein–protein interactions, and entries with >30 chains were not used, because of a program limit (see the Materials and Methods for details). As a result, 68,474 chains were used to identify the sociable proteins. They were clustered by the BLASTCLUST program (Jeong et al. 2000) with a threshold of identity ≥30% and an alignment length ≥80%, which generated 6398 clusters. We will refer to these clusters as sequence families hereafter. One representative entry was selected for each sequence family as the best resolved structure.
Summary of sociable proteins
We identified 86 sociable proteins as the proteins with three or more binding states, and 1013 nonsociable proteins that have only one binding state. The remaining 5291 proteins were classified as marginal ones and were not used. The binding state is an indicator to evaluate the number of different binding modes found in each sequence family, and it was estimated by using the above 68,474 PDB entries. The number of binding states for each sequence family was assigned to each representative protein (see Materials and Methods for details).
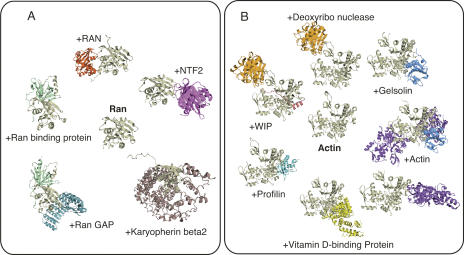
The list of all sociable proteins is provided in the Supplementary Table S1. In brief, as we expected, many of the sociable proteins are related to biological processes such as cell signaling (e.g., Fig. 1A) or transcription, which are usually considered to require transient complexes with many partners. Interestingly, some other proteins were also identified as sociable proteins (e.g., Fig. 1B). The number of sociable proteins is not small, and thus we cannot describe all of them here. Generally speaking, most of them are reasonable ones, but minor problems were caused by the inclusion of crystal contacts and by large missing regions in crystallographic analyses. For the first problem, we used the biological units provided by the PDB as the putative biological complexes, but, as described in some analyses (Henrick and Thornton 1998; Tsuchiya et al. 2006), these annotations are not perfect. However, we think that this problem is not serious, because one incorrect biological unit will only increase the number of binding states by one. The second problem is a little more problematic. In X-ray crystallography, some regions can be missing, because of the poor electron density caused by the high mobility of the regions. If the missing area corresponds to a whole subunit or multiple subunits, then our method will regard it as a different binding state. As a result, the number of binding states can get larger than we expected. Therefore, we manually checked all of the sociable proteins identified automatically (94 proteins), and we found that eight of them were related with the second problem (indicated in Supplementary Table S1). They were supramolecules, and thus we decided not to consider them as sociable proteins but as stable hub proteins, which are described later.
Figure 1.
Examples of sociable proteins. (A) Ran protein: It was found to interact with Ran binding protein (PDB: 1rrp), Ran GAP (PDB: 1k5d), karyopherin β2 (PDB: 1qbk), and nuclear transport factor 2 (PDB: 1a2k), and forms a dimer (PDB: 1byu). In addition, Ran can exist as a monomer (PDB: 1qg4). (B) α-Actin 1 (PDB: 2fxu): It was found to interact with gelsolin segment 1 (PDB: 1p8z), deoxyribonuclease 1 (PDB: 2a40), profilin (PDB: 1hlu), vitamin D binding proteins (PDB: 1ma9), deoxyribonuclease and Wiskott-Aldrich syndrome protein interacting protein (PDB: 2a41), gelsolin and α-actin 1 (PDB: 1mdu), and α-actin 1 forms a dimer (PDB: 1yxq). In addition, α-actin 1 can exist as a monomer (PDB: 1s22).
Intrinsic disorder at the interface of sociable/nonsociable proteins
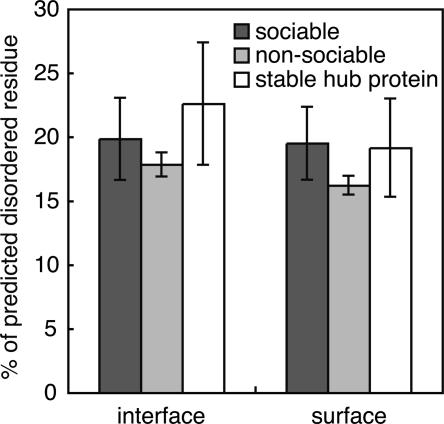
It is now widely assumed that proteins with multiple binding partners, or hub proteins, would have many disordered regions, and that such disordered regions would form the binding interfaces (Wright and Dyson 1999; Liu et al. 2002). If this feature is a strong constraint to define a sociable protein, then sociable proteins would have many disordered regions in their interface regions. To check this possibility, the disordered residues of sociable and nonsociable proteins were predicted, using the PrDOS server (Ishida and Kinoshita 2007) with an 8% false positive rate, which aims to achieve the similar prediction accuracy to the prediction method used in the previous study (Patil and Nakamura 2006). We were surprised to find that there were no significant differences in the average percentage of disordered residues at the interfaces between the sociable and nonsociable proteins (P-value by Wilcoxon rank-sum test is 0.12). However, we found a statistically significant difference between the whole surfaces of sociable proteins and nonsociable proteins (Fig. 2, black and gray bars; P-value of Wilcoxon rank-sum test is 0.011).
Figure 2.
Percentage of predicted disordered residues. The percentages of disordered residues in the interfaces and the whole surfaces of sociable proteins (black bars), nonsociable proteins (gray bars), and stable hub proteins (white bars), respectively. Error bars indicate the 95% confidence intervals of the mean values.
The main difference between this work and previous work exists in the selection of so-called hub proteins and sociable proteins. Therefore, we checked the entire list of hub proteins provided by Patil and Nakamura (2006) and found that about 20% of the hub proteins were the subunits of supramolecular complexes. In addition, we noticed that about half of them were DNA (or RNA) interacting proteins. The self-assembly of a supramolecule is proposed to be coupled with the folding of disordered regions, rather than the assembly of rigid, complementary structures (Namba 2001), and DNA (or RNA) interacting proteins are known to have intrinsically disordered regions at the protein–DNA (RNA) interfaces (Spolar and Record 1994). Therefore, we consider that the abundance of disordered regions in the hub proteins may be the result of the inclusion of supramolecules in the set of hub proteins.
To check this possibility, we selected permanent complexes as nonsociable proteins that interact with multiple partners (three or more different proteins), which we call stable hub proteins. As a result, 46 proteins were identified as stable hub proteins, which were analyzed by the same procedure used for sociable proteins. We found that the stable hub proteins have higher percentages of disordered regions in their interfaces on average (Fig. 2, white bars), and thus we concluded that the higher percentage of disordered regions in the hub proteins is mainly due to the inclusion of stable hub proteins.
Degree of global flexibility of sociable/nonsociable proteins
We have shown that disordered regions may not be the main factor to differentiate sociable proteins and nonsociable ones. In that case, what is the main factor that confers the ability to interact with multiple proteins? In this paper, we addressed the following two aspects: (1) structural flexibility and (2) amino acid propensity. Here we describe the structural flexibility first, and then the amino acid propensity will be discussed in the following sections.
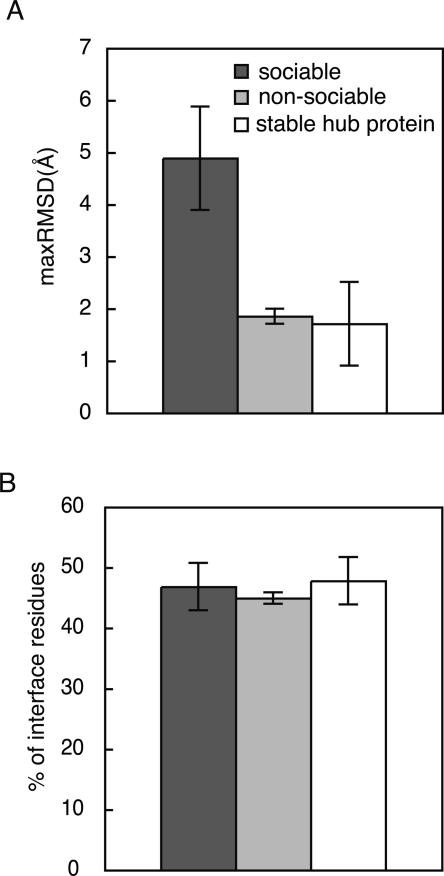
To check the difference in global flexibility between sociable and nonsociable proteins, maximum values of RMSDs (maxRMSDs) for every sequence family were calculated, and the distribution was checked for sociable proteins and nonsociable ones (Fig. 3A). As a result, the sociable proteins were found to exhibit larger conformational changes among the observed structures than those of the nonsociable proteins, on average (P-value by Wilcoxon rank-sum test is virtually 0.0). It may be noteworthy that the average maxRMSD values for sociable proteins are much larger, as compared with the observed structural changes for enzymes (Gutteridge and Thornton 2005). We also checked the maxRMSDs of stable hub proteins (Fig. 3A, white bars), and found again that the stable hub proteins are more likely to be nonsociable proteins. These observations strongly suggested that global flexibility could be a very important factor to allow the sociable proteins to interact with multiple partners.
Figure 3.
Comparison of structural flexibility between sociable and nonsociable proteins. For sociable (black bars), nonsociable (gray bars), and stable hub proteins (white bars), the distribution of maxRMSD (A) and the distribution of secondary structure composition in the interfaces (B) are shown, respectively. Error bars indicate the 95% confidence intervals of the mean values.
The composition of the secondary structural elements (SSEs) at the interfaces can be another indicator to assess the structural flexibility, because loop regions have higher mobility than regular SSEs. Loop regions are not necessarily disordered, because they can have a fixed tertiary structure without a regular secondary structure (Bracken et al. 2004). The SSEs of the interfaces of sociable and nonsociable proteins were defined using DSSP (Kabsch and Sander 1983), and the compositions of the loops at the interfaces were assessed (Fig. 3B). As a result, we could not find any significant differences in the composition of the loops (P-value by Wilcoxon rank-sum test is 0.12). It may be noteworthy that this observation is not consistent with the previous report by another group. Liu et al. (2002) previously reported that proteins with nonregulatory regions (NORS proteins) had a few more interaction partners than proteins without NORS, and that the loop regions at the interfaces mediated the multiple interactions.
Amino acid propensity of sociable/nonsociable proteins
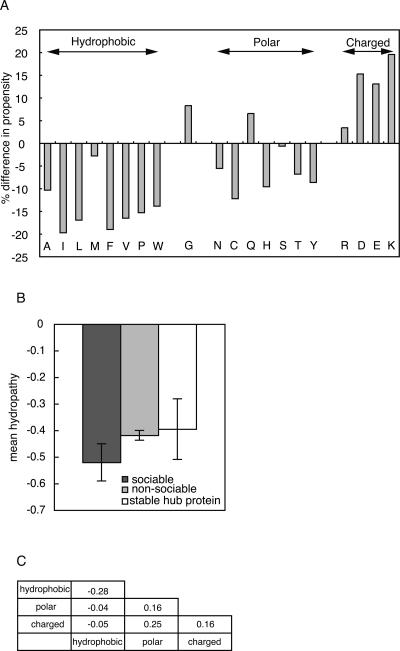
The amino acid propensity (Jones and Thornton 1996) at the interface was compared to characterize the difference between sociable and nonsociable proteins in their amino acid sequences. It can be calculated as the ratio of a fraction of area of a given amino acid in interface to that in the surface region. Figure 4A shows the differences in the amino acid propensity between sociable proteins and nonsociable proteins, where the residues with a positive value indicate that they are preferable in the interfaces of sociable proteins. We found that the interfaces of sociable proteins have more charged residues and fewer hydrophobic residues, as compared with those of nonsociable proteins. A previous study (Patil and Nakamura 2006) also reported similar profiles, but the tendency for the interfaces to prefer hydrophilic residues is much stronger in sociable proteins, as compared with the difference between hub and non-hub proteins. To clarify why sociable proteins prefer hydrophilic residues more strongly than hub proteins, we compared the amino acid profile as the mean hydropathy of sociable proteins, nonsociable proteins, and stable hub proteins (Fig. 4B), where Kyte and Dolittle hydropathy indexes (Kyte and Doolittle 1982) were employed for every residue. As a result, we observed that the interfaces of sociable proteins are more hydrophilic, as compared with those of nonsociable proteins and stable hub proteins (P-value by Wilcoxon rank-sum test is 0.001). Note that a more negative value of the mean hydropathy means that the interfaces are more hydrophilic in Figure 4B. Therefore, as in the other properties, the inclusion of stable complexes as so-called hub proteins weakens the nature of proteins with multiple partners.
Figure 4.
Comparison of sequence propensity between sociable and nonsociable proteins. (A) Percent differences in amino acid propensities are shown. Amino acid names are shown in one-letter codes at the bottom. (B) Mean hydropathy values are shown for sociable (black bar), nonsociable (gray bar), and stable hub proteins (white bar), respectively. Error bars indicate the 95% confidence intervals of the mean values. (C) Observed frequency ratios of atom contacts in sociable and nonsociable proteins in the natural logarithmic scale.
Since we found an abundance of charged residues on the interfaces of sociable proteins, we also compared the residue–residue contacts at the interfaces of sociable proteins and nonsociable proteins to reveal the different natures of their interactions. For this purpose, the amino acid residues in the interface were first classified into three classes, hydrophobic, polar, or charged, as in Figure 4A, and then the frequencies of the contacting residues pairs were calculated. Finally, the ratios of the observed frequency between sociable and nonsociable proteins were obtained in the natural logarithmic scale (Fig. 4C). Therefore, positive values indicate that the contact pairs are more abundantly observed in sociable interfaces. As a result, we found that polar–polar contacts, polar–charged contacts, and charged–charged contacts are more frequently observed at the interfaces of sociable proteins than nonsociable ones. This observation is reasonable from the viewpoint of molecular interactions, because the interfaces of sociable proteins have to be exposed to the solvent when they change the binding partners. It is possible that sociable proteins might have some unfavorable interactions, such as hydrophobic–charged contacts, because it would be difficult to achieve favorable interactions with all of the different partners. However, we did not find an abundance of unfavorable contacts at the interfaces of sociable proteins, as compared with those of nonsociable proteins. In that sense, the interfaces of sociable proteins are well designed. The evolutionary aspects of sociable proteins are beyond the scope of our paper, but it will be interesting to see how the sociable proteins were developed during the course of evolution.
Conclusion
We found that disordered regions do not preferably exist in the interface regions of sociable proteins, as compared with those of nonsociable proteins (Fig. 2), as previously proposed (Dunker et al. 2005; Patil and Nakamura 2006), and that the global flexibility of sociable proteins is the most important differences between sociable and nonsociable proteins (Fig. 3). In addition, we revealed that the inclusion of stable hub proteins is one of the reasons a higher ratio of disordered regions was observed in the so-called hub proteins.
Why are disordered regions not so important in the sociable proteins? From the previous studies on disordered proteins (Spolar and Record 1994; Wright and Dyson 1999; Dyson and Wright 2005), it is now well accepted that structural flexibility, as in the form of disordered regions, is advantageous, as also supported by a model calculation (Shoemaker et al. 2000). According to this commonplace assumption, sociable proteins as well as stable hub proteins should have a higher ratio of disordered regions to interact with many different proteins, regardless of the stability of the complex structure. However, as proposed by Dyson and Wright (2005), disordered regions have some disadvantage. There is a “biological cost” carried by a disordered region (Dyson and Wright 2005) or a weakness to mutation or translocation. If a translocation occurred at a fully ordered region, then a misfolded protein would be synthesized, which would be rapidly degraded during the course of quality control in the cell. On the other hand, translocation in a disordered region would not affect the structures of ordered regions, and thus abnormal proteins may be produced in the cell, which could lead to diseases, as in the case of human leukemias (Goodman and Smolik 2000; Yang 2004). In a similar way, too many disordered regions may enhance the weakness of the disordered regions. In stable hub proteins, once a complex is formed, the complex is stable, and thus the disordered regions will not remain in a disordered state. However, sociable proteins have to repeat the disorder/order transition many times during their transient interactions, and thus the disordered region will remain in the disordered state far longer than a stable hub protein. Although these considerations are just speculation at this time, this may be one possible explanation why the global flexibility of the sociable proteins is more important than the rate of disordered.
Materials and Methods
Definition of interface residues
For each protein chain, the surface residues were first identified as the residues with an accessible surface area (ASA) more than 0.0. The ASA was calculated using the program NSC (Eisenhaber and Argos 1993). Then, the residues in the interfaces were defined as the residues with interacting residues in the other subunit. If a residue pair has one or more contacting atoms in the other residue, then the residue pair is considered as interacting. When the distance between an atom pair is less than 4.0 Å, the atom pair is regarded as in contact.
Mapping of all possible interfaces
The interfaces of protein–protein interactions are generally extracted from a single protein complex in the PDB. This extraction may not be suitable for sociable proteins, because it is expected that sociable proteins interact with many partners, and they are usually solved as different crystal structures. To extract all possible interfaces, we also checked the interactions of closely related proteins found in the PDB. To search the closely related proteins, we clustered the 68,474 proteins (see data set subsection in Results and Discussion) using BLASTCLUST (threshold: identity ≥90% and alignment length ≥80%), and the proteins belonging to the same cluster were regarded as closely related proteins. The interface residues of closely related proteins were then mapped onto the corresponding residues in the other related proteins. For this purpose, the calculation costs of mapping increase as the number of subunits in an entry becomes larger, and the proteins with many subunits are usually stable complexes. Therefore, we neglected the entries with more than 30 subunits in a single entry.
Definition of sociable protein
Sociable proteins are, ideally, proteins that can interact with multiple partners, and the partners can be changed dynamically. Such proteins will be identified as follows: First, the number of observed complexes in the PDB, or the binding states, containing a given protein chain is counted, and the proteins with a large number of binding states are identified as the sociable proteins. For example, if protein A is found in the PDB as the A–B complex and the A–C complex (A, B, and C are different proteins) in different PDB entries, then the number of binding states is defined as two. This approach is based on the naïve assumption that a sufficient number of protein structures have been solved. However, it is true that the PDB data are limited, and a true sociable protein might be regarded as nonsociable when only one structure is available. To reduce the weakness of this point, we also used the homologous proteins in the PDB. In other words, the number of binding states is counted for each sequence family defined above (see the data set subsection in the Results and Discussion section). Therefore, if the protein complex, for example, A′–D is also known (A′ belongs to the same sequence family as the protein A), then the number of binding states of the above example becomes three. Furthermore, to enhance the reliability of the identifications, sequence families with less than five members were not used in the following analyses, and only the family members with more than three binding patterns were considered as sociable. In addition, only the family members with a single binding pattern were used as the nonsociable proteins. It may be noteworthy that the main reason the number of sociable proteins is relatively smaller than the number of all entries in the PDB stems from the limitation about the minimum number of members in the sequence family. Thus, if we reduced the number of limitations, five, then we would find more sociable proteins, but this would increase the error rate of stable complexes being classified as sociable proteins. Therefore, we used the conditions described above.
Degree of structural flexibility
The degree of structural flexibility is defined for each sequence family. All-against-all comparisons of protein structures for each sequence family are carried out, and maxRMSD is used as an indicator to evaluate the degree of structural flexibility of the sequence family. The RMSD value of Cα atoms for a pair of proteins is calculated after a best-fit superimposition (Kearsley 1989), using an alignment obtained by traditional sequence alignment algorithms (Needleman and Wunsch 1970).
Acknowledgments
We are grateful to Dr. Patil for providing the PDB ID list of their hub proteins. This work was partially supported by a Grant-in-Aid for Scientific Research on the Priority Area “Transportsome” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to K.K. Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, at the University of Tokyo.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Kengo Kinoshita, University of Tokyo, 4-6-1, Shirokanedai, Minato-ku, Tokyo 108-0032, Japan; e-mail: kino@ims.u-tokyo.ac.jp; fax: 81-3-5449-5133.
Abbreviations: PDB, Protein Data Bank; RMSD, root mean square deviation.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073196308.
References
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken, C., Iakoucheva, L.M., Romero, P.R., Dunker, A.K. Combining prediction, computation and experiment for the characterization of protein disorder. Curr. Opin. Struct. Biol. 2004;14:570–576. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Dunker, A.K., Cortese, M.S., Romero, P., Iakoucheva, L.M., Uversky, V.N. Flexible nets—The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dyson, H.J., Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Eisenhaber, F., Argos, P. Improved strategy in analytic surface calculation for molecular systems: Handling of singularities and computational efficiency. J. Comput. Chem. 1993;14:1272–1280. [Google Scholar]
- Friedler, A., Veprintsev, D.B., Rutherford, T., von Glos, K.I., Fersht, A.R. Binding of Rad51 and other peptide sequences to a promiscuous, highly electrostatic binding site in p53. J. Biol. Chem. 2005;280:8051–8059. doi: 10.1074/jbc.M411176200. [DOI] [PubMed] [Google Scholar]
- Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., et al. A protein interaction map of Drosophila melanogaster . Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goodman, R.H., Smolik, S. CBP/p300 in cell growth, transformation, and development. Genes & Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Gutteridge, A., Thornton, J. Conformational changes observed in enzyme crystal structures upon substrate binding. J. Mol. Biol. 2005;346:21–28. doi: 10.1016/j.jmb.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Henrick, K., Thornton, J.M. PQS: A protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- Iakoucheva, L.M., Brown, C.J., Lawson, J.D., Obradovic, Z., Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Ishida, T., Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., Sakaki, Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H., Tombor, B., Albert, R., Oltvai, Z.N., Barabasi, A.L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Jeong, H., Mason, S.P., Barabasi, A.L., Oltvai, Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jones, S., Thornton, J.M. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W., Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kearsley, S.K. On the orthogonal transformation used for structural comparisons. Acta Crystallographica A. 1989;45:208–210. [Google Scholar]
- Kriwacki, R.W., Hengst, L., Tennant, L., Reed, S.I., Wright, P.E. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: Conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. A map of the interactome network of the metazoan C. elegans . Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Tan, H., Rost, B. Loopy proteins appear conserved in evolution. J. Mol. Biol. 2002;322:53–64. doi: 10.1016/s0022-2836(02)00736-2. [DOI] [PubMed] [Google Scholar]
- McEwan, I.J., Dahlman-Wright, K., Ford, J., Wright, A.P. Functional interaction of the c-Myc transactivation domain with the TATA binding protein: Evidence for an induced fit model of transactivation domain folding. Biochemistry. 1996;35:9584–9593. doi: 10.1021/bi960793v. [DOI] [PubMed] [Google Scholar]
- Namba, K. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- Needleman, S.B., Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Patil, A., Nakamura, H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Schulz, G.E. Nucleotide binding proteins. In: Balaban M., editor. Molecular mechanism of biological recognition. Elsevier; New York: 1979. pp. 79–94. [Google Scholar]
- Sheinerman, F.B., Honig, B. On the role of electrostatic interactions in the design of protein–protein interfaces. J. Mol. Biol. 2002;318:161–177. doi: 10.1016/S0022-2836(02)00030-X. [DOI] [PubMed] [Google Scholar]
- Shoemaker, B.A., Portman, J.J., Wolynes, P.G. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl. Acad. Sci. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolar, R.S., Record, M.T. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, Y., Kinoshita, K., Nakamura, H. Analyses of homo-oligomer interfaces of proteins from the complementarity of molecular surface, electrostatic potential and hydrophobicity. Protein Eng. Des. Sel. 2006;19:421–429. doi: 10.1093/protein/gzl026. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wright, P.E., Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure–function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Yang, X.J. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]