Abstract
Green fluorescent protein (GFP) was used to tag proteins of the mitochondrial matrix, inner, and outer membranes to examine their sorting patterns relative to mtDNA in zygotes of synchronously mated yeast cells in ρ+ × ρ0 crosses. When transiently expressed in one of the haploid parents, each of the marker proteins distributes throughout the fused mitochondrial reticulum of the zygote before equilibration of mtDNA, although the membrane markers equilibrate slower than the matrix marker. A GFP-tagged form of Abf2p, a mtDNA binding protein required for faithful transmission of ρ+ mtDNA in vegetatively growing cells, colocalizes with mtDNA in situ. In zygotes of a ρ+ × ρ+ cross, in which there is little mixing of parental mtDNAs, Abf2p–GFP prelabeled in one parent rapidly equilibrates to most or all of the mtDNA, showing that the mtDNA compartment is accessible to exchange of proteins. In ρ+ × ρ0 crosses, mtDNA is preferentially transmitted to the medial diploid bud, whereas mitochondrial GFP marker proteins distribute throughout the zygote and the bud. In zygotes lacking Abf2p, mtDNA sorting is delayed and preferential sorting is reduced. These findings argue for the existence of a segregation apparatus that directs mtDNA to the emerging bud.
Keywords: yeast, mitochondria, mitochondrial DNA, zygotes, DNA segregation
Mitochondria are involved in many essential metabolic activities and therefore their inheritance is vital for the survival of progeny cells. However, yeast cells that fail to transmit their wild-type (ρ+) mitochondrial genomes can still give rise to viable progeny, although those cells are respiratory deficient. This essential difference between the inheritance of bulk mitochondria and mtDNA is readily seen in the yeast Saccharomyces cerevisiae where daughter buds that fail to inherit mitochondria are inviable (Yaffe, 1991; Berger and Yaffe, 1996), but daughter buds that do not receive any mtDNA (ρ0 petite mutants) are viable as long as they are provided with a fermentable carbon source. mtDNA is inherited with great fidelity in wild-type yeast cells grown under standard laboratory conditions, but different factors, external as well as genotypic, can lead to an instability of the mitochondrial genome. For example, ρ+ cells that lack the mitochondrial high mobility group (HMG)1 protein, Abf2p, fail to propagate their mtDNA during growth on fermentable carbon sources, a defect which leads to an accumulation of ρ0 petites in the population (Diffley and Stillman, 1991; Megraw and Chae, 1993; Megraw et al., 1994; Zelenaya-Troitskaya et al., 1998).
Although the analysis of the transmission of mtDNA in vegetatively growing cells has proved useful for identifying genes that function directly or indirectly in mtDNA inheritance, studies with zygotes offer some distinct advantages to dissecting the mechanism of mtDNA inheritance. During zygote maturation, diploid buds can arise from either parental end or from the medial region of the zygote. These budding patterns provide topographical landmarks that facilitate the analysis of sorting of mitochondrial constituents not possible with vegetatively growing cells. For example, zygotes formed from haploid parents that have different mitochondrial genotypes or from parents that have been differentially labeled with mitochondrial marker proteins can be analyzed to determine the fate of these mitochondrial constituents. Such experiments have revealed a number of important features related to the sorting and redistribution of different mitochondrial components during zygote maturation and diploid bud formation.
First, it is clear that mitochondria from each parent fuse into an extended mitochondrial reticulum in the zygote. This was suggested from early observations that parental mtDNAs actively recombine in zygotes (Thomas and Wilkie, 1968; Dujon et al., 1974, 1976) and later from experiments that directly showed the redistribution of prelabeled mitochondrial matrix protein from one parental end of the zygote to the other (Azpiroz and Butow, 1993). Second, in ρ+ × ρ+ crosses, there is very limited mixing of parental mtDNAs in the fused mitochondrial reticulum of the zygote; whatever mixing occurs is confined to the neck region of the zygote where the medial bud emerges. End buds tend to be genotypically pure for the parental mtDNA of the parent that gave rise to that end of the zygote, whereas medial buds contain both parental (as well as recombinant) mtDNA molecules (Strausberg and Perlman, 1978; Zinn et al., 1987). Thus, mtDNA segregation in zygotes is not random. Third, in crosses between ρ+ cells and certain ρ− petites (cells whose mitochondrial genomes have suffered large deletions of ρ+ mtDNA), there can be a nearly quantitative suppression of the transmission of ρ+ mtDNAs to the diploid progeny (for reviews see Dujon, 1981 and Piskur, 1994). Petites exhibiting that property are called hypersuppressive and their mtDNA genomes characteristically include a ∼300-bp element as part of their repeating unit referred to as an ori/rep sequence (Blanc and Dujon, 1980, 1982; de Zamaroczy et al., 1981).
Finally, the analysis of the sorting of mtDNA and mitochondrial matrix protein markers in zygotes derived from synchronous matings in ρ+ × ρ+ and ρ+ × ρ0 crosses showed that mtDNA movements are independent of the movements of mitochondrial matrix proteins (Azpiroz and Butow, 1993). For example, in zygotes of a ρ+ × ρ+ cross, where there is little mixing of the parental mtDNAs, matrix marker proteins rapidly and quantitatively mix throughout the zygote and into the diploid buds. Similar conclusions were recently drawn using vital fluorescent dyes and labeled mtDNA by Nunnari et al. (1997). In zygote maturation experiments involving ρ+ × ρ0 crosses, we observed extensive mixing of ρ+ mtDNA in the zygote, as well as mixing of mitochondrial matrix protein markers (Azpiroz and Butow, 1993). Significantly, a number of intermediate zygote forms were detected showing that mtDNA movements could be well resolved both temporally and spatially from those of the matrix protein markers.
Here, we describe experiments measuring mtDNA sorting in zygotes relative to additional mitochondrial markers including inner and outer mitochondrial membrane proteins and a mitochondrial DNA binding protein. These studies show that mtDNA sorting is independent of the sorting of the marker proteins of these different mitochondrial compartments. These studies also reveal a remarkable preferential transmission of mtDNA into the medial diploid bud in ρ+ × ρ0 crosses, strongly suggesting that there is an active segregation apparatus that directs mtDNA into emerging buds.
Materials and Methods
Strains and Growth Conditions
The strains used in this study were PSY142 ρ+ (MATα, leu2, lys2, ura3) and S150-2B ρ+ (MATa, his3Δ1, leu2-113, ura3-52, trp1-289). ρ0 derivatives of these strains were generated by passage of ρ+ cells on YPD medium containing 25 μg/ml ethidium bromide. PSY142 ρ0 and S150-2B ρ+ carrying a deletion mutation in the ABF2 gene, called PSY142 Δabf2 ρ0 and S150-2B Δabf2 ρ+, were generated as described below. Cells were grown at 30°C on YP medium (1% yeast extract and 1% Bactopeptone) containing either 2% dextrose (YPD) or 2% glycerol (YPGly), or on minimal YNB medium (0.67% yeast nitrogen base without amino acids) containing either 2% dextrose (YNBD) or 2% glycerol (YNBGly), and YNB medium containing 1% casamino acids and either 2% dextrose (YNBD + cas), 2% glycerol (YNBGly + cas), 2% galactose (YNBGal + cas), 2% raffinose (YNBR + cas) or 2% galactose and 2% raffinose (YNBGalR + cas) supplemented with nutritional requirements as necessary.
Construction of Green Fluorescent–Fusion Protein Expression Vectors
Plasmid pGAL-CLbGFP was constructed by ligation of a 290-bp BamHI-XhoI fragment containing the 5′-flanking region plus codons 1–52 of the CIT1 gene, together with a 730-bp XhoI-KpnI fragment containing the coding regions of a bright green version of green fluorescent protein (GFP) (bGFP; see below) and a 490-bp KpnI-HindIII fragment containing the 3′- flanking region of CIT1 and ligated into the BamHI-HindIII site of the CEN-URA3 plasmid pGAL68 (Zelenaya-Troitskaya et al., 1998). The CIT1 5′-flanking region plus codons 1–52 was generated by PCR using the primers, 5′-TAAGGGGGATCCTTGCTGTTTAC-3′ and 5′-TGCCTTTGCTCGAGTAATTTCAGC-3′, and digested with BamHI and XhoI. The 3′-flanking region of CIT1 was generated by PCR using the primers, 5′-CGAAAGTAGGTACCAAGGAAAATTTG-3′ and 5′-GTGACATTAAGCTTGAGGTAAGAAC-3′, and digested with KpnI and HindIII. The bGFP contained three amino acid substitutions, F99S, M153T, and V163A (our unpublished materials).
To construct plasmid pGAL-YbGFP, a 2.3-kb fragment containing the coding region of the YTA10 gene was generated by PCR using the primers, 5' TTAACGCAGTCTAGAAATAAAGGCATC-3′ and 5′-CGTTTTATTTCCTCGAGAATTTGTTGC-3′. The PCR product was digested with XbaI and XhoI. A XbaI-XhoI fragment from pG7GAL-CLbGFP, which was constructed by insertion of a 2.1-kb EcoRI-HindIII fragment from pGAL-CLbGFP ligated into the EcoRI-HindIII site of pGEM-7Zf (+) (Promega Corp., Madison, WI), was replaced with the XbaI-XhoI fragment of the YTA10 gene, yielding pG7-YbGFP. A 3.5-kb XbaI-HindIII fragment from pG7-YbGFP was cloned into the XbaI- HindIII site of pGAL68 to generate pGAL-YbGFP.
To construct plasmid pGAL-bGFPT, a 0.2-kb fragment containing the TOM6 (ISP6/MOM8b) gene was generated by PCR using the primers, 5′-AAATAATTGAAATGCATACGGTATGTTTGC-3′ and 5′-AATCTCAACGGTACCAGAACCAAC-3′. The PCR product was digested with NsiI and KpnI. The NH2-terminal in-frame fusion cassette of bGFP, in which the TAG (stop) codon was replaced with a GCA (alanine) codon, was generated by PCR using the plasmid pG7-bGFP as template DNA, and the primers, 5′-ATTTAGGTGACACTATA-3′ and 5′-TTCTACGAATATGCATTGTATAGTTCATCC-3′. The PCR product was digested with BamHI and NsiI. A 1-kb BamHI-KpnI fragment from pG3GAL-CLbGFP, which was constructed by insertion of a 2.1-kb EcoRI-HindIII fragment from pGAL-CLbGFP into the EcoRI-HindIII site of pGEM-3Zf(+), was replaced with a BamHI-NsiI fragment (GFP cassette) and a NsiI-KpnI fragment of the TOM6 gene to yield pG3GAL-bGFPT. A 0.8-kb BamHI-HindIII fragment and a 0.6-kb HindIII fragment from pG3GAL-bGFPT were cloned into the BamHI-HindIII site of pGAL68 to generate pGAL-bGFPT.
Plasmid pGAL-Abf2-GFP was constructed as follows: a 1.6-kb BamHI-HindIII fragment consisting of 0.5-kb BamHI-XhoI fragment of the coding region of the ABF2 gene ligated to 0.73-kb XhoI-KpnI fragment of bGFP, ligated to a 0.4-kb KpnI- HindIII fragment containing the 3′ flanking region of the ABF2 gene, was cloned into the BamHI-HindIII site of pGAL68 to generate pGAL-Abf2-GFP.
Construction of Δabf2 Strains
To construct strain PSY142 Δabf2 ρ0, a 750-bp EcoRI-XhoI fragment containing the 5′- and 3′-flanking regions and entire coding sequence of the ABF2 gene, was obtained from pRS416/ABF2-GFP (Zelenaya-Troitskaya et al., 1998), and cloned into the EcoRI-XhoI site of plasmid pBluescript II KS (+) (Stratagene, La Jolla, CA), creating pBSII-ABF2. The ABF2 coding sequence between the MunI and NheI sites was replaced with a EcoRI-NheI fragment containing the URA3 gene to generate pBSII-abf2:: URA3. Strain PSY142 ρ0 was transformed with a linearized 1.5-kb EcoRI-XhoI fragment from pBSII-abf2::URA3. S150-2B Δabf2 ρ+ was constructed by transforming S150-2B ρ+ with a linearized 1.6-kb EcoRI fragment from plasmid pAM1A::TRP1 (Diffley and Stillman, 1991). Disruption in these strains was verified by Southern blotting.
Fractionation of Mitochondria
PSY142 ρ+ cells transformed with the various GFP constructs described above were grown on YNBGalR + cas medium for 16–20 h (OD600 1.2– 1.6) and then converted to spheroplasts using Zymolyase 100T. The spheroplasts were broken using a loose-fitting Dounce homogenizer and subjected to differential centrifugation to isolate mitochondria as previously described (Zinser and Daum, 1995; Newman et al., 1996). Fractionation of mitochondria, based on published procedures (Pajic et al., 1994; Glick, 1995), was performed as follows: for conversion to mitoplasts, mitochondria (2 mg of protein) were resuspended in 1 ml of hypotonic buffer (20 mM Hepes-KOH, pH 7.4, containing 0.5% BSA), and incubated for 30 min at 4°C with gentle mixing every 5 min. After 25 min, KCl was added to a concentration of 80 mM. For control (nonswelling conditions), isolated mitochondria (1 mg of protein) were incubated in 1 ml of isotonic buffer (0.6 M sorbitol, 20 mM Hepes-KOH, pH 7.4, containing 0.5% BSA), and as indicated in the Results, treated with proteinase K at a concentration of 50 μg/ml for 20 min on ice, followed by a 10-min incubation on ice with 1 mM of PMSF. Mitochondria and mitoplasts were reisolated by centrifugation for 10 min at 12,000 g. Mitochondria were resuspended in 1 ml of SDS sample buffer. For carbonate extraction, mitoplasts were resuspended in 1 ml of extraction buffer (0.1 M Na2CO3 and 1 mM PMSF), and incubated for 30 min. For sonication, mitoplasts were resuspended in 1 ml of sonication buffer (0.1 M NaCl, 20 mM Hepes-KOH, pH 7.4, and 1 mM PMSF) and subjected to three rounds of freeze-thaw at −80°C, followed by sonicating at 70 W for 30 times at 1-s each with a sonifier (model 450; Branson Ultrasonics Corp., Danbury, CT). The samples from carbonate extraction and sonication were centrifuged (Sorvall Ultra 80; Dupont, Newtown, CT) at 226,000 g for 60 min. The pellet fractions were resuspended in 1 ml of SDS sample buffer. All samples were analyzed by SDS-PAGE and Western blotting, probed with a polyclonal antibody of GFP (Clontech Laboratories, Inc., Palo Alto, CA), and detected using a goat anti–rabbit IgG (H+L)-HRP conjugate (Bio-Rad Laboratories, Inc., Hercules, CA) and the enhanced chemiluminescence reagents (Amersham Corp., Arlington Heights, IL).
Induction of Protein–GFP Fusions
PSY142 ρ+ or ρ0 cells transformed with GFP chimeric genes under control of the GAL1-10 promoter in the pGAL68 derivatives were grown on YNBR +cas medium to mid-log phase, inoculated into YNBGalR + cas medium and grown to induce the synthesis of the protein–GFP fusions (12–18 h for CS1–GFP, Yta10p–GFP, and GFP–Tom6p; 45–60 min for Abf2p-GFP). These different induction times were chosen from preliminary experiments to optimize detection of each GFP fusion protein. S150-2B Δabf2 ρ+ transformants precultured on YNBGly +cas medium to mid-log phase, were transferred to YNBGal + cas medium, and then incubated for 2 h to induce the synthesis of CS1–GFP. Cells were collected by centrifugation before synchronized mating.
Sorting Analysis of mtDNA and Protein–GFP Fusion in Zygotes
Synchronized mating was performed as previously described (Azpiroz and Butow, 1995). For fixation, cells from mating mixtures at each time point (1–4 or 6 h) were resuspended in 1 ml of 3.7% formaldehyde solution and incubated for 1 h at 30°C. After three washes in water, the cells were stained by brief incubation in 1 μg/ml 4′,6-diamino-2-phenylindole (DAPI) in water followed by three washes in water. The samples were resuspended in 0.25% low melting agarose solution prewarmed at 37°C, placed on a microscope slide protected by a coverslip, and then kept for 10 min at 4°C. Zygotes were scored according to the previous procedures (Azpiroz and Butow, 1993, 1995). At least 100 zygotes were scored for each time point presented in Figs. 4–7, 9, and Table I.
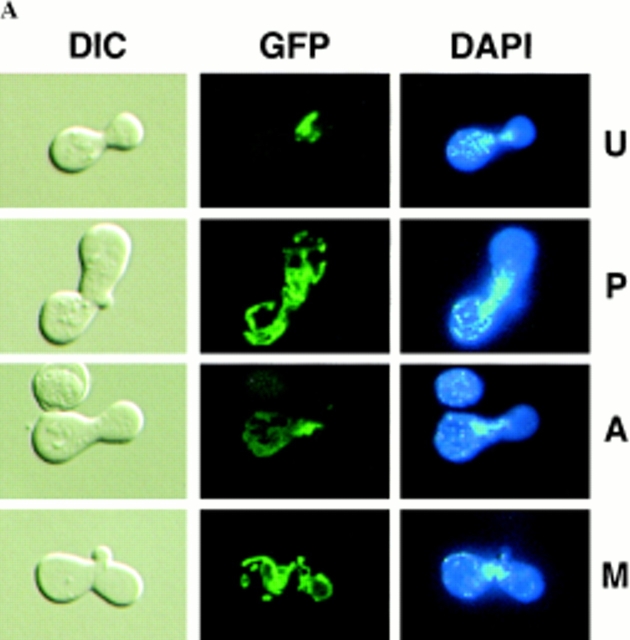
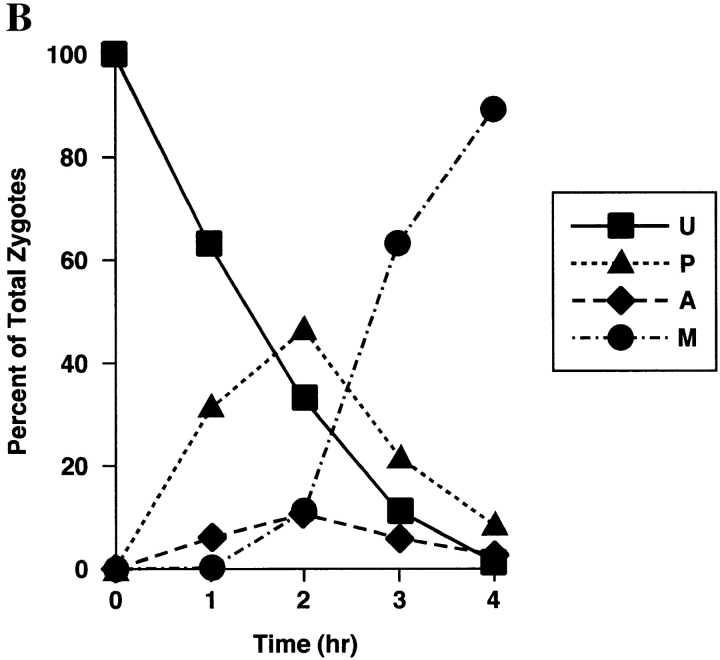
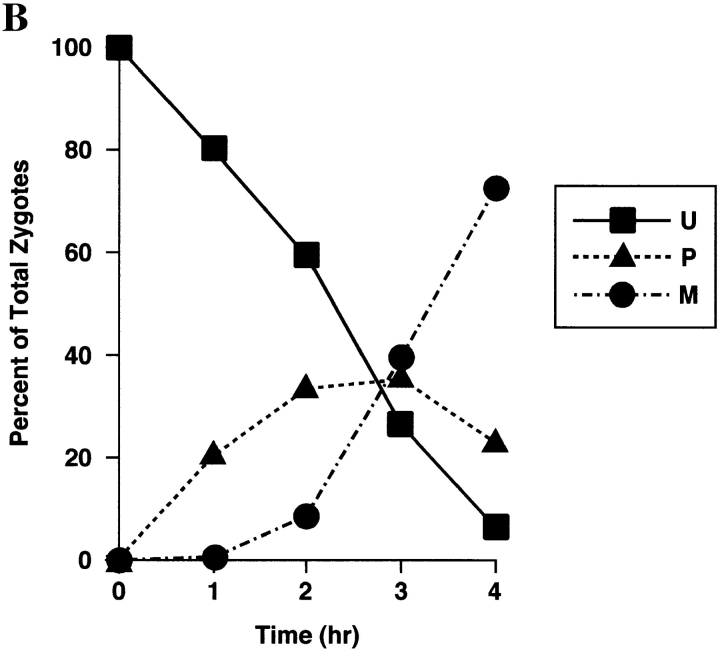
Figure 4.
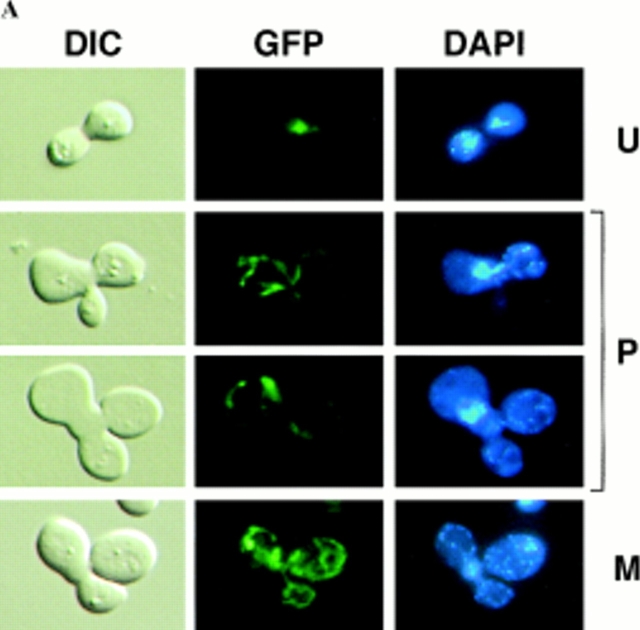
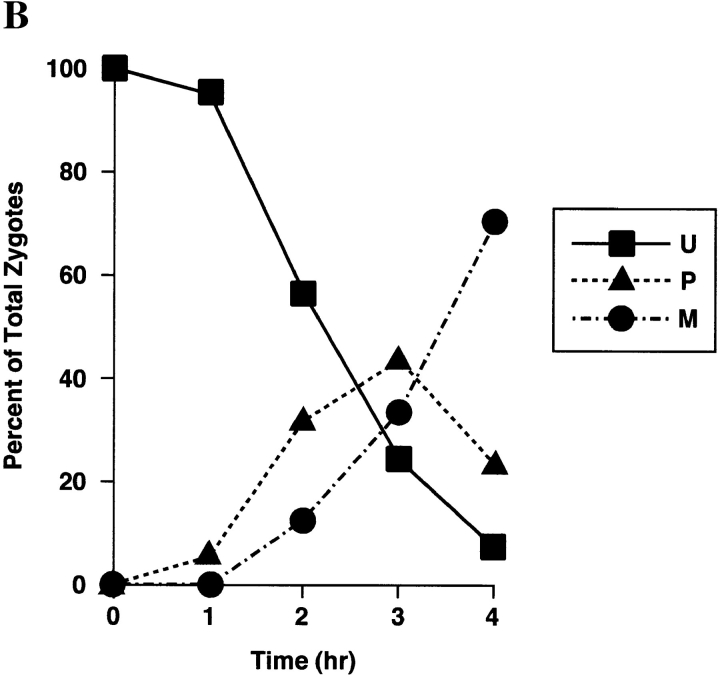
Sorting of CS1–GFP in a PSY142 ρ0 [CS1–GFP] × S150-2B ρ+ cross. (A) Shown are representative examples of the different zygote forms, indicated diagrammatically in Fig. 3, detected in the cross based on the distribution of GFP and DAPI (mtDNA) fluorescence. (B) Kinetics of appearance and disappearance of the different zygote forms.
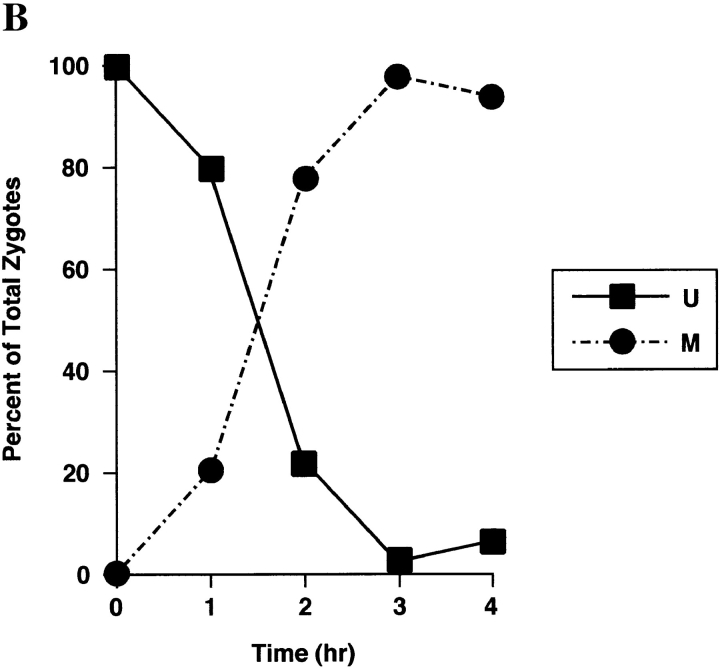
Figure 7.
Sorting of Abf2p–GFP in a PSY142 ρ+ [Abf2p–GFP] × S150-2B ρ+ cross. (A) Representative examples of the U and M form zygotes generated in the cross. In this experiment, PSY142 ρ+ cells were prelabled with Abf2p–GFP before synchronous mating with the S150-2B ρ+ strain. (B) Kinetics of sorting of Abf2p–GFP.
Figure 9.
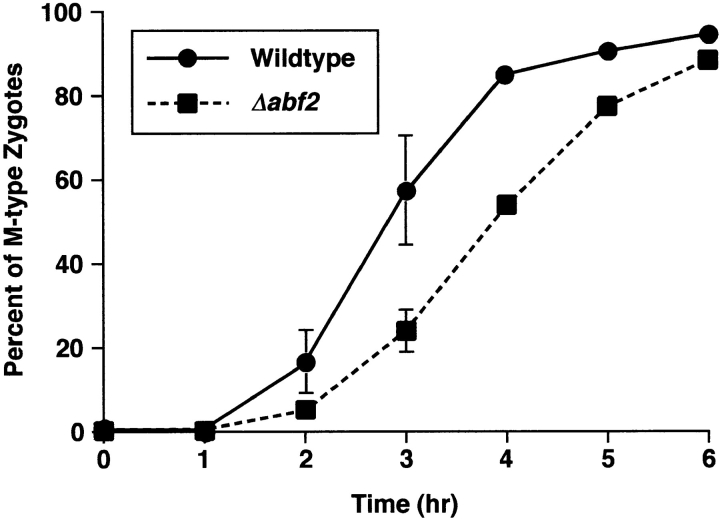
The sorting of CS1–GFP is delayed in a S150-2B Δabf2 ρ+ [CS1–GFP] × PSY142 Δabf2 ρ0 cross. Wild-type and Δabf2 derivatives of S150-2B ρ+ cells transiently expressing CS1–GFP and PSY142 ρ0 wild-type and Δabf2 cells were synchronously mated and the kinetics of appearance of M form zygotes was determined by direct microscopic analysis.
Table I.
mtDNA Is Preferentially Transmitted to the Medial Bud
| Crosses | ||||||
|---|---|---|---|---|---|---|
| ρ+ [CS1-GFP] × ρ0 | 17 | 2 | 81 | |||
| ρ+ [Yta10p-GFP] × ρ0 | 26 | 8 | 66 | |||
| ρ+ [GFP-Tom6p] × ρ0 | 21 | 14 | 65 | |||
Synchronized matings were performed using strain S150-2B ρ+ prelabeled with GFP-tagged fusion proteins and strain PSY142 ρ0 cells as described in Materials and Methods. After formaldehyde fixation, cells from mating mixtures were stained by DAPI and then observed by fluorescence microscopy. Black spots and shaded lines represent mtDNA and GFP fusions in mitochondria, respectively.
100 zygotes with one-third to two-thirds the size of a diploid bud were scored for 2- and 3-h time points after mating.
Microscopy and Image Analysis
The samples were observed using a Leica microscope (model DMRXE; Deerfield, IL) equipped for an HBO 100 W/2 mercury arc lamp, an X100 Plan-Apochromat objective, and epifluorescence with the following filter sets: (a) 340–380-nm band-pass excitation filter; (b) 400-nm dichroic reflector; (c) \>425-nm long-pass emission filter for DAPI, and (d) 450–490, 510, and \>515 for GFP. Differential interference contrast and fluorescence images were captured with a color-chilled three charge-coupled device camera system (model C5810; Hamamatsu Phototonics, Bridgewater, NJ) and then processed using Adobe Photoshop (Adobe Systems, Inc., San Jose, CA).
Results
Mitochondrial Marker Protein–GFP Fusions
To examine the sorting of mitochondrial components during zygote maturation, we prelabeled a ρ0 strain with a mitochondrial marker protein of interest and then synchronously mate those cells to a ρ+ strain lacking the marker protein; fluorescence microscopy is then used to monitor the sorting of the marker protein and DAPI-stained mtDNA during the course of zygote maturation and diploid bud formation. In this study we introduce a new terminology for describing these crosses. Where both markers (i.e., mtDNA and a protein–GFP fusion) originate in the same parent, the experiment is referred to as a cis cross; where each parent introduces a different marker to the zygote, it is referred to as a trans cross. The ρ+ × ρ0 trans crosses are particularly useful for revealing how mitochondrial constituents sort, since the initial separation of the mitochondrial marker protein and mtDNA at opposite ends of the newly formed zygote allows a clear-cut time and spatial resolution of the sorting process (Azpiroz and Butow, 1993).
To extend this analysis to other mitochondrial compartments, we have constructed a series of gene fusions that would direct GFP to the mitochondrial matrix, the inner and outer mitochondrial membranes, and mtDNA. As shown in Fig. 1 A, these gene fusions were constructed to encode chimeric proteins between GFP and (a) 52 amino acids of the NH2-terminal region of the matrix protein, citrate synthase 1 (CS1); (b) the COOH terminus of the full-length inner membrane protease subunit, Yta10p (Tauer et al., 1994; Arlt et al., 1996; Guelin et al., 1996); (c) the NH2 terminus of outer membrane protein, Tom6p (Isp6p), a component of a complex of the outer membrane, protein import apparatus (Kassenbrock et al., 1993; Alconada et al., 1995); and (d) the COOH terminus of the HMG protein, Abf2p, that binds to mtDNA (Diffley and Stillman, 1992; Newman et al., 1996; Zelenaya-Troitskaya et al., 1998). Expression of each of these gene fusions was placed under the control of the GAL1-10 promoter in the plasmid pGAL68 and transformed into PSY142 ρ+ cells. All of these fusion proteins were found to be stable under the conditions of the zygote sorting experiments, and their mRNAs were essentially undetectable by Northern blot analysis of total RNA isolated from cells immediately following the 2-h incubation in the dextrose medium used before the initiation of synchronous mating (data not shown).
Figure 1.
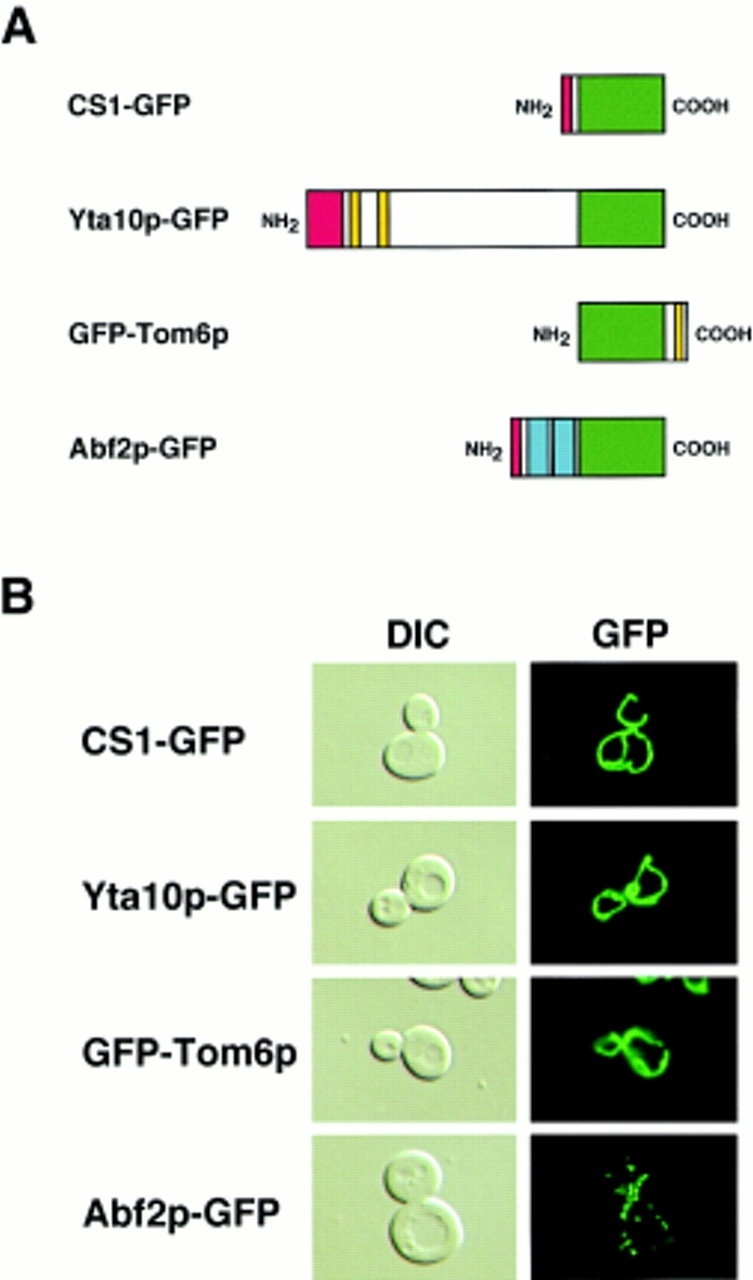
GFP fusion proteins _targeted to mitochondria. (A) Diagrammatic representation of fusion proteins between GFP and all or a portion of the indicated mitochondrial proteins. Green, GFP; red, mitochondrial presequence; yellow, putative transmembrane domains; blue, HMG boxes in Abf2p. (B) Fluorescence pattern of the indicated GFP fusion proteins (right panels) in cultures of PSY142 ρ+ cells grown on YNBGalR + cas medium. Cells were transformed with plasmids encoding either the matrix (CS1-GFP), inner membrane (Yta10p-GFP), outer membrane (GFP-Tom6p), or mtDNA (Abf2p-GFP) marker proteins (refer to Materials and Methods). Left panels, cells visualized by DIC.
To determine whether these fusion proteins are _targeted to mitochondria, cells from log-phase cultures of the transformants grown in YNBGalR + cas medium were examined by epifluorescence microscopy. Fig. 1 B shows that, with the exception of Abf2p–GFP, expression of each of the GFP fusion constructs results in a morphological pattern characteristic of the tubular mitochondrial network located near the periphery of the cell (Fig. 1 B) (Hoffman and Avers, 1973). In contrast, Abf2p–GFP, which complements the mtDNA instability phenotype of a Δabf2 mutant strain (data not shown), shows a distinctly punctate pattern coincident with that of DAPI-stained mtDNA. We showed recently that this Abf2p–GFP fluorescence pattern reflects the association of Abf2p–GFP with mtDNA in vivo (Zelenaya-Troitskaya et al., 1998). In that report, we demonstrated that a GFP derivative of a mutant form of Abf2p containing mutations of key amino acid residues in each of the HMG boxes that inhibit DNA binding in vitro had an in vivo fluorescence pattern that more closely resembled that of a matrix protein than the punctate pattern seen for the wild-type Abf2p–GFP derivative. Moreover, mtDNA nucleoids isolated according to the procedure described in Newman et al. (1996) from cells expressing Abf2p-GFP contained this fusion protein at levels comparable to that of endogenous Abf2p (data not shown).
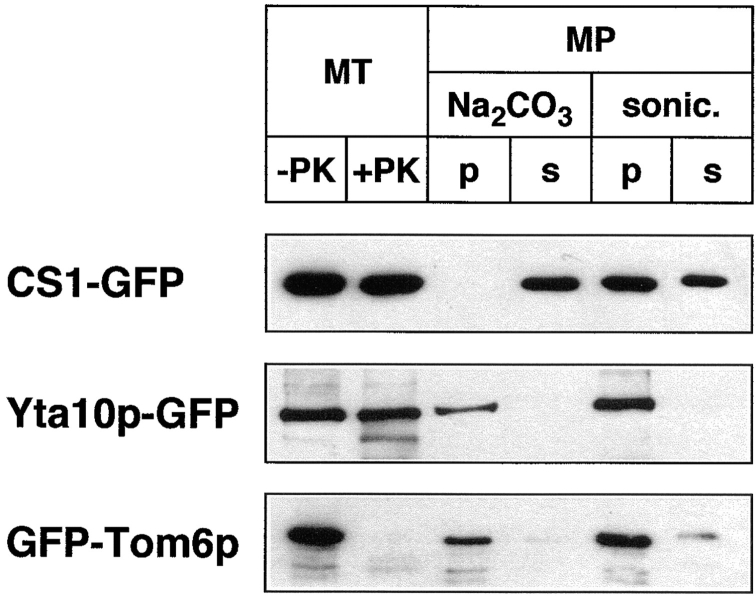
With the exception of Abf2p–GFP, no conclusions can be drawn from the fluorescence patterns alone about the localization of the GFP fusion proteins within specific mitochondrial compartments. Therefore, to verify that each GFP fusion protein is localized to the intended mitochondrial compartment, mitochondria were isolated from strain PSY142 ρ+ expressing each of the GFP fusion proteins and analyzed biochemically to determine the protein localization. In these experiments, the GFP fusion proteins were detected by Western blotting using anti-GFP antiserum. Fig. 2 shows that when mitochondria were treated with proteinase K, the CS1– and Yta10p–GFP fusion proteins were largely resistant to proteolysis, whereas the GFP–Tom6p fusion protein was completely digested, consistent with the known localization and topology of Tom6p in the outer mitochondrial membrane (Kassenbrock et al., 1993). Next, crude mitoplasts were prepared by swelling of isolated mitochondria as described in Materials and Methods, and treated either with Na2CO3, pH 11.5, to release soluble proteins, or sonicated; in both cases, the treated mitoplasts were separated by centrifugation into soluble and pellet fractions. Fig. 2 shows that CS1–GFP was released into the supernatant fraction by treatment with Na2CO3 as well as by sonication. The incomplete release of the CS1–GFP fusion protein by sonication was due to some trapping of the protein by vesicle resealing, which we have verified occurs in parallel experiments with native mitochondrial matrix proteins (data not shown). In contrast to these results, GFP-Tom6p and Yta10p–GFP remained associated with the pellet fractions of the treated mitoplasts. The presence of some signal for GFP–Tom6p is likely due to contamination of the crude mitoplast preparations with outer membrane. Taken together, these results demonstrate that each of these fusion proteins is localized to the intended mitochondrial compartment.
Figure 2.
Localization of GFP fusion proteins in mitochondria. Mitochondria were isolated from PSY142 ρ+ cells grown on YNBGalR + cas medium that were transformed with the plasmid encoding the indicated GFP fusion protein. As described in Materials and Methods, mitochondria (MT) were incubated with or without proteinase K (PK). Crude mitoplasts (MP) were treated with Na2CO3 or sonicated and separated into a pellet (p) and soluble (s) fraction. Proteins were fractionated on a 12% SDS polyacrylamide gel and detected by Western blotting with anti-GFP antiserum.
Sorting Patterns of Mitochondrial Matrix and Membrane Proteins in ρ+ × ρ0 Crosses
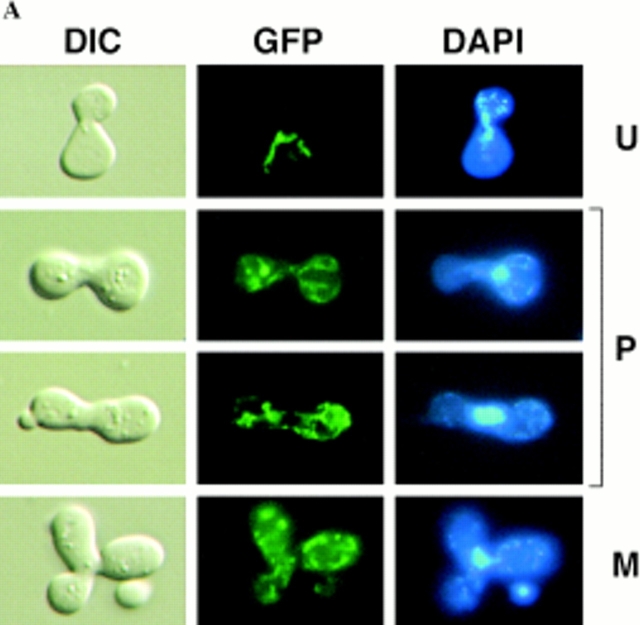
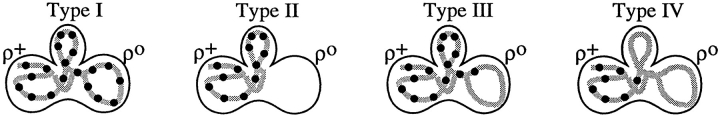
We previously identified four prominent zygote types, shown diagramatically in Fig. 3, that were evident in ρ+ × ρ0 trans crosses when the ρ0 cells were prelabeled with a mitochondrial matrix protein marker (Azpiroz and Butow, 1993). The type U (unmixed) form, in which the mitochondrial protein marker and mtDNA were separated in opposite ends of the zygote, appeared immediately upon zygote formation. This zygote form gradually disappeared with time and was replaced by various intermediate forms, leading eventually to the type M (mixed) zygote in which the marker protein and mtDNA were colocalized and distributed throughout the zygote and into the emerging diploid buds. An unexpected and novel zygote type that appeared early in zygote maturation was the A (asymmetric) form, in which the matrix marker protein had quantitatively translocated from the ρ0 end of the zygote through the fused mitochondrial reticulum into the ρ+ end, well before any movement of mtDNA. Although A form zygotes may not be an obligatory intermediate in the sorting process from U to M zygotes, the mechanism by which that novel zygote type arises remains a mystery, and their presence provides a dramatic illustration of the temporal and spatial resolution of matrix protein and mtDNA within the fused mitochondrial reticulum of the zygote.
Figure 3.
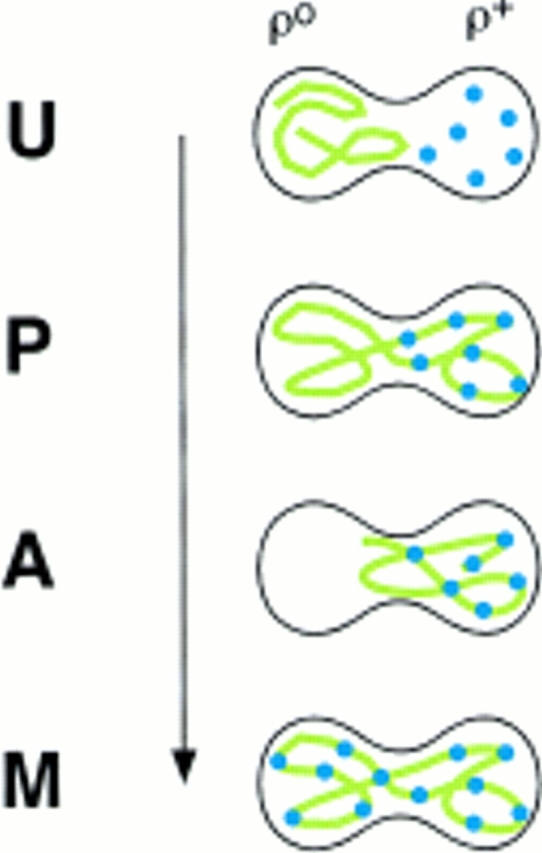
Diagrammatic representation of the different zygote forms detected in synchronous ρ0 × ρ+ crosses with a matrix marker protein prelabeled in the ρ0 parent. Green lines, marker protein in mitochondria; blue dots, mtDNA. U, unmixed; P, partially mixed; A, asymmetric; M, mixed.
To extend these analyses to proteins in other mitochondrial compartments, we followed the time course of sorting of the various GFP fusion proteins and mtDNA in ρ+ × ρ0 trans crosses. The fusion proteins were first expressed in PSY142 ρ0 cells grown in YNBGalR + cas medium, before initiation of synchronous matings to S150-2B ρ+ cells (refer to Materials and Methods). After mating, aliquots of the cells were removed at 1-h intervals and examined by epifluorescence microscopy for the distribution of the GFP fusion proteins and mtDNA by DAPI staining. Representative micrographs of the various zygote types we detected in these crosses are presented in Figs. 4–6, panels A. In Fig. 4–6, panels B, accompanying each micrograph is a plot of the kinetics of appearance and disappearance of the various zygote forms detected.
Figure 6.
Sorting of GFP–Tom6p in a PSY142 ρ0 [GFP–Tom6p] × S150-2B ρ+ cross. (A) Representative examples of the different zygote forms generated in the cross. (B) Kinetics of appearance and disappearance of the different zygote forms.
The sorting patterns observed in the cross with the CS1– GFP matrix marker (Fig. 4) are essentially the same as we previously described for native CS1 and for the chimeric protein OTCase-dihydrofolate reductase (DHFR) (Azpiroz and Butow, 1993). The population of unmixed zygotes, in which CS1–GFP is in the ρ0 end and mtDNA is in the ρ+ end, disappears with time, and by 4 h after zygote formation is replaced almost entirely by the mixed, M-type zygote in which CS1–GFP and mtDNA are distributed to all parts of the cell including the diploid buds. During the course of sorting from the U to M forms, we detected the same two intermediate zygote forms detected previously. One is the P-type zygote, which appears within 1 h of zygote formation, and reaches a maximum of ∼50% of the zygote population by 2 h. In this zygote type some of the CS1–GFP present initially in the ρ0 end of the zygote has moved into the ρ+ end before any appreciable movement of mtDNA. The second intermediate zygote type is the novel A form, which reaches a maximum at 2 h, and accounts for roughly 10% of the zygote population.
Figs. 5 and 6 show the sorting of the inner and outer membrane marker fusion proteins, Yta10p–GFP and GFP– Tom6p, respectively. Similar to matrix protein markers, both of these membrane marker proteins equilibrate throughout the fused mitochondrial reticulum of the zygote and into the diploid buds. There are, however, two notable differences between the sorting patterns of these membrane marker proteins and those of the matrix markers. First, we have never detected any A form zygotes in crosses involving Yta10p–GFP and Tom6p–GFP. Second, the kinetics of sorting of both of the membrane marker proteins is slower than that observed for CS1–GFP (compare Fig. 5 B with Figs. 6 B and 7 B), evident as an ∼1 h delay in the half-time for the disappearance of the U-type zygotes and the time required for appearance of the maximum number of P-type zygotes. These experiments show that, like proteins in the matrix, inner and outer membrane proteins can translocate through the fused mitochondrial reticulum.
Figure 5.
Sorting of Yta10p–GFP in a PSY142 ρ0 [Yta10p–GFP] × S150-2B ρ+ cross. (A) Representative examples of the different zygote forms generated in the cross. (B) Kinetics of appearance and disappearance of the different zygote forms.
Sorting Pattern of Abf2p–GFP in a ρ+× ρ+ Cross
In ρ+ × ρ+ crosses it has long been clear that there is only a very limited mixing of parental mitochondrial genomes, which is confined to the middle or neck region of the zygote where medial buds appear (Strausberg and Perlman, 1978; Zinn et al., 1987; Azpiroz and Butow, 1993). One possibility is that mtDNA, as well as proteins associated with it, represents a compartment of the mitochondrion that exhibits only limited mixing during zygote maturation. To test this notion, we used Abf2p-GFP, which is a functional form of this protein that colocalizes with mtDNA, as a marker for the mtDNA compartment and tested how this protein sorts in zygotes of a ρ+ × ρ+ cross. Possible outcomes are that either Abf2p-GFP, prelabeled in one of the parents, remains with the mtDNA of that parent, or that it is capable of equilibrating among all of the mtDNA molecules in the zygote.
We performed a ρ+ × ρ+ cross in which Abf2p-GFP was prelabeled in PSY142 ρ+ cells in the same manner as described above for prelabeling cells with markers for other mitochondrial membrane compartments, and then synchronously mated to S150-2B ρ+ cells. The results of this experiment (Fig. 7) show that Abf2p-GFP rapidly equilibrates throughout the zygote and into the diploid bud. Moreover, the half-time for disappearance of the U form zygotes is roughly 1 h, similar to that of CS1–GFP U form zygotes (refer to Fig. 4 B). Thus, despite the fact that ρ+ parental mtDNAs do not mix in ρ+ × ρ+ crosses, Abf2p– GFP rapidly redistributes among all of the mtDNA in the zygote. This finding also indicates that although Abf2p– GFP may be a useful marker for mtDNA in static experiments, it is not useful for examining the dynamics of mtDNA movements in this experiment.
Preferential Transmission of mtDNA into Medial Diploid Buds
During the scoring of the various zygote forms generated in the trans crosses described thus far, we noticed that in some cases mtDNA appeared to have been distributed preferentially from the ρ+ end of the zygote into the diploid bud. To examine this potentially important aspect of mtDNA sorting in more detail, we have performed cis ρ+ × ρ0 crosses in which the various mitochondrial marker proteins were transiently expressed in the ρ+ parent before mating with ρ0 cells. In this cis configuration, both mtDNA and the mitochondrial marker protein are initially present in one end of the newly formed zygote, allowing us to observe how these components distribute with time through the fused mitochondrial reticulum to different regions of the zygote and the emerging diploid buds. One possible outcome is that both mtDNA and the marker protein distribute randomly to the other end of the zygote and to the emerging bud; alternatively, the protein and mtDNA may sort differently. In these experiments we have examined zygotes that contain a medial bud between one-third and two-thirds the size of a mature diploid bud. These zygotes, which are most abundant between 2–3 h after mating, have not yet fully equilibrated these mitochondrial components and are thus optimal to detect intermediates in the sorting process (Table I).
Immediately following zygote formation we observed the expected unmixed, unbudded cis zygote type containing mtDNA and the mitochondrial GFP marker protein together in one end of the cell (Fig. 8). A survey of zygotes with a medial bud scored after 2 and 3 h of mating reveals three major zygote types (I–III), representative examples of which are shown in Fig. 8; a quantification of the relative abundance of these zygote types in crosses involving each of the marker proteins is presented in Table I. Type I zygotes are those in which the protein marker and mtDNA have fully equilibrated throughout the zygote and into the diploid buds. These are equivalent to the M form zygotes that are generated in the ρ+ × ρ0 trans crosses described earlier, and they represent between one-fourth and one-fifth of the zygote population for each of the marker proteins (Table I). Type II zygotes are those in which both mtDNA and the marker protein have sorted together and predominately into the diploid bud; they represent the least abundant of the zygote types observed.
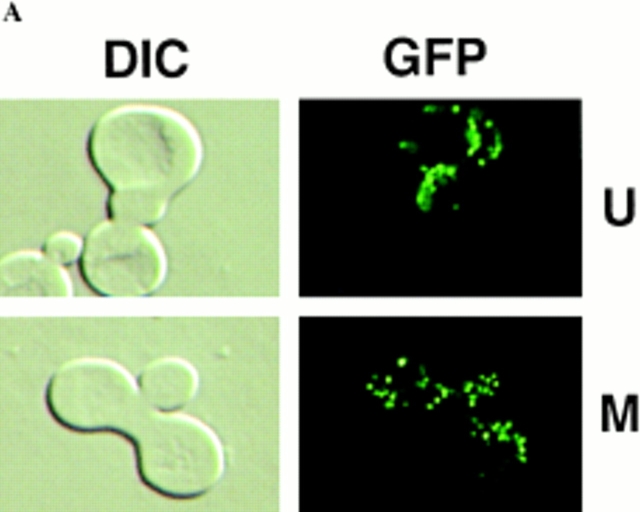
Figure 8.
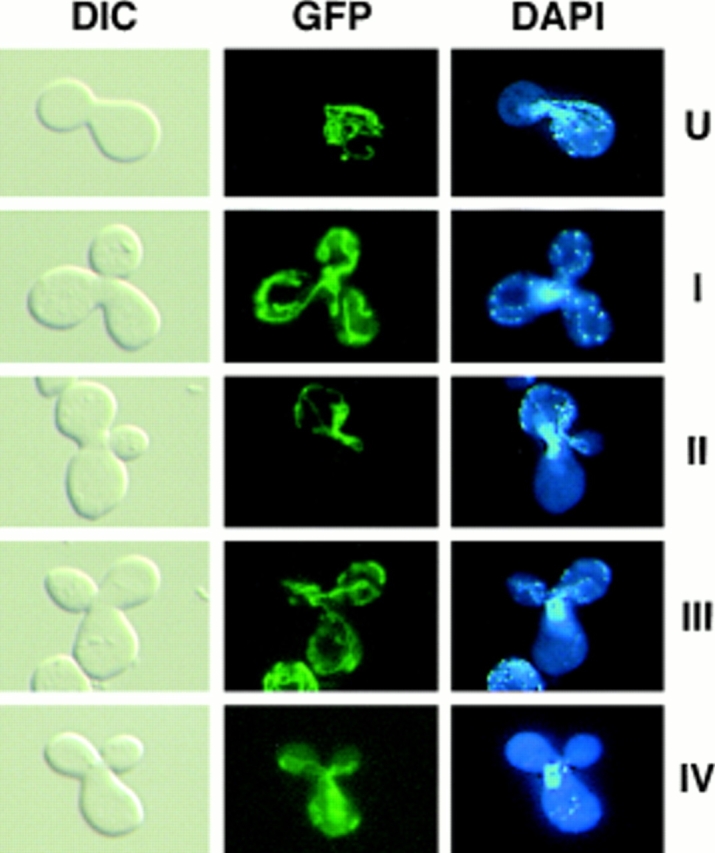
Differential sorting of CS1–GFP and mtDNA in ρ+ [CS1–GFP] × ρ0 crosses. The top four panel sets are representative micrographs of the different zygote types observed in the cross between S150-2B ρ+ [CS1–GFP] and PSY142 ρ0 cells. The U form zygotes appear immediately after mating. Types I–III are the different zygote forms detected in populations of zygotes with medial buds. The type IV zygote is from the cross S150-2B Δabf2 ρ+ [CS1–GFP] × PSY142 Δabf2 ρ0.
By far the most abundant zygote form is type III in which mtDNA appears preferentially in the diploid bud, whereas the protein marker has equilibrated throughout the zygote as well as into the diploid bud. This unique sorting of mtDNA is particularly evident in zygotes expressing CS1-GFP where the type III zygotes account for 81% of the total zygotes in the population (Table I). Type III zygotes are also the majority type with the inner and outer mitochondrial membrane markers, although they represent a somewhat smaller fraction of the total zygote population in those crosses. The reduction in type III zygotes with the membrane markers could be accounted for by a slower mixing of those markers relative to the matrix marker, and a slight increase in the preferential sorting of those membrane marker proteins. An important conclusion from these experiments is that mtDNA appears to sort from the ρ+ end of the zygote preferentially into the emerging diploid bud, whereas the marker proteins, and especially the matrix marker, appear to equilibrate more or less randomly in the zygote with little preference for the medial diploid bud.
Sorting of mtDNA Is Altered in a Homozygous Δabf2 Cross
The mitochondrial HMG protein, Abf2p, is required for the maintenance of ρ+ mtDNA in cells grown on medium containing glucose as the carbon source, whereas ρ+ mtDNA can be maintained indefinitely in such cells if they are grown on medium containing a nonfermentable carbon source. In those cells, mtDNA nucleoids visualized by DAPI staining of cells appear more disorganized than nucleoids in wild-type cells, and some polypeptides present in isolated nucleoids are depleted or missing compared with nucleoids from wild-type cells (Newman et al., 1996). Those findings suggested a relationship between the organization of mtDNA and its stability. Given the results presented thus far, it was of interest to determine whether the sorting of mtDNA is altered in zygotes that lack Abf2p.
To examine this question, a ρ+ × ρ0 cis cross was performed with the CS1–GFP matrix marker using derivatives of strains PSY142 and S150-2B in which the ABF2 gene was deleted. As with Δabf2 haploid cells, mtDNA is also unstable in diploids derived from these cells (data not shown). The ρ+ Δabf2 derivative was prelabeled with CS1– GFP in exactly the same way as detailed in the preceding section for its ABF2 counterpart, except that the cells were maintained on YNBGly + cas medium before induction of CS1–GFP. In preliminary experiments carried out with crosses between these Δabf2 strains, we observed that mtDNA sorting appeared to be slower than in the control, ABF2 × ABF2 crosses. Therefore, we extended the time course of the analysis to examine zygotes with medial buds to include 4- and 5-h time points in addition to scoring at the 2- and 3-h intervals as before. As shown in Fig. 8 and summarized in Table II, in addition to the zygote types (I– III) found in the homozygous ABF2 cross, a new zygote type (IV) is detected in this Δabf2 cross in which the CS1– GFP marker has fully equilibrated in the zygote and into the diploid bud, but mtDNA has not yet moved out of the ρ+ end of the zygote. Type IV zygotes were never observed in ABF2 crosses. This delay in sorting of mtDNA compared with wild-type crosses accounts, in part, for the smaller fraction of type III zygotes observed at the earlier time points. Throughout the time course of the analysis, the fraction of type III zygotes never reaches that observed with the CS1–GFP marker in the wild-type cross (refer to Table I), indicating that the absence of Abf2p not only delays the onset of mtDNA equilibration, but also reduces the preferential sorting into the medial diploid bud.
Table II.
Sorting of mtDNA Is Altered in Zygotes Lacking Abf2p
| % of total zygotes with a medial bud* | ||||||||
|---|---|---|---|---|---|---|---|---|
|
||||||||
| Time | ||||||||
| (h) | ||||||||
| 2 | 8 | 8 | 36 | 48 | ||||
| 3 | 20 | 2 | 42 | 36 | ||||
| 4 | 30 | 4 | 50 | 16 | ||||
| 5 | 64 | 0 | 30 | 6 | ||||
Synchronized matings were performed using strain S150-2B Δabf2 ρ+ prelabeled with CS1-GFP and strain PSY142 Δabf2 ρ0 cells as described in Materials and Methods. Cells were processed and analyzed as described in Table I.
50 zygotes with one-third to two-thirds the size of a diploid bud were scored for each time point after mating.
To show directly that there is a delay in mtDNA sorting in the Δabf2 cross, we carried out a side by side comparison of the kinetics of appearance of M form zygotes in the Δabf2 cross with that of the wild-type (ABF2) cross. The data of Fig. 9 show that the appearance of M-type zygotes in the Δabf2 cross is delayed by ∼1 h relative to the ABF2 control cross. What is clear from these experiments is that the absence of Abf2p results in a reduction of the type III zygotes showing preferential transmission of mtDNA into the diploid bud because that population is replaced with the type IV zygotes.
Discussion
In this paper we show that the sorting of GFP marker proteins located in four different mitochondrial compartments can be spatially and temporally resolved from the sorting of mtDNA in zygotes of synchronously mated cells. In addition, we show that in cis crosses, mtDNA, in contrast to the marker proteins, is first directed preferentially to the medial diploid bud before fully equilibrating to the ρ0 end of the zygote. To carry out these experiments, we constructed a set of gene fusions designed to _target GFP to the matrix, the inner, and outer mitochondrial membranes, as well as to mtDNA. Biochemical experiments verified the specific mitochondrial location of the matrix and membrane-bound GFP fusion proteins, whereas the localization of the Abf2p–GFP fusion protein to mtDNA was evident from direct fluorescence microscopic analysis, which showed a punctate staining pattern identical to that of DAPI-stained mtDNA. In a previous study (Zelenaya-Troitskaya et al., 1998), we demonstrated that the punctate staining pattern of Abf2p–GFP was dependent on the ability of the protein to bind to DNA, since an Abf2p–GFP fusion protein derived from a mutant form of Abf2p with mutations of the two HMG boxes that markedly reduced DNA binding activity in vitro, showed an in vivo staining pattern that was more typical of that observed with a mitochondrial matrix protein than the punctate staining of mtDNA.
The sorting pattern of the CS1–GFP marker in zygotes from a ρ+ × ρ0 trans cross, in which the ρ0 parent was prelabeled with the marker protein, agrees well with the pattern previously described using indirect immunofluorescence methods to detect endogenous CS1 and a foreign protein (DHFR) _targeted to the mitochondrial matrix (Azpiroz and Butow, 1993). In particular, we observed the appearance of the novel A form zygote where, within 2 h after zygote formation, the marker protein initially present in the ρ0 end of the cell quantitatively translocated to the ρ+ end before any significant movement of ρ+ mtDNA. We do not yet understand the mechanism by which A form zygotes are generated or why this unusual intermediate form is only observed in zygotes from ρ+ × ρ0 crosses, independent of whether the ρ+ parent is respiratory competent or not (Azpiroz and Butow, 1993). The only other instance where we failed to observe A form zygotes using a matrix marker protein for the sorting analysis was in ρ+ × ρ0 trans crosses between Δabf2 parental strains (Zelenaya-Troitskaya et al., 1998). A further consideration of the effects of the absence of Abf2p in the sorting patterns in zygotes is discussed below.
Both the inner and outer mitochondrial membrane marker proteins, Yta10p–GFP, and GFP–Tom6p, respectively, translocated with very similar kinetics through the fused mitochondrial reticulum; however, their rates of equilibration were slower than that observed for CS1– GFP. This is not an unexpected result, since the lateral diffusion of proteins through membranes has generally been observed to be slower than diffusion of nonmembrane proteins. The observed diffusion coefficients for membrane proteins can, however, vary greatly depending on the protein or protein complex, the particular membrane and whether there is an interaction between the membrane protein and components such as cytoskeletal elements (Jacobson et al., 1987). For mitochondria, the diffusion properties of respiratory chain complexes of the inner mitochondrial membrane have been studied (Chazotte and Hackenbrock, 1991). Those experiments showed that there were no significant differences in diffusion coefficients of the inner membrane complexes examined and that the observed rates of diffusion were insensitive to membrane folding or the environment immediately adjacent to the membrane. We can conclude from the present studies that when parental mitochondria fuse in zygotes, mitochondrial membrane as well as matrix proteins can exchange throughout the extended mitochondrial reticulum. Finally, besides the slower equilibration rate of Yta10p–GFP and GFP–Tom6p, the other notable difference between the sorting pattern of these membrane proteins and matrix markers is the absence of any detectable A form zygotes. Since we do not know the mechanism by which A form zygote arise in crosses with the matrix markers, it is difficult to speculate on why that novel zygote type is not observed with the membrane markers.
The sorting of Abf2p–GFP was analyzed somewhat differently than the other mitochondrial marker proteins in that it was transiently expressed in one of the parents of a ρ+ × ρ+ cross. Because the parental mtDNAs in zygotes from ρ+ × ρ+ crosses do not mix to any appreciable extent during zygote maturation (Strausberg and Perlman, 1978; Zinn et al., 1987; Azpiroz and Butow, 1993; Nunnari et al., 1997), it was of particular interest to know whether Abf2p–GFP would remain with the parental mtDNA or partition to all of the mtDNA nucleoids of the zygote. We found that Abf2p-GFP rapidly equilibrated throughout the zygote, eventually colocalizing with the ρ+ mtDNA of the unlabeled parent, with kinetics similar to that of CS1-GFP. These results indicate that even though parental ρ+ mtDNAs do not mix in the zygote, the parental mtDNA compartments are nevertheless fully accessible to each other in the fused mitochondrial reticulum.
The finding of preferential transmission of mtDNA to the medial diploid bud provides the strongest evidence yet that mtDNA inheritance is not stochastic. The preponderance of type III zygotes in the ρ+ × ρ0 cis crosses of Table I is particularly significant in that both mtDNA and the marker protein, present initially in the ρ+ end of the zygote, have in principle the same opportunity to segregate through the mitochondrial reticulum to the ρ0 end of the zygote as well as to the medial diploid bud. Yet, for the majority of zygotes, mtDNA first appears in the bud, whereas the marker protein distributes more or less randomly between the bud and the ρ0 end of the zygote. This clear-cut distinction between preferential versus random transmission of mtDNA cannot be made for ρ+ × ρ+ crosses, since the well-documented restriction of mtDNA movement within the body of the zygote means that in either circumstance only the medial buds would receive a sample of both parental mtDNAs (Strausberg and Perlman, 1978; Zinn et al., 1987; Nunnari et al., 1997).
In zygotes lacking Abf2p, the process of mtDNA sorting is altered in two significant ways: first, its movement out of the ρ+ end of the zygote is delayed, as evident by type IV zygotes where the DNA remains in the ρ+ end but the marker protein has equilibrated throughout the zygote. Second, the preferential sorting of mtDNA to the medial bud is reduced. Since in zygotes, mtDNA mixing and sorting to buds are important parameters that can affect the observed frequency of mtDNA recombination in crosses (Strausberg and Perlman, 1978; Zinn et al., 1987), these alterations in sorting probably contribute to the observation that recombination between mtDNA markers is suppressed in homozygous Δabf2 crosses (Zelenaya-Troitskaya et al., 1998). However, we have recently found that Abf2p also affects the level of mtDNA recombination intermediates (Holliday junctions) (MacAlpine et al., 1998), suggesting that this protein has a direct role in recombination.
In the zygote system we have used, it is useful to draw a distinction between sorting within the fused mitochondrial reticulum of the zygote itself, and sorting from the zygote to the emerging diploid bud. In the latter, sorting would be accompanied by a directed and continuous transfer of mitochondria to the bud whereas in the former, sorting would occur within the mitochondrial reticulum of the zygote and be subject only to intracellular mitochondrial dynamics, e.g., fusion and fission events. For bulk mitochondrial inheritance, there is now considerable evidence that the transmission of mitochondria from mother to daughter cell is an active, regulated process requiring a number of gene products (McConnell et al., 1990; McConnell and Yaffe, 1992; Sogo and Yaffe, 1994; Hermann et al., 1997). There is, in addition, evidence to suggest that mitochondrial organization and inheritance in yeast is linked to the actin cytoskeleton (Drubin et al., 1993; Lazzarino et al., 1994; Simon et al., 1995; Smith et al., 1995). There is much less information on how mitochondrial dynamics, and particularly fusion and fission events, are controlled. Recently, a predicted transmembrane GTPase encoded by the fuzzy onion gene in Drosophila has been implicated in mediating mitochondrial fusion during Drosophila spermatogenesis (Hales and Fuller, 1997). It will be of great interest to learn whether yeast require a similar type of protein for mitochondrial fusion.
The process that accounts for the preferential sorting of mtDNA to the zygotic bud may also account for its faithful segregation in vegetatively growing cells; that is, mtDNA segregation might be firmly coupled to mitochondrial transmission. It is intriguing that the absence of Abf2p in zygotes results in a decrease, but not in an elimination, of the preferential sorting of mtDNA to the bud, and that mtDNA transmission is compromised but not blocked altogether in Δabf2 cells (In Δabf2 cells the rate of mtDNA loss is significantly slower than the division time [Zelenaya-Troitskaya et al., 1998]). Perhaps in the absence of Abf2p, mtDNA–protein complexes are altered such that mtDNA is less able to engage the putative segregation apparatus, resulting in an uncoupling between mitochondrial transmission and mtDNA segregation. Studies are currently underway to identify proteins that would both interact with mtDNA and function in the transmission process. Our finding of directed movements of mtDNA warrants drawing an analogy to the mitotic apparatus for chromosomal segregation that insures each cell receives its full chromosome complement during cell division. Although it is unlikely that mtDNA segregation will turn out to be as complicated or as precise as chromosome segregation in mitosis and meiosis, accumulating evidence suggests that it will not be as simple as random sorting.
Acknowledgments
We are grateful to S. Newman (University of Texas Southwestern, Dallas, TX) for making available some of the ABF2 constructs used and for advice and helpful suggestions during the course of this study. We thank other members of the Butow/Perlman lab for helpful discussions.
This work was supported by grants from the National Institutes of Health (GM 33510) and The Robert A. Welch Foundation (I-0642).
Abbreviations used in this paper
- CS1
citrate synthase 1
- DAPI
4′,6-diamino-2-phenylindole
- GFP
green fluorescent protein
- HMG
high mobility group
- Tom
translocase outer membrane
Footnotes
Address all correspondence to Ronald A. Butow, Department of Molecular Biology and Oncology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75235-9148. Tel.: (214) 648-1465. Fax: (214) 648-1488. E-mail: butow@swmed.edu
References
- Alconada A, Kübrich M, Moczko M, Hönlinger A, Pfanner N. The mitochondrial receptor complex: the small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol Cell Biol. 1995;15:6196–6205. doi: 10.1128/mcb.15.11.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Azpiroz R, Butow RA. Patterns of mitochondrial sorting in yeast zygotes. Mol Biol Cell. 1993;4:21–36. doi: 10.1091/mbc.4.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Butow RA. Mitochondrial inheritance in yeast. Methods Enzymol. 1995;260:453–466. doi: 10.1016/0076-6879(95)60158-9. [DOI] [PubMed] [Google Scholar]
- Berger KH, Yaffe MP. Mitochondrial distribution and inheritance. Experentia. 1996;52:1111–1116. doi: 10.1007/BF01952109. [DOI] [PubMed] [Google Scholar]
- Blanc H, Dujon B. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc Natl Acad Sci USA. 1980;77:3942–3946. doi: 10.1073/pnas.77.7.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, H., and B. Dujon. 1982. Replicator regions of the yeast mitochondrial DNA active in vivo and in yeast transformants. In Mitochondrial Genes. P. Slonimski, P. Borst, and G. Attardis, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 279–294.
- Chazotte B, Hackenbrock CR. Lateral diffusion of redox components in the mitochondrial inner membrane is unaffected by inner membrane folding and matrix density. J Biol Chem. 1991;266:5973–5979. [PubMed] [Google Scholar]
- de Zamaroczy M, Marotta R, Faugeron-Fonty G, Goursot R, Mangin M, Baldacci G, Bernardi G. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature. 1981;292:75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX, Stillman B. DNA binding properties of an HMG1- related protein from yeast mitochondria. J Biol Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- Drubin DG, Jones HD, Wertman KF. Actin structure and function–roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B. 1981. Mitochondrial Genetics and Functions. In The Molecular Biology of the Yeast Saccharomyces. J.N. Strathern, E.W. Jones, and J.R. Broachs, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 505–635.
- Dujon B, Bolotin-Fukuhara M, Coen D, Deutsch J, Netter P, Slonimski PP, Weill L. Mitochondrial genetics. XI. Mutations at the mitochondrial locus omega affecting the recombination of mitochondrial genes in Saccharomyces cerevisiae. . Mol Gen Genet. 1976;143:131–165. doi: 10.1007/BF00266918. [DOI] [PubMed] [Google Scholar]
- Dujon B, Slonimski PP, Weill L. Mitochondrial genetics. IX. A model for the recombination and segregation of mitochondrial genes in Saccharomyces cerevisiae. . Genetics. 1974;78:415–437. doi: 10.1093/genetics/78.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS. Pathways and energetics of mitochondrial protein import in Saccharomyces cerevisiae. . Methods Enzymol. 1995;260:224–231. doi: 10.1016/0076-6879(95)60140-6. [DOI] [PubMed] [Google Scholar]
- Guelin E, Rep M, Grivell LA. Afg3p, a mitochondrial ATP- dependent metalloprotease, is involved in degradation of mitochondrially- encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS (Fed Eur Biochem Soc) Lett. 1996;381:42–46. doi: 10.1016/0014-5793(96)00074-9. [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H, Avers CJ. Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science. 1973;181:749–751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- Jacobson K, Ishihara A, Inman R. Lateral diffusion of proteins in membranes. Annu Rev Physiol. 1987;49:163–175. doi: 10.1146/annurev.ph.49.030187.001115. [DOI] [PubMed] [Google Scholar]
- Kassenbrock CK, Cao W, Douglas MG. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO (Eur Mol Biol Organ) J. 1993;12:3023–3034. doi: 10.1002/j.1460-2075.1993.tb05971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarino DA, Boldogh I, Smith MG, Rosand J, Pon LA. Yeast mitochondria contain ATP-sensitive, reversible actin-binding activity. Mol Biol Cell. 1994;5:807–818. doi: 10.1091/mbc.5.7.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Perlman PS, Butow RA. The high mobility group protein, Abf2p, influences the level of yeast mitochondrial DNA recombination intermediates in vivo. Proc Natl Acad Sci USA. 1998;95:6739–3743. doi: 10.1073/pnas.95.12.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J Cell Biol. 1992;118:385–395. doi: 10.1083/jcb.118.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Chae CB. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J Biol Chem. 1993;268:12758–12763. [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Chae CB. The mitochondrial histone HM: an evolutionary link between bacterial HU and nuclear HMG1 proteins. Biochimie. 1994;76:909–916. doi: 10.1016/0300-9084(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Newman SM, Zelenaya-Troitskaya O, Perlman PS, Butow RA. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiaethat lacks the mitochondrial HMG-box protein, Abf2p. Nucl Acids Res. 1996;24:386–393. doi: 10.1093/nar/24.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiaeis determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic A, Tauer R, Feldmann H, Neupert W, Langer T. Yta10p is required for the ATP-dependent degradation of polypeptides in the inner memebrane of mitochondria. FEBS (Fed Eur Biochem Soc) Lett. 1994;353:201–206. doi: 10.1016/0014-5793(94)01046-3. [DOI] [PubMed] [Google Scholar]
- Piskur J. Inheritance of the yeast mitochondrial genome. Plasmid. 1994;31:229–241. doi: 10.1006/plas.1994.1025. [DOI] [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MG, Simon VR, O'Sullivan H, Pon LA. Organelle-cytoskeletal interactions: actin mutations inhibit meiosis-dependent mitochondrial rearrangement in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1381–1396. doi: 10.1091/mbc.6.10.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Perlman PS. The effect of zygotic bud position on the transmission of mitochondrial genes in Saccharomyces cerevisiae. . Mol Gen Genet. 1978;163:131–144. doi: 10.1007/BF00267404. [DOI] [PubMed] [Google Scholar]
- Tauer R, Mannhaupt G, Schanll R, Pajic A, Langer T, Feldmann H. Yta10p, a member of a novel ATPase family in yeast, is essential for mitochondrial function. FEBS (Fed Eur Biochem Soc) Lett. 1994;353:197–200. doi: 10.1016/0014-5793(94)01045-5. [DOI] [PubMed] [Google Scholar]
- Thomas DY, Wilkie D. Recombination of mitochondrial drug resistance factors in Saccharomyces cerevisiae. . Biochem Biophys Res Commun. 1968;30:368–372. doi: 10.1016/0006-291x(68)90753-5. [DOI] [PubMed] [Google Scholar]
- Yaffe M. Organelle inheritance in the yeast cell cycle. Trends Cell Biol. 1991;1:160–164. doi: 10.1016/0962-8924(91)90017-4. [DOI] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya O, Newman SM, Okamoto K, Perlman PS, Butow RA. Functions of the HMG box protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. . Genetics. 1998;148:1763–1776. doi: 10.1093/genetics/148.4.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn AR, Pohlman JK, Perlman PS, Butow RA. Kinetic and segregational analysis of mitochondrial DNA recombination in yeast. Plasmid. 1987;17:248–256. doi: 10.1016/0147-619x(87)90033-3. [DOI] [PubMed] [Google Scholar]
- Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. . Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]