SUMMARY
Solving the biological roles of covalent histone modifications, including monoubiqutination of histone H2A, and the molecular mechanisms by which these modifications regulate specific transcriptional programs, remains a central question for all eukaryotes. Here, we report that the N-CoR/HDAC1/3 complex specifically recruits a previously-unidentified specific histone H2A ubiquitin ligase, 2A-HUB/hRUL138, to a subset of regulated gene promoters. 2A-HUB catalyzes monoubiquitination of H2A at lysine 119, functioning as a combinatoric component of the repression machinery required for specific gene regulation programs. Thus, 2A-HUB mediates a selective repression of a specific set of chemokine genes in macrophages, critically modulating migratory responses to TLR activation. H2A monoubiquitination acts to prevent FACT recruitment at the transcriptional promoter region, blocking RNA polymerase II release at the early stage of elongation. We suggest that distinct H2A ubiquitinases, each recruited based on interactions with different corepressor complexes, contribute to effective distinct transcriptional repression programs.
INTRODUCTION
Packaging of DNA into chromatin within eukaryotic cells inhibits all aspects of transcription that are carried out by RNA polymerase II (RNAPII) (reviewed in Li et al., 2007). As nucleosomes present a physical barrier to Pol II during transcriptional initiation and elongation, histone-modifying enzymes, chromatin remodeling factors and numerous other factors play important roles in both transcriptional initiation and elongation in a chromatin environment (reviewed in Li et al., 2007). The covalent modifications of specific residues of histones, including acetylation, methylation, phosphorylation, ubiquitination, and sumoylation, have been suggested to provide interaction surfaces for proteins that either increase or decrease chromatin accessibility and therefore impact on the regulation of transcription, replication, and chromosome behavior (Fischle et al., 2003; Jenuwein and Allis, 2001; Margueron et al., 2005; Peterson and Laniel, 2004; Strahl and Allis, 2000).
Histone H2B-K123 (K120 in vertebrates) monoubiquitination has been linked to gene activation and transcription elongation (Henry et al., 2003; Kao et al., 2004; Kim et al., 2005a; Osley, 2004; Pavri et al., 2006; Tanny et al., 2007; Zhu et al., 2005). Histone H2B-K123 monoubiquitination is mediated by the E2-conjugating enzyme Rad6 and the E3 ligase Bre1 in yeast (Hwang et al., 2003; Robzyk et al., 2000; Wood et al., 2003). Importantly, histone H2B-K123 monoubiquitination is a prerequisite for histone H3-K4 and H3-K79 methylation (Sun and Allis, 2002; Wood et al., 2003). Interestingly, deubiquitination of H2B-K123 is required for histone H3-K36 methylation (Henry et al., 2003). In vertebrates, histone H2B is monoubiquitinated at lysine 120 (Thorne et al., 1987) by the human homolog of yeast Bre1 E3 ubiquitin ligase hBRE1 or RNF20/RNF40 (Kim et al., 2005a; Zhu et al., 2005). BRE1 can interact directly with p53 and serve as a transcriptional cofactor for p53 (Kim et al., 2005a). Moreover, Hox genes are also regulated by the level of histone H2B-K120 monoubiquitination (Zhu et al., 2005). Recently, the p53 binding protein Mdm2 was also reported to function as the E3 ligase for histone H2B monoubiquitination in mammals (Minsky and Oren, 2004).
H2A ubiquitination which occurs at lysine 119 in many but not all higher eukaryotes has been estimated to comprise between 5% and 15% of the available H2A and is a relatively abundant modification (Goldknopf et al., 1975; Nickel and Davie, 1989). There is contradictory evidence concerning its function. While some studies have pointed to an association of uH2A with transcriptionally active chromatin (Levinger and Varshavsky, 1982; Nickel et al., 1989), others have failed to demonstrate such a link (Dawson et al., 1991; Huang et al., 1986; Parlow et al., 1990). Recent studies show that RING2/Ring1b in the hPRC1L (Polycomb repressive complex 1-like) complex monoubiquitinates nucleosomal histone H2A at lysine 119 and links H2A ubiquitination to Polycomb silencing (Wang et al., 2004) as well as to X chromosome inactivation (de Napoles et al., 2004; Fang et al., 2004). More recent studies suggested that both Ring1A and another component of PRC1, Bmi-1, play an important role in H2A ubiquitination and Hox gene silencing and demonstrate the sequential events of H3-K27 methylation by methyltransferase EZH2 and H2A-K119 ubiquitination (Cao et al., 2005). However, the mechanism of H2A ubiquitination leading to gene silencing or repression is unknown.
Here, we report the identification and characterization of an alternative RING domain-containing protein that functions as a histone H2A-K119 E3 ubiquitin ligase, referred to as 2A-HUB. 2A-HUB is specifically recruited by the N-CoR/HDAC1/3 complex to a subset of regulated chemokine gene promoters, promoting H2A monoubiquitination on these promoters, a function required for repressing a specific chemokine gene expression program. A central function of H2A monoubiquitylation proves to be prevention of FACT recruitment to the promoter regions of these genes, inhibiting RNA Pol II-dependent early elongation events.
RESULTS
Identification of a RING Finger Protein 2A-HUB that Functions as a Transcriptional Repressor
We identified a histone H2A monoubiquitination ligase, the hypothetical product of the KIAA0675 clone [Genbank Accession Number AB014575], which we refer to as 2A-HUB. 2A-HUB/KIAA0675 was initially identified from a yeast two-hybrid screen that we conducted using Pitx2 (Amendt et al., 1998; Baek et al., 2003; Kioussi et al., 2002; Lu et al., 1999) as a bait, modulating Pitx2-mediated repression function (data not shown). The human KIAA0675 has been previously identified as an RNA-binding RING-dependent ubiquitin protein ligase (hRUL138) (Kreft and Nassal, 2003). The 2A-HUB protein contains an intact RING (‘really interesting new gene’) motif of the RING-H2 type defined by the consensus sequence CX2CX9-39CX1-3HX2-3C/HX2CX4-48CX2C with eight cysteines and two histidines that coordinate two zinc ions (Borden, 2000), and lysine rich domain in the middle (Figure 1A). Two major bands reflecting the alternatively spliced 2A-HUB, as estimated by molecular weight determination, were detected by Western blot analysis using specific antibodies against 2A-HUB from nuclear extracts of C2C12 cells (Figure S1A). 2A-HUB exhibited a primarily nuclear localization by immunostaining with specific anti-2A-HUB IgG in C2C12 cells (Figure S1B).
Figure 1. 2A-HUB Functions as a Transcriptional Repressor.
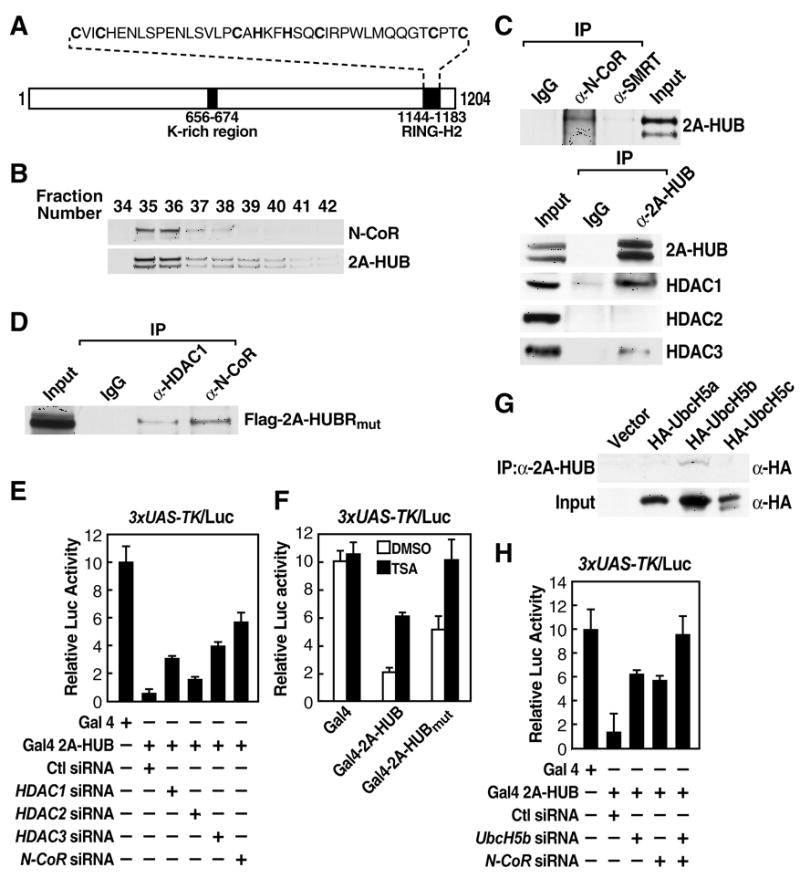
(A) Schematic representation of the domain organization of 2A-HUB protein.
(B) 2A-HUB cofractions with N-CoR. The fractions (34–42) were analyzed by Western blot analysis to detect N-CoR and 2A-HUB.
(C) Endogenous co-immunoprecipitation of 2A-HUB with N-CoR and HDACs. Nuclear extracts of C2C12 cells were immunoprecipitated using control IgG, anti-N-CoR, anti-SMRT, or anti-2A-HUB antibodies and co-immunoprecipitated proteins were detected by Western blot analysis using indicated antibodies.
(D) 2A-HUBmut interacts with N-CoR and HDAC1. Cell lysates from HEK293T cells transfected with Flag-2A-HUBmut was incubated with IgG or anti-HDAC1 or anti-N-CoR antibody, and immunoprecipitated proteins were analyzed by anti-Flag antibodies. 5% of the input was loaded.
(E) Reporter assays using a Gal4-dependant luciferase reporter in HEK293T cells. Cells were first transfected with control siRNA or siRNAs against HDAC1, HDAC2, HDAC3 and N-CoR, and then transfected with 3XUAS-TK-luciferase reporter in combination with vector expressing Gal4, Gal4-2A-HUB.
(F) Reporter assays using a Gal4-dependant luciferase reporter in HEK293T cells. Cells were transfected with 3XUAS-TK/luciferase reporter in combination with vector expressing Gal4 or Gal4-2A-HUB or Gal4-2A-HUBmut and then treated with DMSO or TSA.
(G) Endogenous 2A-HUB coimmunoprecipitated with HA-UbcH5b in vivo. Cell lysates from HEK293T cells transfected with a HA-UbcH5a, HA-UbcH5b or HA-UbcH5c expression plasmid was incubated with anti-2A-HUB antibody and immunoprecipitated proteins were analyzed by anti-HA antibodies. 5% of the input was loaded.
(H) Reporter assays using a Gal4-dependant luciferase reporter in HEK293T cells. Cells were first transfected with control siRNA or siRNAs against UbcH5b or N-CoR as indicated, and then transfected with 3XUAS-TK-luciferase reporter in combination with vector expressing Gal4, Gal4-2A-HUB. In (E), (F) and (H), the activity observed from Gal4 was arbitrarily set as 10. Data represent mean ± SEM from three independent experiments.
To investigate the possibility that 2A-HUB is a component of a co-repressor complex, we performed gel filtration assays using nuclear extract from C2C12 cells and demonstrated that 2A-HUB eluted in a ~600 kDa fractions (Figure 1B). Intriguingly, analysis of candidate co-repressor molecules that might be present in this complex demonstrated that 2A-HUB coimmunoprecipitated with N-CoR, which also eluted in a ~600 kDa fractions (Figure 1B and 1C), but not with SMRT (Figure 1C). It also coimmunoprecipitated with HDAC1 and HDAC3, but not with HDAC2 (Figure 1C). Interestingly, an enzymatic domain-deficient mutation, 2A-HUBmut (Kreft and Nassal, 2003) was still able to interact with N-CoR and HDAC1 (Figure 1D). To further understand the mechanism of 2A-HUB-mediated transcriptional repression, we fused full length 2A-HUB to the DNA binding domain (DBD) of Gal4. While Gal4-2A-HUB functioned as a repressor on Gal4RE luciferase reporter (Figure 1E), this 2A-HUB-dependent repression could be partially overcome by knockdown of N-CoR, HDAC1 or HDAC3, but not HDAC2 (Figure 1E and Figure S2). While Gal4-2A-HUB repression activity was only partially attenuated in cells treated with the HDAC inhibitor trichostatin A (TSA), the repression activity of Gal4-2A-HUBmut was completely abolished, suggesting that both HDAC and ubiquitination activities are involved in 2A-HUB dependant-repression (Figure 1F). To further distinguish the ubiquitination activity of 2A-HUB mediated repression function, we performed a Gal4-2A-HUB reporter assays by knocking down the E2 enzyme UbCH5b, which exhibits specific interactions with 2A-HUB in vivo (Figure 1G and Figure S2E), observing that the repressive activity of Gal4-2A-HUB was markedly decreased (Figure 1H). More strikingly, the repressive activity of 2A-HUB was totally lost upon depletion both UbcH5b and N-CoR (Figure 1H). Taken together, these data suggest that the monoubiquitination activity of 2A-HUB exerts a combinatoric function with the NCoR/HDAC1/3 complex in mediating repression.
2A-HUB Monoubiquitinates Histone H2A in Vitro and in Vitro
RING domain-containing proteins have recently been shown to be integral parts of a second major class, in addition to HECT domain proteins, of ubiquitin ligase (or E3 enzyme) (Borden, 2000). We next investigated whether 2A-HUB can be an E3 ligase by testing its capability of self-ubiquitination. In vitro translated 35Met 2A-HUB with a reaction mix containing ATP, His-tagged ubiquitin, recombinant E1 and E2 ubiquitin-conjugating enzyme UbcH5 exhibited self-ubiquitination activities (Figure2A, lane2), which was dependent on the RING finger (Figure 2A, lane 4).
Figure 2. 2A-HUB Monoubiquitinates Histone H2A in Vitro.
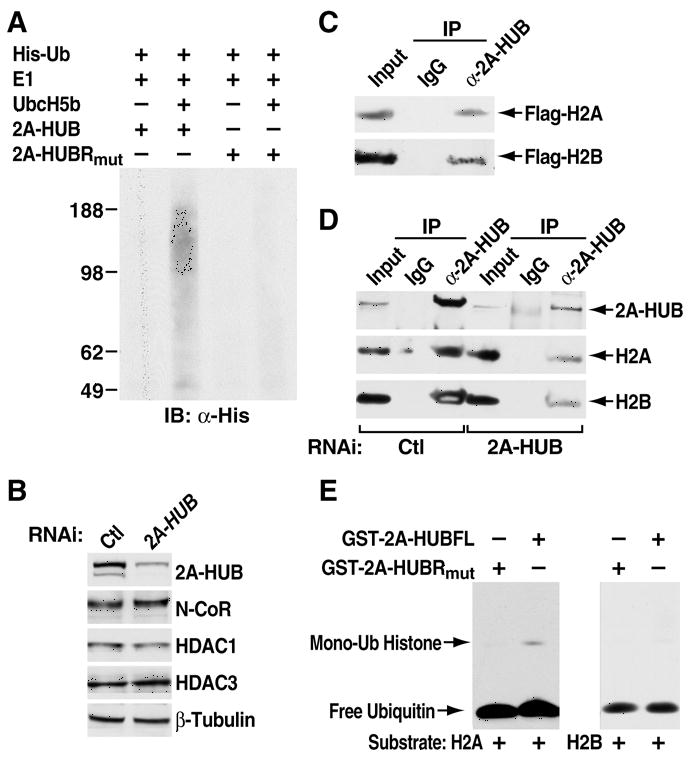
(A) In vitro ubiquitination assay for 2A-HUB. In vitro ubiquitination reactions were performed in the presence of recombinant E1, recombinant E2 (UbcH5b), recombinant His-ubiquitin and in vitro-translated 35S labeled 2A-HUB or 2A-HUBRmut. Reaction products were resolved by SDS-PAGE, and subject to Western blot analysis with anti-His antibodies to visualize ubiquitin conjugates.
(B) Western blot analysis of 2A-HUB, N-CoR, HDAC1, HDAC3 protein levels after knockdown of 2A-HUB. Equal loading was confirmed using anti-β-tubulin antibodies.
(C) Endogenous 2A-HUB coimmunoprecipitated with Flag-H2A or Flag-H2B in vivo. Soluble chromatin extracted from HEK293T cells transfected with a Flag-H2A or Flag-H2B expression plasmid was incubated with anti-2A-HUB antibody and immunoprecipitated proteins were analyzed by anti-Flag antibodies. 5% of the input was loaded.
(D) Endogenous 2A-HUB associates with endogenous H2A and H2B in vivo. Soluble chromatin extracts from either HEK293T transfected with control siRNA or 2A-HUB siRNA was incubated with IgG or anti-2A-HUB antibody, and immunoprecipitated proteins were analyzed by anti-H2A or anti-H2B antibodies. 5% of the input was loaded.
(E) In vitro ubiquitination assay for H2A or H2B. In vitro ubiquitination reactions were performed in the presence of recombinant E1, recombinant E2(UbcH5b), recombinant His-ubiquitin, histone H2A or H2B and purified recombinant GST-fused wt 2A-HUB or recombinant GST-fused 2A-HUBRmut as indicated. Reaction products were probed with antibodies against His to visualize ubiquitin conjugates.
To investigate the potential substrates of 2A-HUB, we examined the 2A-HUB interacting proteins. We did not observe any changes of the protein levels of N-CoR, HDAC1, and HDAC3 by knockdown of 2A-HUB in HEK293T cells (Figure 2B). In light of the ability of 2A-HUB to function as a transcriptional repressor, it appeared plausible that 2A-HUB might affect histone modifications (Jenuwein and Allis, 2001; Zhang, 2003). We therefore examined 2A-HUB and histone interactions in vivo by employing transfected Flag-tagged human H2A or H2B in HEK293T cells. Substantial amounts of 2A-HUB was coimmunoprecipitated with Flag-H2A or Flag-H2B, but was not precipitated with control IgG (Figure 2C). Furthermore, interactions at physiological levels suggested by the finding that the endogenous 2A-HUB specifically brought down endogenous H2A or H2B from HEK293T in coimmunoprecipitation assays (Figure 2D). Together, these observations imply that 2A-HUB can interact with H2A and H2B in vivo. To test whether this factor could promote H2A or H2B ubiquitination, purified histones H2A or H2B were incubated with bacterially-expressed recombinant GST-2A-HUB or 2A-HUBRmut and His-tagged ubiquitin, under conditions appropriate for in vitro ubiquitination. As seen in Figure 2E lane 2, a His-tagged band migrating at ~25 kDa was obtained only for histone H2A, but not for histone H2B (lane 4). The size is consistent with mono His-ubiquitinated H2A. Monoubiquitination was not observed in the presence of recombinant GST-2A-HUB carrying a RING domain mutation, which abolishes E3 activity (Figure 2A). Thus, 2A-HUB is capable of promoting histone H2A monoubiquitination in vitro in a RING domain-dependent fashion.
To confirm that 2A-HUB can monoubiquitinate histone H2A in vivo, we ectopically expressed 2A-HUB in HEK293T cells, finding that histone H2A ubiquitination level was increased significantly, as detected by western blot using a specific antibody recognizing the monoubiquitinated form of H2A (uH2A) (Wang et al., 2004) (Figure 3A). In contrast, over-expression of 2A-HUBRmut did not affect H2A ubiquitination (Figure 3A). To further investigate 2A-HUB, an ubiquitin E3 ligase that catalyzes monoubiquitination of H2A in vivo, cell extracts from HEK293T cells transfected with Flag-H2A, together with His6-ubiquitin and 2A-HUB or 2A-HUBRmut, expression plasmids were subjected to immunoprecipitation using Ni-NTA beads under denaturing conditions, and immunoblotted with anti-Flag antibodies to detect all forms of Flag-H2A in the precipitate. While cells transfected only with Flag-H2A and his-ubiquitin displayed barely detectable levels of monoubiquitinated Flag-H2A, presumably reflecting activity of endogenous E3 ligases, the expression of 2A-HUB resulted in a pronounced increase in ubiquitinated Flag-H2A both in whole cell lysates and in precipitates pulled down by Ni-NTA beads (Figure 3B, lane 3). In order to test whether K119 is the only 2A-HUB-dependent ubiquitination site in mammalian H2A, 2A-HUB FL or 2A-HUBRmut expression vectors were co-transfected with Flag-H2A or Flag-H2A (K119R) (H2A harboring a mutation in the major monoubiquitinated residue) and a His6-ubiquitin expression plasmid into HEK293T cells. While wt Flag-H2A can be monoubiquitinted by the expression of 2A-HUB (Figure 3C, lane 3), the ubiquitinated form of Flag-H2A (K119R) could not be detected in the same condition (Figure 3C, lane 7). This result suggests that 2A-HUB was not able to promote H2A monoubiquitination at sites other than the K119 residue (Figure 3C). We also examined whether 2A-HUB might promote histone H2B ubiquitination in vivo. His6-ubiquitin plasmid and 2A-HUB or 2A-HUBRmut were transfected into HEK293T cells and cell extracts were subjected to immunoprecipitation of his-tagged ubiquitin using Ni-NTA beads under denaturing conditions. Western blot analysis with anti-H2B antibodies was employed to detect all forms of H2B in the precipitates. Cells co-transfected with His6-ubiquitin, and 2A-HUB or 2A-HUBRmut displayed highly comparable levels of monoubiquitinated H2B (Figure 3D). Together, these data indicate that 2A-HUB can specifically promote H2A, but not H2B, monoubiquitination both in vitro and vivo.
Figure 3. 2A-HUB Monoubiquitinates Histone H2A in Vivo.
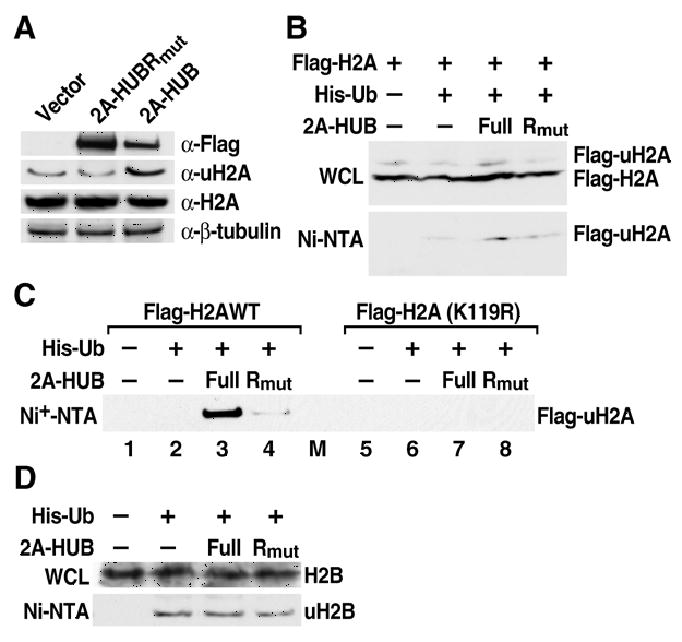
(A) Western blot analysis endogenous uH2A and H2A. Soluble chromatin extracts from HEK293T transfected with Flag-2A-HUB or Flag-2A-HUBRmut were probed with anti-Flag, anti-uH2A, anti-Histone H2A and anti-β-tubulin antibodies.
(B) In vivo ubiquitination of Histone H2A by 2A-HUB. HEK293T cells were transfected with the indicated combinations of expression plasmids. The top panel shows the whole cell lysates (WCL) were probed with anti-Flag antibodies. The lower panel shows Ni-NTA agarose precipitates of the lysates of transfected cells, which were probed with anti-Flag antibodies.
(C) In vivo ubiquitination assays using wt Flag-H2A- or Flag-H2A(K119R)-expressing plasmid. HEK293T were transfected with the indicated combinations of expression plasmids. The Ni-NTA agarose precipitates of the lysates of transfected cells were probed with anti-Flag antibodies.
(D) In vivo ubiquitination of Histone H2B by 2A-HUB. The experiment was carried out as described in Figure 3B.
2A-HUB and N-CoR Complex are Required for Repression of a Subset of Chemokine Genes Expression
N-CoR acts as an active transcriptional repressor for many nuclear receptors, NF-κB and AP-1 _target genes that regulate diverse biological processes including inflammation, cell migration, and collagen catabolism, with loss of N-CoR in macrophages, resulting in a partially activated-phenotype (Ogawa et al., 2004; Pascual et al., 2005; Perissi et al., 2004). To identify a potential cohort of genes co-regulated by N-CoR and 2A-HUB, we performed an RNA profiling analysis in RAW 264.7 (a murine leukaemic monocyte/macrophage cell line) cells treated with either control siRNA or 2A-HUB siRNA that specifically knocked down 2A-HUB protein (Figure S3A). Knockdown of endogenous 2A-HUB expression revealed a number of genes that were directly or indirectly regulated by 2A-HUB. Analysis of biological process annotations defined by the GO Consortium (Gene Ontology Consortium, 2001) associated with each differentially expressed gene, indicated statistically-significant over-representation of genes involved in several categories, including immune response, cell cycle, and pattern specification. A subset of the chemokine genes that were selectively derepressed by knockdown of 2A-HUB in RAW 264.7 cells included those previously identified as N-CoR _target genes (Figure S3B) (Ogawa et al., 2004). To verify that these identified candidate genes were differentially expressed in RAW 264.7 cells treated by control siRNA when compared to those treated by siRNA to 2A-HUB or N-CoR, quantitative real-time RT-PCR (qRT-PCR) analysis was performed. qRT-PCR from different biological samples revealed, for example, that Cxcl2/MIP2(macrophage inflammatory protein-2), Ccl5/RANTE, (regulated on activation normal T cell expressed and secreted) and Cxcl10/IP-10(interferon-inducible protein-10) were indeed derepressed in RAW 264.7 cells by knockdown of 2A-HUB using different siRNA oligo nucleotides specific for 2A-HUB, as well as by knockdown of N-CoR (Figures 4A, B and Figure S4A). These genes were also depressed in thioglycollate-elicited macrophages after siRNA knockdown of 2A-HUB (Figure S4B). In contrast, the Cxcl1/KC gene expression was increased in cells with knockdown of N-CoR, but remained unchanged in cells with knockdown of 2A-HUB (Figure 4B).
Figure 4. 2A-HUB and N-CoR Regulate a Subset of Chemokine Gene Expression.
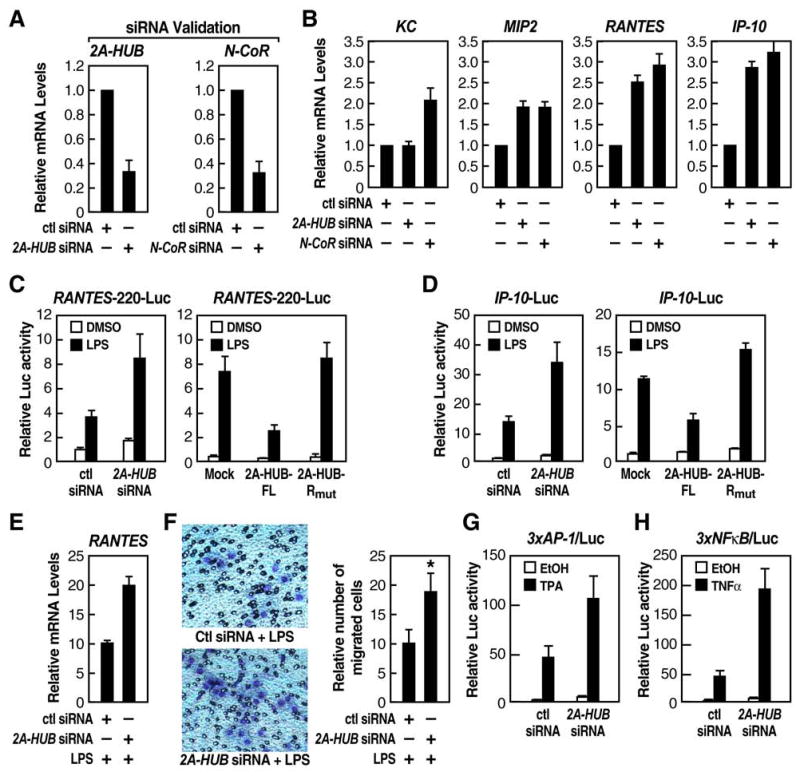
(A) Real-time RT-PCR (RT-qPCR) analysis was performed to document efficiency of 2A-HUB 1# siRNA, N-CoR siRNAs to diminish endogenous 2A-HUB and N-CoR.
(B) RT-qPCR analysis of several endogenous 2A-HUB/N-CoR _target genes upon 2A-HUB 1# siRNA or N-CoR siRNA transfection. In (A) and (B), values (normalized to corresponding values of internal control gene Hprt) are the average of three replicates ± SEM.
(C&D) Reporter assays using a RANTES-200-Luc reporter (C), IP-10-Luc reporter (D), in RAW264.7 cells. Cells were transfected with the reporters, siRNA or different expression plasmids as indicated. 24 hours after transfection, cells were treated with DMSO or LPS for 12 hours.
(E) RT-qPCR analysis of RANTES genes expression upon control siRNA or 2A-HUB 1# siRNA transfection. 48 hours after transfection, cells were treated with LPS for 8 hours. Values (normalized to corresponding values of internal control gene Hprt) are the average of three replicates ± SEM.
(F) Migration of RAW 264.7 cells was increased in response to the supernatant from LPS-treated 2A-HUB knockdown macrophages (see Supplemental Experimental Procedure). Values are means ± SEM from three independent experiments. *P<0.05 (versus control, Student t test).
(G&H) Reporter assays using a 3xAP-1-Luc reporter (G), 3xNF-κB-Luc reporter (H), in RAW264.7 cells. Cells were transfected with the reporters, control siRNA and 2A-HUB siRNA as indicated. 24 hours after transfection, cells were treated with TPA or TNF-α for 12 hours. In (C), (D), (G) and (H), values are the relative values to normalized activity from basal, reporter-only transfected cells (bar 1, set as 1) and are expressed as average of three independent experiments ± SEM.
To begin to test whether 2A-HUB directly regulates RANTES and IP-10 gene expression at the promoter level, we performed transient transfection assays using luciferase reporter constructs under control of the regulatory promoter sequences from either RANTES or IP-10. The basal activity of the 220bp RANTES promoter reporter was increased by two-fold in RAW 264.7 cells treated with siRNA against 2A-HUB, in comparison to the control/scramble siRNA (Figure 4C, left panel). Induction of the RANTES promoter reporter was also increased significantly after stimulation with LPS in RAW 264.7 cells treated with siRNA against 2A-HUB (Figure 4C, left panel). In contrast, induction of the RANTES promoter reporter was significantly reduced in RAW 264.7 cells, in which full length 2A-HUB was overexpressed, but not in which the 2A-HUBRmut was overexpressed (Figure 4C, right panel). We next examined the effect of knockdown or over-expression of 2A-HUB on the IP-10 promoter reporter activity. The activity of this reporter gene was increased both at the basal level and upon LPS treatment in response to siRNA depletion of 2A-HUB (Figures 4D, left panel). In contrast, induction of the IP-10 promoter reporter was reduced significantly in the presence of over-expressed full length 2A-HUB, while the mutant 2A-HUBRmut had no effect (Figure 4D, right panel). Consistent with these data, the induction of endogenous RANTES or IP-10 gene upon LPS treatment was also significantly increased in RAW 264.7 cells after knockdown of 2A-HUB (Figure 4E and data not shown). Collectively, these results suggested that 2A-HUB can mediate active repression of RANTES, IP-10 and other chemokine gene expression in the presence and absence of activation stimuli. IP-10 has been shown to be a chemoattractant for activated T-lymphocytes (Rollins, 1997). RANTES, plays key role in inflammation by chemo-attracting various cell types, including T cells, macrophages, and eosinophils to inflammatory sites (Zlotnik and Yoshie, 2000). Therefore, we were interested in determining whether the increased expression of RANTES and other _targets by siRNA depletion of 2A-HUB would affect monocyte or macrophage migration. We performed cell migration experiments using supernatants taken from LPS treated RAW 264.7 cells either transfected with control siRNA or 2A-HUB siRNA. RAW 264.7 cells were strongly attracted by the source of supernatant taken from 2A-HUB knockdown macrophage stimulated with LPS compared to control siRNA treated cells (Figure 4F). These results indicated that 2A-HUB is a critical regulator of chemokine expression and chemokine-mediated cell migration.
2A-HUB negatively regulates AP-1-dependant or NF-κB dependant transcription units, consistent with the presence of common elements of AP-1 and NF-κB consensus sequences within the promoter regions of Cxcl2, Cxcl5 and Cxcl10 genes (Doyle et al., 2002). Both AP-1-luc or NF-κB-luc reporter gene activities was increased in the basal level as well as in response to TPA or TNF-α treatment in cells depleted of 2A-HUB by siRNA treatment (Figure 4G & 4H). We also examined the functional role of 2A-HUB in other N-CoR complex-regulated transcriptional events, finding that 2A-HUB negatively regulates retinoic acid receptor (RA) and estrogen receptor (ER)-dependent transcription, as revealed by reporter assays and by analysis of mRNA expression of endogenous gene _targets (Figure S5A-S5B). In contrast, PSA, a known _target gene of androgen receptor (AR), was not affected by 2A-HUB (Figure S5C). Collectively, these results reveal that 2A-HUB, along with the nuclear receptor corepressor (N-CoR), play important combinatoric roles in active repression of a cohort of repressed _target genes.
ChIP experiments revealed that both 2A-HUB and uH2A were present on the promoter regions of the endogenous MIP2, RANTES and IP-10 transcription units, but not on the KC promoter region (Figure 5A). Both 2A-HUB and uH2A were dismissed from the promoter region of MIP2, RANTES and IP-10 upon LPS stimulation (Figure 5A). We next performed a two-step chromatin immunoprecipitation experiment, first using α-N-CoR IgG and subsequently α-2A-HUB IgG. We were able to observe 2A-HUB recruitment on the N-CoR+ RANTES promoter, but not on the N-CoR+ KC promoter (Figure 5B and 5C). These data suggest that 2A-HUB is selectively required for repression a specific cohort of N-CoR- repressed chemokine genes. To determine how 2A-HUB is recruited to its _target gene promoter, we performed ChIP analysis in cells treated N-CoR siRNA. Recruitment of 2A-HUB on the RANTES promoter was significantly decreased following knockdown of N-CoR (Figure 5D), suggesting that 2A-HUB recruitment is dependent on the presence of N-CoR. Together these results indicate that H2A monoubiquitination, mediated by 2A-HUB, along with N-CoR complexes, directly regulates RANTES, IP-10 and other chemokine gene expression, and plays a role in maintaining these inflammatory response genes in a repressed state in the absence of an inflammatory stimulus.
Figure 5. The Recruitment of 2A-HUB and uH2A on the _target Gene Promoters.
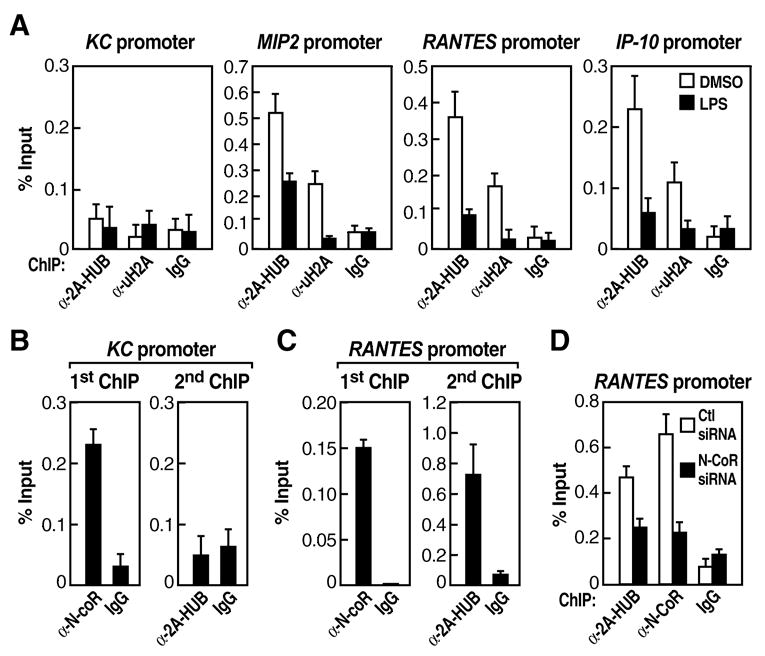
(A)ChIP analyses were performed after cells were stimulated with LPS for 30 minutes. Soluble chromatin was immunoprecipitated by the antibodies indicated, the bound DNA was analyzed by quantitative PCR using a primer pair flanking the regulatory sequence of the promoter of KC, MIP2, RANTES or IP-10. The ChIP was performed as described in Methods.
(B) Two-step ChIP assays were performed, showing that 2A-HUB is not recruited to the N-coR+ KC promoter.
(C) Two-step ChIP assays were performed, showing that 2A-HUB is recruited to the N-coR+ RANTES promoter.
(D)ChIP analyses of the occupancy of 2A-HUB on the RANTES promoter after depletion of N-CoR by siRNA in RAW 264.7 cells. Values (ratios of ChIP to corresponding inputs) in (A)–(D) are means ± SEM of at least two independent experiments.
H2A Ubiquitination Mediated by 2A-HUB Negatively Regulates Transcriptional Elongation by Preventing FACT Recruitment
FACT (for facilitates chromatin transcription), a dimeric protein that comprises human SPT16 and SSRP1 (Orphanides et al., 1998; Orphanides et al., 1999), acts to relieve chromatin-mediated inhibition of transcriptional elongation by binding and displacing the H2A/H2B dimer from the core nucleosomes (reviewed in Reinberg and Sims, 2006). Histone H2B monoubiquitination occurs at both promoters and ORFs, facilitating FACT function, thereby enhances the rate of transcription elongation on chromatin templates (Kao et al., 2004; Kim et al., 2005a; Pavri et al., 2006; Zhu et al., 2005). In contrast, histone H2A monoubiquitination primarily occurs at promoters region, repressing _target gene expression (Cao et al., 2005; Wang et al., 2004). To test the hypothesis that H2A ubiquitination might prevent FACT function, in direct opposition to the ubiquitination function of H2B, we examined the potential interaction between one of the FACT subunits, Spt16, with histone H2A or uH2A, as well as with H2B or uH2B. We observed that Spt16 specifically interacted to H2A but not with uH2A, in HEK293 cells stably expressing Flag-H2A (Figure 6A, middle panel). In contrast, Spt16 bound both histone H2B and uH2B with the same efficiency in HEK293 cells stably expressing Flag-H2B (Figure 6A, bottom panel).
Figure 6. H2A Ubiquitination by 2A-HUB Prevents FACT Recruitment.
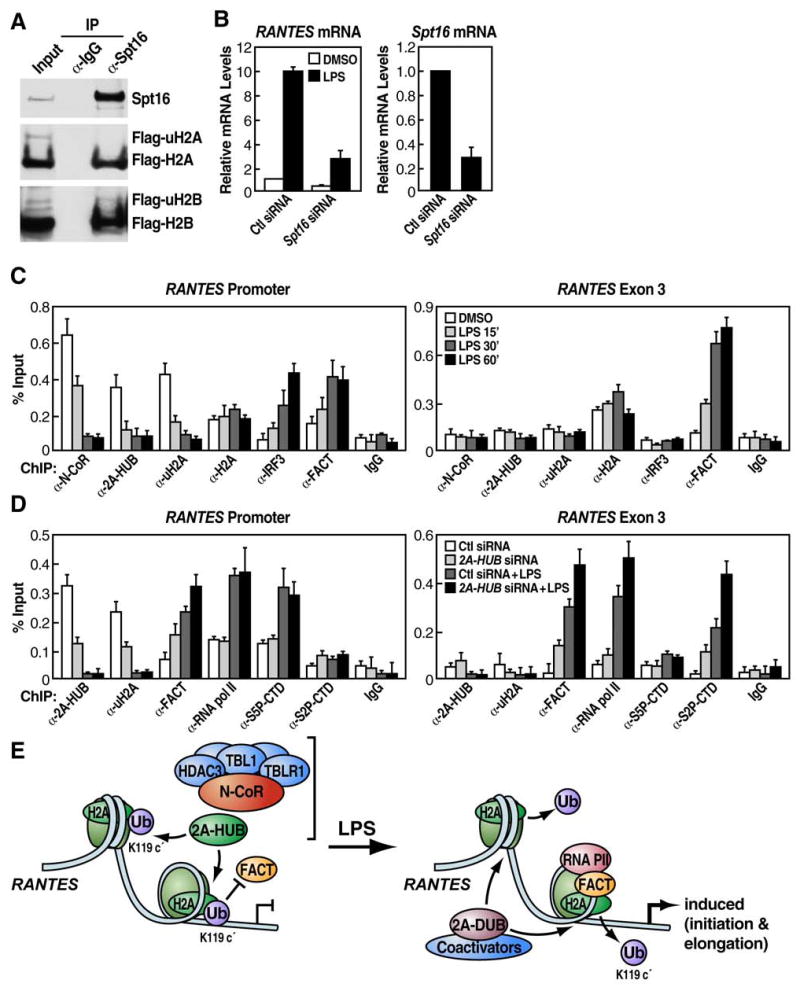
(A) Coimmunoprecipitation of Spt16 and H2A/H2B. Top panel, immunoprecipitation of endogenous Spt16. Middle panel, co-immunoprecipitation of endogenous Spt16 with unmodified Flag-H2A, but not Flag-uH2A, in HEK293 cells stably expressing Flag-H2A. Bottom panel, co-immunoprecipitation of endogenous Spt16 with both unmodified Flag-H2B and Flag-uH2B in HEK293 cells stably expressing Flag-H2B.
(B) RT-qPCR analysis of endogenous RANTES gene expression upon LPS treatment after depletion Spt16 by siRNA. Values (normalized to corresponding values of internal control gene Hprt) are the average of three replicates ± SEM.
(C) ChIP in RAW246.7 cells treated with LPS (time courses as indicated) using a panel of antibodies as shown. PCR primers amplifying the promoter region and a downstream region corresponding to exon 3 of the RANTES gene were used.
(D) ChIP was performed in RAW264.7 cells, first treated with control or 2A-HUB siRNA and then induced by LPS for 30 minutes. A panel of antibodies was used as shown. Values (ratios of ChIP to corresponding inputs) in (C) and (D) are the mean ± SEM of at least two independent experiments.
(E) Model: H2A monoubiquitination mediated by 2A-HUB inhibits the early step of RNA Pol II transcriptional elongation by blocking the recruitment of FACT and maintains gene in a repressed status. Upon gene activation by LPS, removing ubiquitin from H2A by H2A deubiquitinase (2A-DUB) increases FACT recruitment and facilitates RNA polymerase II transcriptional elongation.
In accord with this finding, RANTES gene induction by LPS was found to be impaired after depletion of Spt16 in RAW 246.7 cell (Figure 6B), suggesting that Spt16/FACT, is functionally required for RANTES gene transcription. ChIP analysis of the endogenous RANTES promoter and exon 3 regions in RAW 264.7 cells revealed that N-CoR, 2A-HUB and uH2A were all present on promoter under basal conditions and dismissed following LPS treatment (Figure 6C, left panel). Conversely, FACT and IRF3 were recruited to the promoter only after LPS treatment (Figure 6C, left panel). Importantly, FACT recruitment increased both in the promoter and in the coding region upon LPS treatment (Figure 6C). Specificity of the ChIP was demonstrated by the inability to detect N-CoR, 2A-HUB or IRF3 within the transcribed region (Figure 6C, right panel). We did not observe dramatic alternation on histone H2A recruitment under these conditions.
These studies suggest that histone H2A ubiquitination might regulate Spt16/FACT recruitment and influence RNA Pol II transcriptional elongation events. In order to investigate this possibility, we performed ChIP analysis with siRNA knockdown of 2A-HUB in RAW 264.7 in combination with LPS treatment. Under basal conditions, recruitment of 2A-HUB and uH2A at the RANTES gene promoter was decreased significantly in cells treated with 2A-HUB but not with control siRNA, indicating that 2A-HUB is crucial in maintaining the H2A ubiquitination status (Figure 6D, left panel). Consequently, recruitment of FACT to the RANTES promoter and exon 3 region was increased in cells treated with 2A-HUB siRNA (Figure 6D). Following dephosphorylation of Ser5 and phosphorylation of Ser2 of the CTD of RNA PolII by CtK1 kinase (Cdk9/P-TEFb in mammals), the Pol II enzyme is converted into a fully productive, elongation-engaged form and transits to the 3′ end (Komarnitsky et al., 2000). We therefore investigated RNA Pol II phosphorylation status in RANTES gene promoter and exon 3 region upon LPS treatment after depletion of 2A-HUB by siRNA. ChIP analysis using antibodies against Ser5- and Ser2-phosphorylated CTD (S5P-CTD and S2P-CTD) revealed significantly increased S5P signals in response to LPS at the promoter region, but not in the exonic region (Figure 6D). In contrast, S2P-CTD signal in response to LPS increased at exonic region (Figure 6D). While S5P-CTD signals in response to LPS exhibited little alternation in cells previously treated with 2A-HUB siRNA, FACT and Pol II signal, in particular, S2P-CTD signals in response to LPS in the exon region increased significantly in cells in which 2A-HUB was depleted (Figure 6D, right panel). These results suggest that H2A ubiquitination mediated by 2A-HUB plays an important role in blocking FACT recruitment, inhibiting RNA Pol II-dependent transcriptional elongation and repressing those chemokine genes expression (Figure 6E).
DISCUSSION
H2A Ubiquitination and Gene Repression
Here, we have identified and characterized 2A-HUB, an ubiquitin E3 ligase that catalyzes monoubiquitination of histone H2A in vivo, as well as in vitro. 2A-HUB functions as a transcriptional corepressor and 2A-HUB-dependent transcriptional repression requires its E3 ubiquitin ligase activity. We documented that H2A monoubiquitination-mediated by 2A-HUB is involved in repression of specific gene cohorts, such as those involved in inflammatory response in macrophages. Because there is no significant change of general H2A ubiquitination after 2A-HUB depletion (data not shown), we suggest that the role of 2A-HUB is to modulate a specifically-localized histone modification, rather than a global effect. To begin to understand the functions related to the roles of other proteins capable of catalyzing monoubiquitination of H2A, we tested whether another well known H2A E3 ligase Ring1b might be involved in transcriptional regulation of those chemokine genes regulated by 2A-HUB. In contrast to the requirement for 2A-HUB, no effect on these chemokine genes expression was incurred by depletion of Ring1b by siRNA in RAW 246.7 cells (data not shown). These results suggest that regulation of H2A ubiquitination occurs in a gene- and enzyme-specific fashion, while RANTES gene promoter region is modified by 2A-HUB, reflecting recruitment by the N-CoR complex (Figure 6E).
The fact that the RING domain of 2A-HUB is suggested to elicit efficient transcriptional repression when tethered to a promoter region implicates that 2A-HUB may contribute to transcriptional repression in gene-specific manner via local induction of histone ubiquitination. Indeed, the activity of 2A-HUB may serve to dampen the transcriptional activity, of at least some _target genes, under nonstimulated conditions. Because 2A-HUB does not exist in the biochemically purified N-CoR core complexes (Li et al., 2000; Zhang et al., 2002), it can be suggested that 2A-HUB interacts with N-CoR complexes dependent upon the specific promoter context. Interestingly, 2A-HUB was recruited by the N-CoR complex to the _target gene promoters to tightly control gene expression at transcriptional elongation level, supporting the concept that histone ubiquitination by 2A-HUB facilitates gene repression programs. While ubiquitination of other components of the transcriptional machinery by 2A-HUB is not excluded, we suggest that repression is due, at least in part, to enhanced histone H2A ubiquitination. The recruitment of at least three distinct classes of histone-modifying enzymes, histone deacetylases, E3 ubiquitin ligases and JmjC domain-containing histone demethylases by N-CoR suggests that the unique combination of epigenetic modifications is utilized to direct gene repression by N-CoR, implying that histone ubiquitination, deacetylation and demethylation are coordinated during gene transcriptional repression regulation (Klose et al., 2006; Whetstine et al., 2006; Zhang et al., 2005).
In contrast, another histone H2A E3 ligase Ring1b was found in polycomb repressive complex (PRC1), which ubiquitinates H2A, playing an important role in X-chromosome inactivation (de Napoles et al., 2004), polycomb _target gene silencing (Wang et al., 2004) and Hox gene expression (Cao et al., 2005). However, the mechanism of how H2A ubiquitination may lead to gene silencing or repression has been largely unknown. It has been proposed that H2A monoubiquitination may directly or indirectly block recruitment of the basal transcriptional machinery (Cao et al., 2005). Recent studies have suggested that PRC1 may repress transcription via direct interaction with components of the basal transcription machinery due to the presence of TFIID subunits in PRC1 (Breiling et al., 2001; Saurin et al., 2001). Furthermore, TBP and Pol II are occupied on promoters that were repressed by PRC1, suggesting that PRC1 selectively interferes with events downstream of Pol II recruitment (Breiling et al., 2001; Dellino et al., 2004). Conversely, the Drosophila trithorax group protein Kismet facilitates RNA polymerase II transcriptional elongation (Srinivasan et al., 2005). In addition, Ring1b was identified as the co-repressor BCoR complex and promoted monoubiquitinated H2A at BCL6 _target gene promoters, to direct gene repression (Gearhart et al., 2006). It is interesting that FBXL10, a JmjC-domain containing protein that has recently been identified as histone H3K36 demethylase (Klose et al., 2006) also co-purified with BCoR, suggesting that BCoR mediated _target gene repression may need a combination of histone ubiquitination and histone demethylation events. Because histone H3K36 methylation was mainly coupled with active gene elongation (Eissenberg and Shilatifard, 2006), it is possible that BCoR also represses _target gene expression by negatively regulating transcriptional elongation. Taken together, we suggest that distinct H2A E3 ligases, each recruited by different corepressor complexes, contribute to effective repression of distinct transcriptional programs.
Histone H2A Ubiquitination Negative Regulates Transcriptional Elongation
During the transcription into a nucleosomal template, RNA Polymerase II pauses at certain sites which may relate to the strength or nature of the histone-DNA contacts (Bondarenko et al., 2006; Kireeva et al., 2005). Given the fact that the transcribing RNA Pol II stalls at the first nucleosome, this and the subsequent nucleosome barriers must be relieved by FACT (Pavri et al., 2006). Here, we have found that histone H2A monoubiquitination forms a physical barrier in the nucleosome to prevent FACT recruitment, and causes RNA Pol II to pause at the promoter-proximal region. Promoter-proximal pausing is a wide spread phenomenon at many genes in eukaryotes (Kim et al., 2005b) and constitutes an important step in the regulation of Pol II transcriptional elongation. The RNA Pol II can be released rapidly from the pause into the productive elongation phase, and this highly dynamic level of regulation enables the rapid alteration of transcriptional output (Peterlin and Price, 2006; Saunders et al., 2006). In this regard, we propose that removing ubiquitin from H2A substantially increases the interaction between H2A and FACT and facilitates RNA Pol II elongation upon gene activation (Figure 6E). Therefore, we suspect that the H2A deubiquitination process is required for transcription initiation or elongation by Pol II. In light of our recent finding that histone H2A deubiquitination mediated by newly identified MPN+/JAMM domain–containing histone H2A deubiquitinase (2A-DUB) occurs during transcriptional initiation and elongation and 2A-DUB participates in transcriptional initiation/elongation events in androgen receptor-dependant gene activation (Zhu et al., 2007), we speculate that various H2A ubiquitinases/deubiquitinases will be discovered to regulate distinct transcriptional programs (Figure 6E).
While there is considerable knowledge with regard to transcription initiation in a chromatinized context, little is known about the subsequent post initiation events associated with transcription elongation. Our findings suggest the model that RNA Pol II pausing by histone H2A monoubiquitination might provide a checkpoint to assess whether the Pol II is correctly prepared for productive elongation, and allows rapid regulation of gene expression. Biologically, 2A-HUB appears of particular, specific important in regulating a subset of chemokine genes, modulating macrophage response to TLR signaling. Because chemokines and their receptors play a central role in recruitment, activation, and mobilization of leukocytes to sites of infection, eliminating infectious pathogens while at the same time avoiding immuno-mediated tissue damages (Rollins, 1997; Zlotnik and Yoshie, 2000), it is intriguing that 2A-HUB, along with the N-CoR complex, have evolved to combinationally attenuate expression of a cohort of chemokine genes normally induced by TLR signaling, critical for maintaining intact innate immunity. The requirement for a histone H2A monoubiquitination-dependent checkpoint is suggested to permit the appropriate dynamic range of transcriptional responses to inflammatory stimuli that regulate diverse immune responses and homeostatic processes.
EXPERIMENTAL PROCEDURES
Materials and Reagents
A GST-tagged 2A-HUB fragment (residues 1006–1193) was expressed in bacteria, affinity purified, and used as an antigen to produce anti-2A-HUB polyclonal antibody (Harland). Commercially available cell lines, antibodies and siRNAs are described in the Supplemental Data.
In Vivo and in Vitro Histone Ubiquitination Assays
The in vivo and in vitro ubiquitination experimental procedures are described in the Supplemental Data.
Transient transfection assay
Transfections were carried out in RAW 264.7 or HEK293T cells using Lipofectamin™ 2000 (Invitrogen), following manufacture’s protocol.
ChIP Assays
ChIP assay was conducted as previously described (Baek et al., 2003). Further details are available in Supplemental Data.
Supplementary Material
Acknowledgments
We thank T. Maniatis for RANTES-Luciferase and IP-10-luciferase reporters, M. Oren for Flag-H2A and Flag-H2B constructs and Dr. Kreft for human 2A-HUB expression plasmids. We thank Xiangting Wang, Jie Zhang, Chuck Nelson for their technical assistance. We also thank J. Hightower and M. Fisher for figure and manuscript preparation. P.Z. is supported by a postdoctoral fellowship from the Human Frontier Science Program (HFSP). M.G.R. is an investigator with the Howard Hughes Medical Institute. This work was supported by grants from NIH and NCI to C.K.G and M.G.R. and the PCF and PCRP to M.G.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. The Journal of biological chemistry. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- Baek SH, Kioussi C, Briata P, Wang D, Nguyen HD, Ohgi KA, Glass CK, Wynshaw-Boris A, Rose DW, Rosenfeld MG. Regulated subset of G1 growth-control genes in response to derepression by the Wnt pathway. Proc Natl Acad Sci U S A. 2003;100:3245–3250. doi: 10.1073/pnas.0330217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Molecular cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Molecular cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Dawson BA, Herman T, Haas AL, Lough J. Affinity isolation of active murine erythroleukemia cell chromatin: uniform distribution of ubiquitinated histone H2A between active and inactive fractions. J Cell Biochem. 1991;46:166–173. doi: 10.1002/jcb.240460210. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V. Polycomb silencing blocks transcription initiation. Molecular cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O’Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr Opin Genet Dev. 2006;16:184–190. doi: 10.1016/j.gde.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. The Journal of biological chemistry. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 _targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. The Journal of biological chemistry. 1975;250:7182–7187. [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & development. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SY, Barnard MB, Xu M, Matsui S, Rose SM, Garrard WT. The active immunoglobulin kappa chain gene is packaged by non-ubiquitin-conjugated nucleosomes. Proc Natl Acad Sci U S A. 1986;83:3738–3742. doi: 10.1073/pnas.83.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Molecular cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes & development. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Molecular cell. 2005a;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005b;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, et al. Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Molecular cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & development. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft SG, Nassal M. hRUL138, a novel human RNA-binding RING-H2 ubiquitin-protein ligase. J Cell Sci. 2003;116:605–616. doi: 10.1242/jcs.00261. [DOI] [PubMed] [Google Scholar]
- Levinger L, Varshavsky A. Selective arrangement of ubiquitinated and D1 protein-containing nucleosomes within the Drosophila genome. Cell. 1982;28:375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Molecular cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Nickel BE, Allis CD, Davie JR. Ubiquitinated histone H2B is preferentially located in transcriptionally active chromatin. Biochemistry. 1989;28:958–963. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- Nickel BE, Davie JR. Structure of polyubiquitinated histone H2A. Biochemistry. 1989;28:964–968. doi: 10.1021/bi00429a007. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci U S A. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Osley MA. H2B ubiquitylation: the end is in sight. Biochim Biophys Acta. 2004;1677:74–78. doi: 10.1016/j.bbaexp.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Parlow MH, Haas AL, Lough J. Enrichment of ubiquitinated histone H2A in a low salt extract of micrococcal nuclease-digested myotube nuclei. The Journal of biological chemistry. 1990;265:7507–7512. [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. The Journal of biological chemistry. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Rollins BJ. Chemokines. Blood. 1997;90:909–928. [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes & development. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne AW, Sautiere P, Briand G, Crane-Robinson C. The structure of ubiquitinated histone H2B. Embo J. 1987;6:1005–1010. doi: 10.1002/j.1460-2075.1987.tb04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Molecular cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Zhang D, Yoon HG, Wong J. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2) Mol Cell Biol. 2005;25:6404–6414. doi: 10.1128/MCB.25.15.6404-6414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Molecular cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes & development. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Molecular cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A Histone H2A Deubiquitinase Complex Coordinating Histone Acetylation and H1 Dissociation in Transcriptional Regulation. Molecular cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


