Abstract
Intrauterine growth retardation (IUGR) has been linked to the onset of diseases in adulthood, including type 2 diabetes, and has been proposed to result from altered gene regulation patterns due to epigenetic modifications of developmental genes. To determine whether epigenetic modifications may play a role in the development of adult diabetes following IUGR, we used a rodent model of IUGR that expresses lower levels of Pdx1, a pancreatic and duodenal homeobox 1 transcription factor critical for β cell function and development, which develops diabetes in adulthood. We found that expression of Pdx1 was permanently reduced in IUGR β cells and underwent epigenetic modifications throughout development. The fetal IUGR state was characterized by loss of USF-1 binding at the proximal promoter of Pdx1, recruitment of the histone deacetylase 1 (HDAC1) and the corepressor Sin3A, and deacetylation of histones H3 and H4. Following birth, histone 3 lysine 4 (H3K4) was demethylated and histone 3 lysine 9 (H3K9) was methylated. During the neonatal period, these epigenetic changes and the reduction in Pdx1 expression could be reversed by HDAC inhibition. After the onset of diabetes in adulthood, the CpG island in the proximal promoter was methylated, resulting in permanent silencing of the Pdx1 locus. These results provide insight into the development of type 2 diabetes following IUGR and we believe they are the first to describe the ontogeny of chromatin remodeling in vivo from the fetus to the onset of disease in adulthood.
Introduction
Intrauterine growth retardation (IUGR), a common complication of pregnancy, has been linked to the later development of diseases in adulthood such as type 2 diabetes (1). It has been hypothesized that the molecular mechanisms underlying this phenomenon may in part be related to epigenetic modulation of expression of key developmental genes (2, 3). Epigenetic modifications provide a mechanism that allows the stable propagation of gene activity states from 1 generation of cells to the next. Epigenetic states can be modified by environmental factors, which may contribute to the development of abnormal phenotypes. In mammals, DNA methylation and histone modifications represent the major epigenetic mechanisms implicated in the regulation of gene transcription.
We have developed an animal model of IUGR caused by uteroplacental insufficiency, which limits the supply of critical substrates and hormones to the fetus (4, 5). This abnormal metabolic intrauterine milieu affects the development of the fetus by permanently modifying gene expression and function of susceptible cells, such as the β cell (4–6), and leads to the development of diabetes in adulthood (4–6).
Pdx1 is a pancreatic and duodenal homeobox 1 transcription factor that regulates pancreas development and β cell differentiation.
Both genetic and acquired reductions in Pdx1 expression in humans and in animal models have been shown to cause type 2 diabetes, β cell dysfunction (7–14), and impaired islet compensation in the presence of insulin resistance (7, 8). In IUGR rats, we have observed that Pdx1 mRNA levels are reduced by more than 50% in IUGR fetuses (6). At birth, β cell mass is normal but Pdx1 mRNA levels are decreased in IUGR rats (6). In IUGR adult rats, β cell mass is markedly decreased and Pdx1 expression is nearly absent (6).
IUGR leads to the permanent suppression of Pdx1 in islets, suggesting that an epigenetic mechanism may be responsible via changes in DNA methylation, histone modifications, or chromatin protein binding (15). The region spanning the proximal 5ι promoter and all of exon 1 of Pdx1 contains a highly conserved CpG island. Deletion of the proximal promoter completely abolishes Pdx1 promoter activity (16). This region is also heavily acetylated at histone H3 and H4 in a β cell tumor line (17). However, the epigenetic characteristics of the Pdx1 gene have not been examined in primary islet tissue in normal or disease states.
In the present study, we hypothesized that epigenetic changes involving histone modifications, DNA methylation, chromatin remodeling, and transcription were responsible for Pdx1 silencing during the transition from IUGR to diabetes in adults. We determined that in early postnatal life, prior to the onset of diabetes, the IUGR state induces deacetylation of histones H3 and H4, which is facilitated by recruitment of histone deacetylase 1 (HDAC1) and Sin3A to the proximal promoter of Pdx1. Loss of acetylation is accompanied by loss of binding of the key transcription factor, USF-1. As the disease state progresses, histone 3 lysine 4 (H3K4) is demethylated and histone 3 lysine 9 (H3K9) is methylated. Finally, once diabetes occurs, methylation of the CpG island in the proximal promoter of Pdx1 ensues, locking in marked suppression of Pdx1. These are the first studies to our knowledge to describe the ontogeny of chromatin remodeling in vivo from the fetus to the onset of a disease in adulthood.
Results
Pdx1 transcription is reduced in IUGR islets.
In previous studies, we found that Pdx1 mRNA levels were decreased by 50% in fetuses and 2-week-old pups and 80% in adult IUGR animals (6). To determine whether this reduction in Pdx1 mRNA levels was secondary to a reduction in transcription of the Pdx1 gene or to an increase in mRNA stability, we induced IUGR in Pdx1-LacZ in vivo transcriptional reporter mice. The transgene was regulated by the 4.6-kb XbaI-XbaI fragment containing 4.5 kb of the mouse Pdx1 promoter and most of the 5ι untranslated region (UTR). Most of the important Pdx1 enhancers described to date are included within this promoter fragment (17–19). Endogenous Pdx1 mRNA levels are reduced by 50.4% and transgene LacZ mRNA is reduced in parallel by approximately 57.1% (P < 0.05 versus control). This indicates that IUGR regulates Pdx1 mRNA by reducing the activity of transcriptional promoter or enhancer elements in the proximal 4.5-kb Pdx1 5ι region.
Methylation status of the Pdx1 gene promoter in IUGR and control islets.
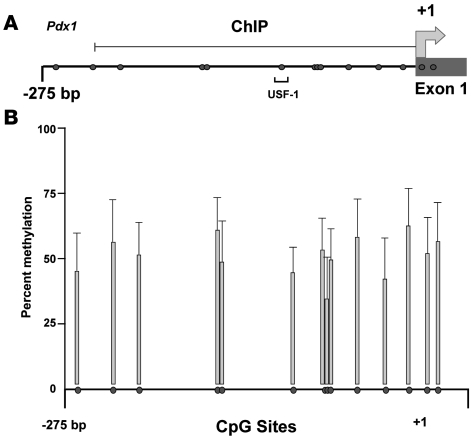
The proximal 5ι flanking region of the Pdx1 gene and its first exon include a highly conserved CpG island that encompasses nucleotides –360 to +200 relative to the translational start site (Figure 1A). Active promoters are associated with unmethylated CpG islands and open chromatin structure, whereas inactive promoters are typically characterized by a repressed chromatin structure and hypermethylated CpGs. We determined the DNA methylation status of the portion of the CpG island that is located in the Pdx1 promoter in islets of IUGR and control animals using bisulfite treatment and pyrosequencing analyses. Evaluation of DNA from islets of 5 IUGR and 5 control animals revealed that at age 2 weeks none of the 14 CpG sites in the Pdx1 promoter were methylated in islets from either IUGR or control animals. At 6 months of age (n = 5 animals per group), Pdx1 DNA methylation levels across the CpG island averaged 51.3% ± 10.3% compared with no CpG methylation in controls (P < 0.05 vs. controls). No single CpG site was consistently methylated (Figure 1B).
Figure 1. Methylation status of proximal promoter of Pdx1.
(A) Map of Pdx1. The proximal promoter of Pdx1 is part of a more extensive CpG island. Filled circles represent CpG dinucleotides, and numbers indicate positions relative to the transcriptional start site. The position of the E-box bound by USF-1 is indicated. (B) Methylation of CpG dinucleotides within the promoter of Pdx1. Islets were isolated from 5 IUGR and 5 control animals at 6 months of age, and Pdx1 DNA methylation was quantified by pyrosequencing of bisulfite-treated DNA. We did not detect methylation at any CpG of Pdx1 in controls. The shaded bars represent data obtained and averaged from 5 IUGR animals and presented as percent methylation at each CpG site. Data are ± SEM.
Binding of DNA methyltransferases at Pdx1 in IUGR islets.
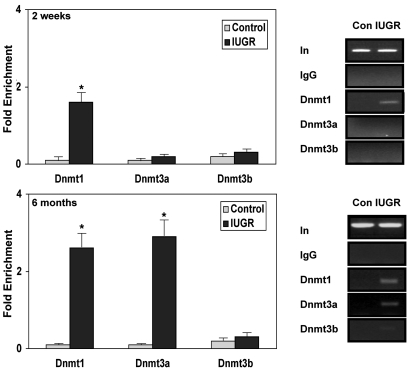
Mammalian CpG methylation is mediated by 3 active DNA methyltransferases (Dnmts), Dnmt1, Dnmt3a, and Dnmt3b. Dnmt1 mediates replication-coupled maintenance of DNA methylation patterns, whereas Dnmt3a and Dnmt3b are considered to be de novo DNA methylases (20, 21). Surprisingly, we observed some association of Dnmt1 to Pdx1 in islets of 2-week-old IUGR pups, despite the lack of DNA methylation of CpGs (Figure 2). However, there was no binding of Dnmt3a or Dnmt3b to Pdx1 at this age. At 6 months of age, when DNA methylation at Pdx1 is increased in IUGR islets, Dnmt1 binding was significantly enhanced (Figure 2) and moderate levels of Dnmt3a, but not Dnmt3b, were also associated with Pdx1 (Figure 2). No binding of Dnmt1 or Dnmt3a and -3b to Pdx1 was detected in control islets at either age.
Figure 2. Quantitative analysis of Dnmt1, Dnmt3a, and Dnmt3b bound at the Pdx1 promoter region.
ChIP analysis of cross-linked chromatin from islets of IUGR and control animals at 2 weeks and 6 months of age IP with antibody to Dnmt1, Dnmt3a, and Dnmt3b. Control (Con) and IUGR samples were run on the same gel. Input DNA (In) represents PCR products without prior IP. The IgG IP showed negligible PCR product, indicating little or no IP in the absence of primary antibody. The relative amount of Dnmt1-, Dnmt3a-, and Dnmt3b-bound Pdx1 promoter was measured by genomic quantitative real-time PCR (Q-PCR) and normalized to input DNA. Data are represented as percent of control values. n = 3 experiments, data are ± SEM; *P < 0.05 versus controls.
Characterization of the histone code at the Pdx1 promoter.
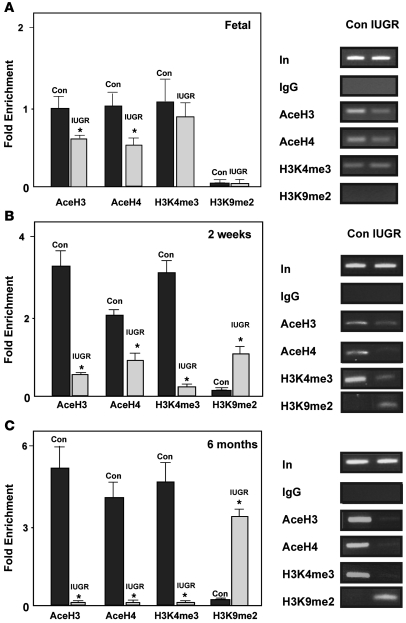
Whereas DNA methylation is associated with transcriptional silencing, histone acetylation is associated with active chromatin, and deacetylation results in repressed chromatin structure (15). The impact of IUGR on the acetylation status of core histones H3 and H4, which are physically associated with the Pdx1 promoter, was determined by ChIP analyses using primers that encompass the region of the CpG island at Pdx1, shown in Figure 1A. Similar to findings in a β cell tumor line (17), this region of Pdx1 was heavily acetylated at H3 and H4 in islets from control animals (Figure 3). IUGR significantly reduced the abundance of acetylated H3 and H4 and deacetylation progressed with age. By 6 months of age, H3 and H4 acetylation was absent (Figure 3).
Figure 3. Histone acetylation and methylation at the Pdx1 promoter.
ChIP analysis of cross-linked chromatin from islets of IUGR and control animals at fetal day 21 (A), 2 weeks (B), and 6 months of age (C) IP with antibody to acetylated H3 (AceH3), acetylated H4 (AceH4), H3K4me3, and H3K9me2. Input DNA represents PCR products without prior IP. The IgG IP showed negligible PCR product, indicating little or no IP in the absence of primary antibody. The relative amount of acetylated H3–, acetylated H4–, H3K4me3-, and H3K9me2-bound Pdx1 promoter was measured by Q-PCR and normalized to input DNA. Data are represented as percent of control values. n = 3 experiments, data are ± SEM; *P < 0.05 versus controls.
Trimethylation of lysine 4 at H3 (H3K4me3) preferentially marks promoters, is associated with active genes, and is influenced by density of H3 acetylation (22, 23). In islets from control animals at all ages, there was a high association of H3K4me3 at Pdx1. In contrast, despite the marked decrease in histone acetylation in IUGR fetuses, there was no difference in the methylation status of H3K4 between IUGR and control fetal animals. However, in IUGR animals at 2 weeks of age, the abundance of H3K4me3 was significantly reduced, and by 6 months of age, H3K4me3 was absent at the Pdx1 proximal promoter region (Figure 3).
Loss of H3K4 trimethylation can initiate dimethylation of lysine 9 at H3 (H3K9me2), a histone modification that is involved in the establishment and maintenance of silent heterochromatin regions (24). ChIP assays demonstrated that by 2 weeks of age, H3K9me2 at the Pdx1 promoter was present in IUGR islets and H3K9me2 binding increased with age (Figure 3). We did not observe methylation of H3K27, another silencing methyl mark (data not shown) in IUGR or control islets at any age.
Histone modifications inhibit USF-1 binding at Pdx1.
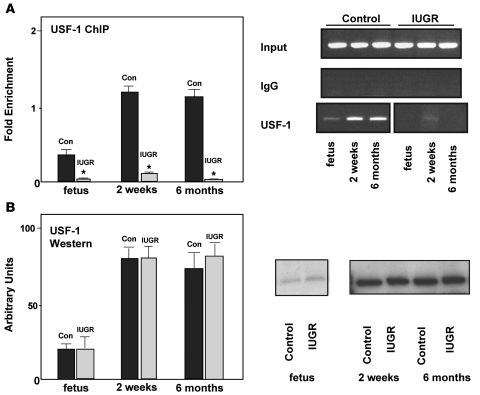
A number of studies have demonstrated that USF-1 binds to and functionally regulates the Pdx1 gene promoter (25, 26). A reduction in endogenous USF-1 binding leads to a significant decrease in Pdx1 promoter activity, which, in turn, results in marked reductions in Pdx1 mRNA and protein levels (25). The USF-1 binding site is highly conserved and is located at a CpG site within the CpG island (Figure 1A). As expected, we observed strong association of USF-1 at Pdx1 in control islets at all ages examined (Figure 4A). In contrast, USF-1 binding was abolished in IUGR islets at all ages. Western blot analyses showed no difference in USF-1 protein abundance between IUGR and controls indicating that decreased USF-1 association with Pdx1 was not due to a decrease in USF-1 protein levels in IUGR islets (Figure 4B).
Figure 4. USF-1 in IUGR and control islets.
(A) ChIP analysis of cross-linked chromatin from islets of IUGR and control animals at fetal day 21, 2 weeks, and 6 months of age IP with antibody to USF-1. Input DNA represents PCR products without prior IP. The IgG IP showed negligible PCR product, indicating little or no IP in the absence of primary antibody. The relative amount of USF-1 bound Pdx1 promoter was measured by Q-PCR and normalized to input DNA. Data are represented as percent of control values. n = 3 experiments, data are ± SEM; *P < 0.05 versus controls. (B) Western blot and densitometric analyses of islets isolated from IUGR and control rats at fetal day 21, 2 weeks, and 6 months of age. Blots were probed with anti–USF-1 and stripped and probed with anti-actin as a control. Data are ± SEM.
Recruitment of transcription repressors to the Pdx1 promoter in IUGR islets.
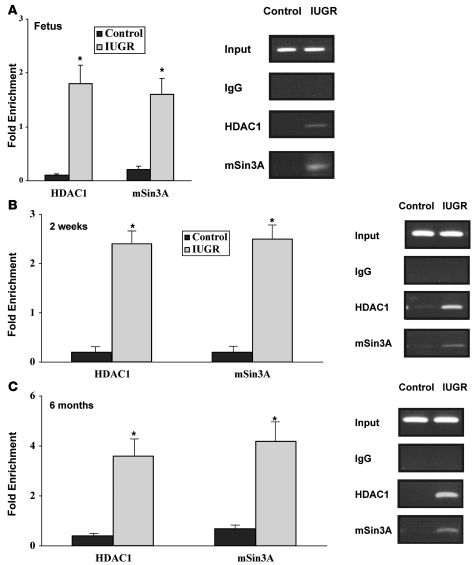
The reduction of histone H3 and H4 acetylation in IUGR islets suggested that HDACs might be involved in facilitating the observed histone modifications. In fetal islets from IUGR animals, there was modest binding of HDAC1, but not mSin3A, at Pdx1. Neither protein was associated with the proximal promoter of Pdx1 in control fetal islets (Figure 5A). At 2 weeks and 6 months of age, we observed a robust association of HDAC1 and mSin3A at Pdx1, which was concomitant to complete deacetylation of H3 and H4 in IUGR islets (Figure 5, B and C). There was no binding of HDAC2 at Pdx1 in IUGR or control islets at any age (data not shown).
Figure 5. Recruitment of repressor complex at Pdx1 in IUGR islets.
ChIP analysis of cross-linked chromatin from islets of IUGR and control animals at fetal day 21 (A), 2 weeks (B), and 6 months of age (C) IP with antibody to HDAC1 and mSin3A. Input DNA represents PCR products without prior IP. The IgG IP showed negligible PCR product, indicating little or no IP in the absence of primary antibody. The relative amount of HDAC1 and mSin3A bound Pdx1 promoter was measured by Q-PCR and normalized to input DNA. Data are represented as percent of control values. n = 3 experiments, data are ± SEM; *P < 0.05 versus controls.
Inhibition of HDAC partially restores Pdx1 expression.
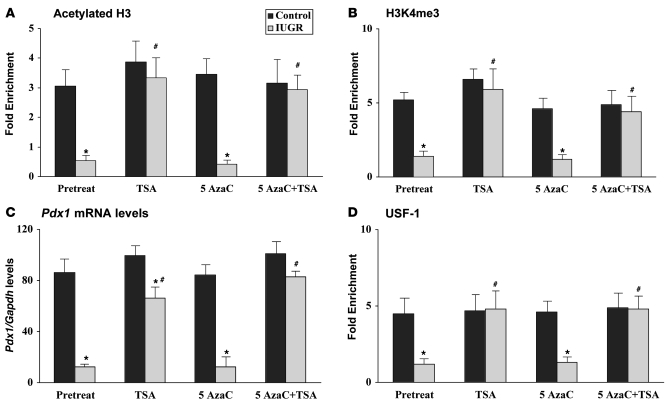
To determine whether either DNA methylation or histone deacetylation is sufficient for silencing of Pdx1 expression in IUGR animals, islets were isolated from 2-week-old IUGR and control animals. At this age (as opposed to later in life), there is significant replication of β cells in rodents making it the ideal age to determine whether stable propagation of histone modifications occurs during cell division and whether these changes can be reversed (27). Islets from IUGR and control animals were cultured for 5 days in the presence of 5-aza-2′-deoxycytidine (5-AzaC) and/or trichostatin A (TSA), which inhibit Dnmts and HDACs, respectively. TSA treatment alone (and in combination with 5-AzaC) normalized acetylation of H3 and restored methylation of H3K4 at the Pdx1 promoter in IUGR islets, further substantiating the dependence of H3K4 methylation upon levels of H3 acetylation (Figure 6, A and B). However, Pdx1 expression (Figure 6C) was largely but not completely restored in IUGR islets, indicating that other relatively minor mechanisms of repression might exist in addition to the recruitment of a histone deacetylation complex. While we did not observe any DNA methylation at the CpG island in IUGR islets, it is possible that other CpGs located in distal enhancer regions could contribute to Pdx1 repression. However, treatment with 5-AzaC alone did not alter Pdx1 mRNA levels in either IUGR or control islets (Figure 6C), suggesting that DNA methylation does not contribute to Pdx1 silencing at this young age.
Figure 6. Reversal of histone modifications partially restores Pdx1 expression in IUGR islets.
The relative amount of acetylated histone H3 (A), H3K4me3 (B), and Pdx1 mRNA (C) levels measured by Q-PCR and normalized to Gapdh. n = 3 experiments, data are ± SEM; *P < 0.05 versus controls; #P < 0.05 versus pretreated IUGR islets. (D) USF-1–bound Pdx1 promoter, as detected by ChIP, was measured by Q-PCR and normalized to input DNA. n = 3 experiments, data are ± SEM; *P < 0.05 versus controls; #P < 0.05 versus pretreated IUGR islets.
Surprisingly, TSA, but not 5-AzaC, treatment resulted in normalization of USF-1 binding to Pdx1 in IUGR islets (Figure 6D). Thus, reversal of histone modifications by TSA normalized association of USF-1 to Pdx1 in the IUGR animal. Neither agent altered USF-1 protein levels (data not shown). USF-1 is a methylation-sensitive transcription factor and the USF site contains an evolutionary, conserved CpG. However, our data suggest that histone modifications, and not DNA methylation, are responsible for loss of USF-1 association with the proximal promoter of Pdx1 in IUGR islets.
Discussion
Pdx1 is a critical regulator of β cell growth and function in both fetal and postnatal developmental stages, and even a relatively modest decrease in expression impairs the compensatory response to insulin resistance (8, 28). Decreased Pdx1 expression plays a pivotal role in the development of diabetes in IUGR animals, as normalization of Pdx1 expression is associated with long-term maintenance of β cell mass and normal glucose homeostasis (6). Uteroplacental insufficiency, the most common cause of IUGR, limits the supply of critical substrates such as oxygen, glucose, and amino acids to the fetus, resulting in an altered redox state and oxidative stress (4, 5). Here we have shown that this altered metabolic milieu decreases Pdx1 transcription by mediating a cascade of epigenetic modifications culminating in silencing of Pdx1. These current studies are the first to our knowledge to characterize the histone code at Pdx1 in vivo in primary islets and link the progression of epigenetic modifications at a key gene to the development of diabetes.
Chromatin modification mechanisms serve a critical function in affecting the transcriptional status of genes. Our data demonstrate that the open chromatin domain marked by histone H3 and H4 acetylation at the proximal promoter of Pdx1 is essential for transcription. Robust Pdx1 expression in islets from control animals is coincident with the presence of acetylated histones H3 and H4 as well as H3K4me3. Loss of these marks results in Pdx1 silencing and reversal of IUGR-induced epigenetic modifications normalizes Pdx1 expression. These data suggest that histone modifications can be stably propagated throughout life.
The first epigenetic mark that is modified in β cells of IUGR animals is histone acetylation (Figure 7). Islets isolated from IUGR fetuses show a significant decrease in H3 and H4 acetylation at the proximal promoter of Pdx1. These changes in H3 and H4 acetylation are associated with a loss of binding of USF-1 to the proximal promoter of Pdx1. USF-1 is a critical activator of Pdx1 transcription, and decreased binding markedly decreases Pdx1 transcription (25, 26). After birth, histone deacetylation progresses and is followed by a marked decrease in H3K4 trimethylation and a significant increase in H3K9me2 in IUGR islets (Figure 7). Progression of these histone modifications parallels the progressive decrease in Pdx1 expression as glucose homeostasis deteriorates and oxidative stress increases in IUGR animals. Nevertheless, in the IUGR pup (at 2 weeks of age), these silencing histone modifications alone suppress Pdx1 expression, since there is no appreciable methylation in the CpG island and reversal of histone deacetylation in IUGR islets (in the presence of active β cell replication) is sufficient to nearly normalize Pdx1 mRNA levels.
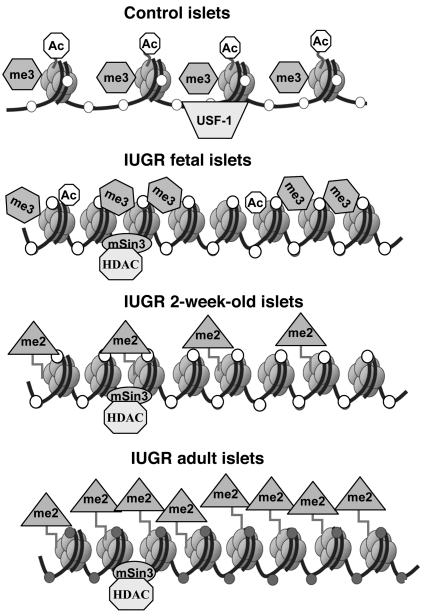
Figure 7. Summary of epigenetic changes at Pdx1 in IUGR rats during the development of type 2 diabetes.
In pancreatic β-cells (top row), the Pdx1 proximal promoter is normally found in an unmethylated (open circles) open chromatin state, allowing access to transcription factors such as USF-1 and associated with nucleosomes characterized by acetylated (Ac, octagons) histones H3 and H4 and with H3K4me3 (Me3, hexagons). In IUGR fetal and 2 week islets (second and third rows), histone acetylation is progressively lost through association with a mSin3A-HDAC1-DNMT1 repressor complex, with H3K4me3 disappearing and H3K9me2 (Me2, triangles) appearing after birth. IUGR adult islets (fourth row) are characterized by inactive chromatin with H3K9me2 and extensive DNA methylation (filled circles) locking in the transcriptionally silent state of Pdx1.
The incomplete restoration of Pdx1 mRNA levels associated with the complete reversal of histone deacetylation in newborn IUGR islets suggests that there may be an additional repressor protein(s) such as SIRT4 or a microRNA like miR9 (both are expressed in islets; refs. 29, 30) that may be involved in chromatin silencing. Identifying such factors will require extensive future experiments. Alternatively, remodeling of active chromatin may only occur in β cells capable of replication, and there may be a minority of cells for which this does not occur, accounting for the extensive but incomplete reactivation of Pdx1 expression.
The initial mechanism by which IUGR silences Pdx1 is by recruitment of corepressors, including HDAC1 and mSin3A, which catalyze histone deacetylation — the first repressive mark observed at Pdx1 in IUGR islets. Binding of these deacetylases in turn facilitated loss of H3K4me3, further repressing Pdx1 expression (Figure 7). Our observation that inhibition of HDAC activity by TSA treatment normalized H3K4me3 levels at Pdx1 in IUGR islets suggests that the association of HDAC1 at Pdx1 in IUGR islets likely serves as a platform for the recruitment of a demethylase, which catalyzes demethylation of H3K4. Lysine demethylases in the Jumonji class remove H3Kme3 and H3Kme2 (reviewed in ref. 31), while LSD1 removes H3Kme1/2 (32). Klose and coworkers have recently demonstrated that the retinoblastoma binding protein, RBP2, contains a JmjC domain, which can specifically demethylate H3K4me3 (33). However, enzymes in the Jumonji class may not catalyze H3K4me3 demethylation in IUGR islets, as activity of these proteins is dependent upon the presence of an iron-binding domain (34, 35), which would be inactivated under conditions of oxidative stress that are present in IUGR islets (5). These results imply that another class of histone demethylases may exist in β cells.
Loss of H3K4me3 was concomitant with a marked increase in H3K9me2 at Pdx1 in 2-week-old IUGR animals, suggesting that K4 methylation precludes methylation at lysine 9. These in vivo findings support several in vitro studies showing that active chromatin states are maintained by H3K4 methylation, which opposes the lysine methylations that characterize inactive chromatin (36, 37). Since restoration of histone acetylation by TSA treatment of IUGR islets reversed H3K9me2, this also demonstrates that histone acetylation prevents methylation of H3K9. Thus, IUGR-induced histone modifications are mutually reinforcing and interdependent.
DNA methylation of a CpG island in the promoter is a key mechanism for silencing gene expression. Most CpG islands remain unmethylated in normal cells; however, under some normal circumstances, such as for imprinted genes, genes on the inactive X chromosome in females, and for some disease processes such as cancer (38) and oxidative stress (39), CpG islands can become methylated de novo. This is particularly relevant to type 2 diabetes, as there are now substantial data that show that oxidative stress plays a significant role in the progression of β cell deterioration (40–44). Further, IUGR induces mitochondrial dysfunction in the β cell leading to increased production of ROS and oxidative stress (5). It is not known why particular CpG islands are susceptible to aberrant methylation. A recent study by Feltus et al. (45) suggests that there is a “sequence signature associated with aberrant methylation.” Of particular relevance to this study is their finding that Pdx1 and a flanking gene, Cdx2 (also encoding a homeobox protein), were 2 of only 15 genes (a total of 1,749 genes with CpG islands were examined) that were methylation susceptible under conditions of increased methylation induced by overexpression of DNMT1.
The molecular mechanism responsible for DNA methylation in IUGR islets is likely to involve H3K9 methylation. A number of studies have shown that methylation of H3K9 precedes DNA methylation (21, 46). It has also been suggested that Dnmts may act only on chromatin that is methylated at lysine 9 on histone H3 (H3K9) (47). Histone methyltransferases bind to the DNA methylases DNMT3A and DNMT3B, thereby initiating DNA methylation (21). Surprisingly, we found that DNMT1 was associated with Pdx1 prior to CpG methylation. Since DNMT1 can be recruited by interaction with a HDAC such as HDAC1 (48), which is already associated with the Pdx1 promoter in fetal IUGR islets, we suggest that this occurs in the IUGR islet prior to the alterations in histone methylation that only occur after birth (Figure 7). As H3K9 methylation occurs during postnatal life in IUGR islets, this would then allow recruitment of the de novo methyltransferase DNMT3A (21) (but not DNMT3B). Subsequent to onset of DNA methylation, DNMT1 would then be positioned to maintain the methylated state, locking in Pdx1 silencing in adult IUGR islets (Figure 7).
In conclusion, our results demonstrate that IUGR induces a self-propagating epigenetic cycle, in which the mSin3A/HDAC complex is first recruited to the Pdx1 promoter, histone tails are subjected to deacetylation, and Pdx1 transcription is repressed. At the neonatal stage, this epigenetic process is reversible and may define an important developmental window for therapeutic approaches. However, as H3K9me2 accumulates, DNMT3A is recruited to the promoter and initiates de novo DNA methylation, which locks in the silenced state in the IUGR adult pancreas, resulting in diabetes. We believe our studies indicate novel mechanisms of epigenetic regulation of gene expression in vivo, which link gene silencing in the β cell to the development of type 2 diabetes and suggest therapeutic agents that we believe are novel for the prevention of common diseases with late-onset phenotypes.
Methods
Animal model.
Surgical methods have been previously described (4, 5, 49). In brief, time-dated Sprague-Dawley pregnant rats were individually housed under standard conditions and allowed free access to standard rat chow and water. On day 18 of gestation (term is 22 days), pregnant rats were anesthetized with intraperitoneal xylazine (8 mg/kg) and ketamine (40 mg/kg), and both uterine arteries were ligated. This results in an approximately 50% reduction in uteroplacental blood flow and induces IUGR (4, 49). Controls underwent sham surgery. Rats recovered within a few hours and had ad libitum access to food and water. The pregnant rats were allowed to deliver spontaneously, and the litter size was randomly reduced to 8 at birth to assure uniformity of litter size between IUGR and control litters. IUGR and control pups were fostered to unoperated normal female rats and remained with their foster mothers until they were weaned. Only male animals were studied to avoid the potentially confounding hormonal variables associated with female rats. While it is possible that the changes that we observed may differ in female rats as gender specific effects on histone acetylation and DNA methylation have been reported (reviewed in refs. 50, 51), we have not observed gender specific effects on β cell function in IUGR rats. Thus, it is unlikely that epigenetic modifications at Pdx1 may differ between male and female animals.
Islets were harvested from 10 litters each of 2-week-old IUGR and control animals, and islets were pooled from a litter. Islets were also harvested from 10 IUGR and 10 control animals at 6 months of age (each animal was from a different litter). In rats, β cells compose 95% of the cellular mass of the islet (52).
These studies were approved by the Animal Care Committee of the Children’s Hospital of Philadelphia and the University of Pennsylvania.
Pdx1 reporter gene analysis.
The generation and characterization of the Pdx1-LacZ transgenic reporter mouse was previously reported (53). A modification of the IUGR procedure was performed on gestation day 17 in maternal pregnant Pdx1-LacZ mice. Pancreatic islets were isolated from transgene-positive offspring at 3 weeks and RNA was isolated for real-time PCR quantification of Pdx1 and LacZ mRNA levels.
RNA isolation and real-time PCR.
Total RNA was extracted from islets using RNAzol B (Tel-Test Inc.). Real-time PCR was carried out using an ABI 7900HT Real-Time PCR system with SYBR Green Master Mix from Applied Biosystems. Results were normalized to glyceraldehyde-3-phosphate dehydrogenase expression (primers are listed in Supplemental Table 1).
Methylation analysis of the Pdx1 CpG island.
Genomic DNA was extracted from islets of 2-week-old (n = 8 litters of 6 pups for each group), 6-month-old IUGR (n = 10), and control (n = 8) rats by phenol-chloroform-isoamyl alcohol and was collected by ethanol precipitation. We denatured 10 μg aliquots of XbaI-digested genomic DNA in 0.3 M NaOH at 37°C. Sulfonation and hydrolytic deamination reactions were then carried out by adding 2.5 M sodium bisulfite (Na2S2O5)/10 mM hydroquinone solution (pH 5.0) to the digested genomic DNA and then incubating at 50°C for 4 hours in the dark. Bisulfite-converted DNA was purified using a QIAquick spin purification kit (QIAGEN) and eluted with tris/EDTA buffer (TE). Desulfonation reactions were performed in 0.3 M NaOH at 37°C for 15 minutes, and DNA was precipitated with 3 M sodium acetate/ethanol and resuspended in water.
The PCR reaction buffer for the pyrosequencing of Pdx1 contained 2.5 mM MgCl2, 0.2 mM each of deoxyribonucleotide triphosphate (dNTP), 0.2 mM each of primer, and 2 U AmpliTaq Gold (Applied Biosystems) in a 25 ml reaction mixture. The PCR amplification was done for 50 cycles with an annealing temperature of 55°C (primers are listed in Supplemental Table 1). All the reactions were constructed as recommended by the manufacturer’s instructions (Biotage AB). The pyrosequencing reaction was performed automatically using a PSQ 96MA system along with a SNP Reagent kit. As a control, plasmid DNA, which is unmethylated, was amplified and then subjected to the same analysis in parallel in order to confirm the completion of the bisulfite reaction.
ChIP assays.
Islets were trypsin digested and suspensions were fixed in phosphate-buffered saline containing 1% formaldehyde and protease inhibitor cocktail. Sonication was performed to yield DNA fragments ranging in size from 200–1,000 bp, followed by centrifugation for 10 minutes. Soluble chromatin (150 μg) was diluted in IP buffer and protease inhibitors. Supernatants were collected and cleared by incubation with protein A–sepharose, sonicated salmon sperm DNA, and 10 μl of preimmune serum. An aliquot of supernatant was collected and used as the input DNA. IP was carried out overnight with IgG (negative control) or primary antibody to the modified histone (acetylated histone H3 and H4, trimethyl-histone H3 [Lys4], acetylated H3K4, and dimethyl-histone H3 [Lys9]), HDAC1 (Upstate Biotech), USF-1, Dnmt1, Dnmt3a, and Dnmt3b (Abcam), and mSin3A (Santa Cruz Biotechnology Inc.). Immune complexes were isolated by incubation with protein A–agarose, and the complexes were then serially washed in low-salt buffer, high-salt buffer, LiCl buffer, and finally TE. Bound complexes were eluted from protein A with a 1% SDS buffer and cross-links were reversed in the presence of Rnase A and extensively digested with proteinase K. The released DNA was extracted, precipitated, and resuspended in water. DNA sequences in the “input” and the IP samples were quantified relative to each other by real-time PCR (primers are listed in Supplemental Table 1).
To determine whether reversal of epigenetic modifications would restore Pdx1 expression in IUGR animals, islets were harvested from 2-week-old IUGR and control pups and cultured for 5 days in RPMI 1640 culture medium containing 11 mM glucose, supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 50 IU/ml penicillin, and 50 mg/ml streptomycin (Gibco BRL) in the presence and absence of 10 μM TSA and/or 1 μM 5-AzaC, which inhibited HDACs and Dnmt1, respectively.
Statistics.
Statistical analyses were performed using analysis of variance and the 2-tailed Student’s unpaired t test. A P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
This study was supported by the NIH grant DK55704 (to R.A. Simmons) and DK062965 (to R.A. Simmons and D.A. Stoffers). We thank Hongshun Niu for his expert technical assistance.
Footnotes
Nonstandard abbreviations used: 5-AzaC, 5-aza-2′-deoxycytidine; Dnmt, DNA methyltransferase; HDAC, histone deacetylase; H3K4, histone 3 lysine 4; H3K9, histone 3 lysine 9; H3K4me3, trimethylation of lysine 4 at H3; H3K9me2, dimethylation of lysine 9 at H3; IUGR, intrauterine growth retardation; TSA, trichostatin A.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:2316–2324 (2008). doi:10.1172/JCI33655
References
- 1.Barker D.J. The fetal origins of type 2 diabetes mellitus. Ann. Intern. Med. 1999;130:322–324. doi: 10.7326/0003-4819-130-4-199902160-00019. [DOI] [PubMed] [Google Scholar]
- 2.Gluckman P.D., Hanson M.A. Living with the past: evolution, development, and patterns of disease. Science. 2004;305:1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- 3.Gluckman P.D., Hanson M.A. Developmental plasticity and human disease: research directions. J. Intern. Med. 2007;261:461–471. doi: 10.1111/j.1365-2796.2007.01802.x. [DOI] [PubMed] [Google Scholar]
- 4.Simmons R.A., Templeton L., Gertz S., Niu H. Intrauterine growth retardation leads to type II diabetes in adulthood in the rat. Diabetes. 2001;50:2279–2286. doi: 10.2337/diabetes.50.10.2279. [DOI] [PubMed] [Google Scholar]
- 5.Simmons R.A., Suponitsky-Kroyter I., Selak M.A. Progressive accumulation of mitochondrial DNA mutations and decline in mitochondrial function lead to beta-cell failure. J. Biol. Chem. 2005;280:28785–28791. doi: 10.1074/jbc.M505695200. [DOI] [PubMed] [Google Scholar]
- 6.Stoffers D.A., Desai B.M., Ng D.D., Simmons R.A. Neonatal exendin-4 prevents the development of diabetes mellitus in the intrauterine growth retarded rat. Diabetes. 2003;52:734–740. doi: 10.2337/diabetes.52.3.734. [DOI] [PubMed] [Google Scholar]
- 7.Brissova M., et al. Reduced PDX-1 expression impairs islet response to insulin resistance and worsens glucose homeostasis. Am. J. Physiol. Endocrinol. Metab. 2005;288:E707–E714. doi: 10.1152/ajpendo.00252.2004. [DOI] [PubMed] [Google Scholar]
- 8.Kulkarni R.N., et al. PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J. Clin. Invest. 2004;114:828–836. doi: 10.1172/JCI21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland A.M., Gonez L.J., Naselli G., MacDonald R.J., Harrison L.C. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54:2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson J., Gonez L.J., Naselli G., MacDonald R.J., Harrison L.C. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 11.Offield M.F., et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. . 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 12.Hui H., Perfetti R. Pancreas duodenum homeobox-1 regulates pancreas development during embryogenesis and islet cell function in adulthood. Eur. J. Endocrinol. 2002;146:129–141. doi: 10.1530/eje.0.1460129. [DOI] [PubMed] [Google Scholar]
- 13.Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoffers D.A., Zinkin N.T., Stanojevic V., Clarke W.L., Habener J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 15.Jaskelioff M., Peterson C.L. Chromatin and transcription: histones continue to make their marks. Nat. Cell Biol. 2003;5:395–399. doi: 10.1038/ncb0503-395. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S., et al. Pancreatic islet expression of the homeobox factor STF-1 relies on an E-box motif that binds USF. J. Biol. Chem. 1996;271:2294–2299. doi: 10.1074/jbc.271.4.2294. [DOI] [PubMed] [Google Scholar]
- 17.Gerrish K., Van Velkinburgh J.C., Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. . Mol. Endocrinol. 2004;18:533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- 18.Gannon M., Gamer L.W., Wright C.V. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. . Dev. Biol. 2001;238:185–201. doi: 10.1006/dbio.2001.0359. [DOI] [PubMed] [Google Scholar]
- 19.Stoffers D.A., Stanojevic V., Habener J.F. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. J. Clin. Invest. 1998;102:232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bird A.P., Wolffe A.P. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 21.Li H., et al. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Biol. Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- 22.Liang G., et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nightingale K.P., et al. Cross talk between histone modifications in response to HDAC inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J. Biol. Chem. 2007;282:4408–4416. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y., Fang J., Bedford M.T., Zhang Y., Xu R.M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 25.Qian J., Kaytor E.N., Towle H.C., Olson L.K. Upstream stimulatory factor regulates Pdx-1 gene expression in differentiated pancreatic β-cells. . Biochem. J. 1999;341:315–322. doi: 10.1042/0264-6021:3410315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S., et al. Pancreatic islet expression of the homeobox factor STF-1 (Pdx-1) relies on an E-box motif that binds USF. . J. Biol. Chem. 1996;271:2294–2299. doi: 10.1074/jbc.271.4.2294. [DOI] [PubMed] [Google Scholar]
- 27.Scaglia L., Cahill C.J., Finegood D.T., Bonner-Weir S. Apoptosis participates in the remodeling of the endocrine pancreas in the neonatal rat. Endocrinology. 1997;138:1736–1741. doi: 10.1210/en.138.4.1736. [DOI] [PubMed] [Google Scholar]
- 28.Del Guerra S., et al. Functional and molecular defects of pancreatic islets in human type 2 diabetes. Diabetes. 2005;54:727–735. doi: 10.2337/diabetes.54.3.727. [DOI] [PubMed] [Google Scholar]
- 29.Plaisance V., et al. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 30.Haigis M.C., et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y., Whetstine J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Forneris F., et al. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J. Biol. Chem. 2006;281:35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- 33.Klose R.J., et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Trewick S.C., Henshaw T.F., Hausinger R.P., Lindahl T., Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 35.Cloos P.A., et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 36.Litt M.D., Simpson M., Gaszner M., Allis C.D., Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 37.Noma K., Grewal S.I. Histone H3 lysine 4 methylation is mediated by Set and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He B., et al. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14133–14138. doi: 10.1073/pnas.2232790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerda S., Weitzman S.A. Influence of oxygen radical injury on DNA methylation. Mutat. Res. 1997;386:141–152. doi: 10.1016/S1383-5742(96)00050-6. [DOI] [PubMed] [Google Scholar]
- 40.Ihara Y., et al. Hyperglycemia causes oxidative stress in pancreatic beta-cells of GK rats, a model of type 2 diabetes. Diabetes. 1999;48:927–932. doi: 10.2337/diabetes.48.4.927. [DOI] [PubMed] [Google Scholar]
- 41.Silva J.P., et al. Impaired insulin secretion and β-cell loss in tissue specific knockout mice with mitochondrial diabetes. Nat. Genet. 2000;26:336–340. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 42.Kaneto H., et al. Activation of the hexosamine pathway leads to deterioration of pancreatic beta-cell function through the induction of oxidative stress. J. Biol. Chem. 2001;276:31099–31104. doi: 10.1074/jbc.M104115200. [DOI] [PubMed] [Google Scholar]
- 43.Sakuraba H., et al. Reduced beta-cell mass and expression of oxidative stress-related DNA damage in the islet of Japanese Type II diabetic patients. Diabetologia. 2002;45:85–96. doi: 10.1007/s125-002-8248-z. [DOI] [PubMed] [Google Scholar]
- 44.Sakai K., et al. Mitochondrial reactive oxygen species reduce insulin secretion by pancreatic β-cells. Biochem. Biophys. Res. Commun. 2003;300:216–222. doi: 10.1016/S0006-291X(02)02832-2. [DOI] [PubMed] [Google Scholar]
- 45.Feltus F.A., Lee E.K., Costello J.F., Plass C., Vertino P.M. Predicting aberrant CpG island methylation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachman K.E., et al. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/S1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- 47.Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/S0959-437X(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 48.Fuks F., Burgers W.A., Brehm A., Hughes-Davies L., Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 49.Ogata E.S., Bussey M., Finley S. Altered gas exchange, limited glucose, branched chain amino acids, and hypoinsulinism retard fetal growth in the rat. Metabolism. 1986;35:950–977. doi: 10.1016/0026-0495(86)90060-0. [DOI] [PubMed] [Google Scholar]
- 50.Kaminsky Z., Sun-Chong W., Petronis A. Complex disease, gender, and epigenetics. Ann. Med. 2006;38:530–544. doi: 10.1080/07853890600989211. [DOI] [PubMed] [Google Scholar]
- 51.Ke X., et al. Uteroplacental insufficiency affects epigenetic determinants of chromatin structure in brains of neonatal and juvenile IUGR rats. Physiol. Genomics. 2006;25:16–28. doi: 10.1152/physiolgenomics.00093.2005. [DOI] [PubMed] [Google Scholar]
- 52.Finegood D.T., Scaglia L., Bonner-Weir S. Dynamics of beta-cell mass in the growing rat pancreas. Estimation with a simple mathematical model. Diabetes. 1995;44:249–256. doi: 10.2337/diabetes.44.3.249. [DOI] [PubMed] [Google Scholar]
- 53.Stoffers D.A., Heller R.S., Miller C.P., Habener J.F. Developmental expression of the homeodomain protein IDX-1 in mice transgenic for an IDX-1 promoter/lacZ transcriptional reporter. Endocrinology. 1999;140:5374–5381. doi: 10.1210/en.140.11.5374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.