Abstract
Prostate cancer cell lines provide ideal in vitro systems for the identification and analysis of prostate tumor suppressors and oncogenes. A detailed characterization of the architecture of prostate cancer cell line genomes would facilitate the study of precise roles of various genes in prostate tumorigenesis in general. To contribute to such a characterization, we used the GeneChip 500K single nucleotide polymorphic (SNP) array for analysis of genotypes and relative DNA copy number changes across the genome of 11 cell lines derived from both normal and cancerous prostate tissues. For comparison purposes, we also examined the alterations observed in the cell lines in tumor/normal pairs of clinical samples from 72 patients. Along with genome-wide maps of DNA copy number changes and loss of heterozygosity for these cell lines, we report previously unreported homozygous deletions and recurrent amplifications in prostate cancers in this study. The homozygous deletions affected a number of biologically important genes, including PPP2R2A and BNIP3L identified in this study and CDKN2A/CDKN2B reported previously. Although most amplified genomic regions tended to be large, amplifications at 8q24.21 were of particular interest because the affected regions are relatively small, are found in multiple cell lines, are located near MYC, an oncogene strongly implicated in prostate tumorigenesis, and are known to harbor SNPs that are associated with inherited susceptibility for prostate cancer. The genomic alterations revealed in this study provide an important catalog of positional information relevant to efforts aimed at deciphering the molecular genetic basis of prostate cancer.
Introduction
Prostate cancer (PCa) cell lines have been widely used not only as homogeneous tumor genomes for the identification of genetic alterations including deletion, amplification, translocation, and single nucleotide mutation but also as in vitro systems for studying the expression and function of various candidate tumor suppressor genes (TSGs) and oncogenes. To fully evaluate the expression and/or function of a specific (group of) gene(s) in a tumor genome, it is important to know the relative status of the overall genetic integrity across the genome because most genes and gene products interact with others to impose a biologic effect resulting in a specific phenotype. Various cytogenetic methods, including G-banding, spectral karyotyping and fluorescent in situ hybridization (FISH), and comparative genomic hybridization (CGH) such as metaphase (or conventional) and BAC/oligo/cDNA/single nucleotide polymorphic (SNP; or array-based) hybridization, have been used in the characterization of various PCa cell lines, primary and metastatic tumors, resulting in the identification of a number of recurrent losses and gains in the PCa tumor genome [1–11].
Some losses and/or gains of genomic DNA may result from a generalized genomic instability combined with long-term culture and associated proliferation of cancer cell lines and may not reflect alterations of genes “driving” the cancer initiation and progression process. However, a number of TSGs and oncogenes, such as PTEN and MYC, have been identified through the analysis of deletion and amplification of genetic material in tumor genomes and have been verified through the functional analysis of the genes in appropriate cell lines or in animal model systems.
Although single-copy losses and one- to two-extra-copy gains of DNA are common, complete genetic loss of both alleles and more extensive amplification of specific DNA sequences have been only rarely observed in PCa. This may be, in part, due to the limited resolution of the methods used in the identification of these copy number alterations. Using cDNA array CGH with a resolution ∼570 kb, Clark et al. [5] identified two novel regions of complete loss at 17q21.31 and 10q23.1 in the PC3 cell line. The sizes of homozygous deletions in general are much smaller than typical hemizygous deletions affecting only one allele, presumably due to the fatal effect of large homozygous losses on the survival of the cancer cells. Therefore, analysis of these homozygous deletions can make a more direct contribution to the discovery of new genes involved in tumorigenesis. As the resolution of these analyses increases, additional homozygous deletions of novel TSG are expected to be revealed.
Analyzing loss of heterozygosity (LOH) in the cancer genome has also contributed to the identification of TSGs. Loss of heterozygosity due to hemizygous deletion may or may not be accompanied by duplication of the remaining allele. An analysis of both copy number changes and genotyping to determine allele-specific alterations is necessary to differentiate these possibilities. Although a number of CGH platforms with various resolutions have been used for mapping most DNA copy number alterations in the genome of PCa cell lines, SNP array is currently the only whole-genome approach for simultaneously mapping copy number alteration and LOH in a single analysis. To date, this type of high-resolution SNP arrays has not been used to analyze DNA copy number changes and LOH simultaneously for prostate cell lines.
In addition to the deletion of TSGs and gain of oncogenes, gene fusion events between androgen-regulated genes and the ETS family of oncogenes have been recently demonstrated in most PCa [12,13]. These fusions result from balanced and more commonly nonbalanced genetic translocations and deletions [13,14]. Using a combined cytogenetic and tiling array BAC CGH with a resolution of ∼78 kb, Watson et al. [10] have demonstrated that a majority of balanced translocations are actually imperfectly balanced and are associated with focal deletions and duplications in PCa cell lines. Therefore, mapping the breakpoints of these translocations, which requires very high resolution, can provide valuable information characterizing gene fusion events in PCa [15].
In this study, we simultaneously analyzed DNA copy number alteration and LOH using the GeneChip 500K SNP array with an average resolution of ∼5.8 kb in 11 prostate cell lines. In this report, we uncover some previously unreported homozygous deletions and amplifications with gains of three or more extra copies of DNA in PCa, along with a comprehensive map of copy number alteration and LOH and the related copy number data of more than 500K SNP probes for each of the cell lines. These data provide important positional information relevant to efforts aimed at deciphering the underlying molecular pathogenesis of PCa.
Materials and Methods
Cell Lines and DNA Isolation
PrECs are nonimmortalized early-passage human normal prostate epithelial cells maintained in serum-free defined medium (i.e., PrEC complete media). The cell and medium were obtained from Lonza Walkerville, Inc. (Walkerville, MD). The human DU145, LNCaP, PC3, and VCaP PCa cell lines were purchased from American Type Culture Collection (Manassas, VA). LNCaP is hypotetraploid, whereas DU145, PC3, LAPC4, and VCaP are near-triploid cell lines (American Type Culture Collection) [6]. BPH1 is a nontumorigenic, SV-40-immortalized, human prostate epithelial cell line generously provided by Simon Hayward [16]. CWR22Rv1 is derived from a human prostatic carcinoma xenograft that was serially propagated in mice after castration-induced regression and relapse of the parental, androgen-dependent CWR22 xenograft and was generously provided by Thomas G. Pretlow [17]. E006AA is a hypertriploid cell line established from primary PCa cells from an African-American patient, which demonstrates androgen-sensitive growth in culture; it was generously provided by Shahriar Koochekpour [18]. 975E/hTERT is a human telomerase reverse transcriptase-immortalized cell line derived from normal prostate cells generated from a radical prostatectomy (RP) specimen of a patient with a family history of PCa [19]. All of the cell lines were cultured in the RPMI-1640 medium with 10% fetal bovine serum or as previously described [19]. PC82 is a hormone-dependent transplantable tumor, which was maintained by serial xenograft transplantation in nude mice [20]; it was generously supplied by Fritz Schroeder [21]. We isolated genomic DNA using a DNA isolation kit from Gentra (Qiagen, Valencia, CA).
Clinical Samples
The clinical samples used in this study include 67 primary tumors and 5 lymph node metastases. All primary tumors were from PCa patients undergoing RP for treatment of clinically localized disease at Johns Hopkins Hospital. For the analysis of somatic DNA copy number alterations in these tumors, we selected cases from which genomic DNA of sufficient quantity (>5 µg) and purity (>70% cancer cells for cancer specimens; no detectable cancer cells for normal samples) could be obtained by macrodissection of matched nonmalignant (hereafter referred to as normal) and cancer-containing areas of prostate tissue as determined by histologic evaluation of hematoxylin and eosin-stained frozen sections of snap-frozen RP specimens. These include 9 with Gleason 6 tumors, 33 with Gleason 7 tumors, 6 with Gleason 8 tumors, 17 with Gleason 9 tumors, 1 with Gleason 10 tumors, and 1 having no Gleason score available.
GeneChip Mapping 500K SNP Assay
The GeneChipMapping 500K set comprisedNsp (∼262,000 SNPs) and Sty (∼238,000 SNPs) arrays. The median physical distance between SNPs is ∼2.5 kb, and the average distance between SNPs is ∼5.8 kb. The 500K SNP arrays were purchased from Affymetrix, Inc., Santa Clara, CA. All of the reagents used for the assay were obtained from manufacturers recommended by Affymetrix. We labeled and hybridized the 500K SNP mapping arrays according to the manufacturer's instructions [15]. We washed and stained the arrays using an Affymetrix Fluidics Station 450 and scanned the arrays using a GeneChip Scanner 3000 7G (Affymetrix, Inc.). The genotype of each SNP was generated by the GeneChip Genotyping analysis software (GTYPE) of Affymetrix.
DNA Copy Number, LOH, and Classification of Deletions and Gains
A database containing probe intensity and genotype data sets generated from the 500K SNP array using more than 200 DNA samples from both blood and normal prostate cells was used as reference samples for DNA copy number analysis in the cell lines. Because the ploidy levels vary among different cell lines, we normalized the data of log2 ratio using most of the chromosomes as the baseline within each of the tumor genomes based on the ploidy levels of the cell lines. DNA copy number and LOH were calculated based on allele intensity using two different software packages: Copy Number Analyzer for Affymetrix GeneChip (CNAG2.0 [22]) and dChip analyzer (dChip [23]). The physical positions of the detected deletions were determined based on the Human hg17 Assembly (NCBI Build 35). Deletions and gains are deduced from the average of log2 ratio with a smoothing window of 10-SNP probes using CNAG2.0 and dChip. The breakpoint SNPs were identified based on the median log2 ratio with a smoothing window of five SNP probes.
To distinguish which allele is deleted or gained in the clinical samples, we used allele-specific analysis as described by Nannya et al. [22]. The algorithm of allele-specific analysis takes advantage of genotype information and allele-specific intensities from paired samples to estimate DNA copy numbers for each heterozygous SNP. Allelespecific analysis can also minimize the effect of normal DNA contamination from nonmalignant cells in the tumors on the identification of DNA copy number changes, because the probe intensity of the other allele can be used as an internal reference for the detection of hemizygous deletion or gain. When the allele intensities of the match nontumor DNA are used as references, the homozygous deletion or gain can also be detected by allele-specific analysis.
We used the criteria described previously to define deletions and gains [24]. Hemizygous deletion refers to the loss of one of the alleles, whereas homozygous (biallelic) deletion refers to the loss of both alleles identified by allele-specific analysis in the clinical samples. In cell lines, homozygous deletions were identified by CNAG2.0 with a copy number of the probes equal to zero in default settings. The absolute log2 ratios of one- and two-copy losses varied depending on the ploidy levels of the cell lines but can be clearly distinguished according to the criteria described previously [24]. For example, in near-triploid cell lines, we used log2 ratios <-0.2 with an average of ∼-0.25 in the 10-SNP genomic smoothed data to define one-copy loss, whereas we used log2 ratios <-0.5 with an average of ∼-0.7 to define twocopy loss and log2 ratios <-0.9 with an average of ∼-1.8 to define three-copy (complete) loss.We used the same method to define gains. For example, in near-triploid cell lines, we used log2 ratios >0.2 with an average of ∼0.24 in the 10-SNP genomic smoothed data to define one extra copy gain, whereas we used log2 ratios >0.4 with an average of ∼0.6 to define two-extra-copy gain and >0.8 with an average of ∼1 to define three-extra-copy gain or more. Where the gains were three or more extra copies, we define these gains as amplifications. We used both quantitative polymerase chain reaction (PCR) and FISH data to validate and refine the criteria for the copy number estimations.
Quantitative PCR
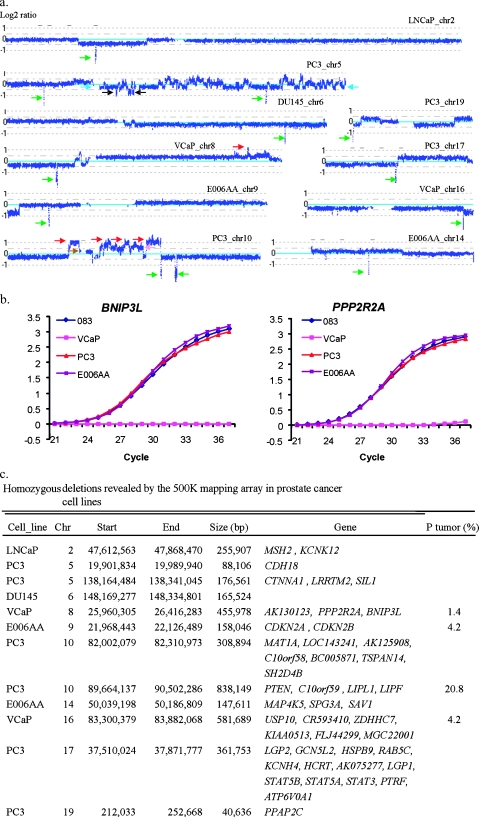
On the basis of the 500K SNP array analysis, we selected two normal DNA samples, PrEC from a primary culture of prostate epithelial cells and 083 from the blood of an unaffected individual, as reference samples for quantitative PCR (qPCR). The reaction was performed using the ABI Prism 7500 Sequence Detection System (Foster City, CA). Primers were designed using PrimerQuest software and were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Amplicons were designed against the putatively homozygous-deleted and -amplified loci in the tumor cells and a control locus of known normalDNAcopy number. The sequences of the primers for BNIP3L, PPP2R2A, CDKN2A, CDKN2B, RAB20, ING1, OTUB1, MARK1, and a locus at 8q24.11 are available on request. Polymerase chain reaction kinetics at the control locus with normal DNA copy number was used to control for sample-to-sample differences in genomic DNA purity and concentration. Polymerase chain reaction conditions and determination of deletion and gain were described previously [15].
Fluorescent In Situ Hybridization
Fluorescent in situ hybridization was performed to analyze the DNA copy number of the androgen receptor gene (AR) in E006AA cells. CEP x (DX21) spectrum green probe (for centromere control) and the AR (Xq12) spectrum orange probe were purchased from Vysis, Inc. (Downers Grove, IL). Cell preparation and hybridization were carried out according to the manufacturer's protocol. Both interphase and metaphase analyses were performed to estimate DNA copy number of AR in relationship to centromere in E006AA and cells from a normal male as control. Fluorescent in situ hybridization analysis was performed using fluorescent microscopy with the appropriate filters to visualize the probes. Slides were blinded, then for each sample, a total of 100 interphase nuclei were scored and a ratio of red signals (AR) to green signals (Xcen) was calculated.
Results
Copy Number Alterations and LOH Revealed by the 500K SNP Array in PCa Cell Lines
DNA copy number alterations in each of the 11 genomes for each chromosome are presented in Figures W1 to WX. The deletions and gains are inferred from the 10-SNP genomic smoothed log2 ratios of the hybridization intensity for each of the 500K probes analyzed (presented in a supplementary data set for each of the cell lines, http://www1.wfubmc.edu/Genomics/Publications+and+Data). Among the cell lines analyzed, PC3 and VCaP harbor the most DNA copy number alterations. Within these two cell lines, chromosome 5 harbors the most changes (Figure W5). In contrast, there were no large DNA copy number aberrations found in PrEC, although some small copy number variations were observed, for example, on chromosomes 14q and 15q (Figures W14 and W15, red arrows) in these presumably normal prostate epithelial cells. We also documented deletions and gains in prostate cell lines that had not been analyzed previously by array CGH (e.g., 975E, E006AA, and PC82; Figures W1–WX).
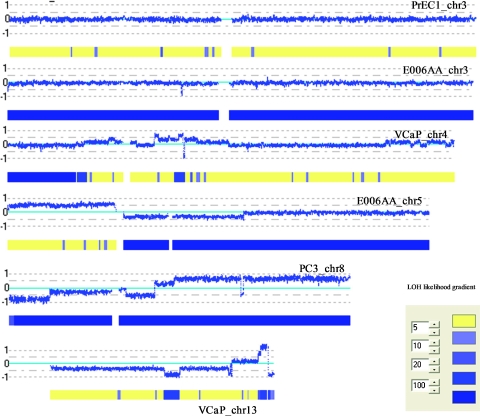
Because the SNP array also generates genotypes at each of the 500K SNPs, it was possible to analyze LOH across the whole genome for each of the cell lines using CNAG2.0 (Figures W1–WX). As would be predicted from similar previous studies, some regions of LOH cover the entire chromosomes in some of the cell lines. These include chromosomes 3, 7, 9, 13, and 14 in E006AA, chromosome 13 in DU145, chromosomes 8, 9, 10, 16, and 22 in PC3, and chromosome 18 in VCaP, reducing hundreds to thousands of genes to homozygosity in these cell lines. Because some LOH events were not accompanied by copy number changes, LOH in these instances apparently occurred along with gain of the remaining chromosomal copy, either completely or partially (Figure 1). In addition, we analyzed the minimum common regions (MCRs) of recurrent LOH and listed the six that occurred in at least four cell lines in Table 1. Among the many genes affected by these recurrent alterations are known and suggested TSGs such as TUSC1, PTEN, and KLF5 and genes involved in cellular senescence (MORF4), DNA repair (HMGB2 and EPC1), and oxidative stress (MGST1).
Figure 1.
Examples of DNA copy number changes and LOH identified by CNAG2.0. Log2 ratios (horizontal dotted lines) are labeled for each of the chromosomes on the left as 1, 0, and -1, with 0 being the baseline (light blue). Dark blue curve indicates 10-SNP genomic smoothed log2 ratios of Nsp probes. Blue bars superimposed on the yellow bars represent LOH likelihood with the likelihood gradients labeled on the left.
Table 1.
Examples of Recurrent LOH Revealed by the 500K Mapping Array in Prostate Cancer Cell Lines.
| Cell Line | Chromosome | Start | End | Size (bp) | Gene |
| 975E, CWR22Rv1, DU145, E006AA, PC3 | 4 | 171,890,559 | 176,165,998 | 4,275,439 | AK128523, SAP30, GALNT7, HMGB2, SCRG1, MORF4, FBXO8, HPGD, GLRA3 |
| DU145, E006AA, PC3, PC82 | 9 | 22,021,005 | 25,996,565 | 3,975,560 | AF109294, DMRTA1, ELAVL2, TUSC1 |
| BPH1, DU145, PC3, PC82 | 10 | 30,398,784 | 34,803,394 | 4,404,610 | 19 genes, MAP3K8, TCF8, EPC1 |
| E006AA, LNCaP, PC3, PC82 | 10 | 88,889,330 | 90,573,072 | 1,683,742 | 11 genes, MINPP1, PTEN, LIPF |
| DU145, LAPC4, LNCaP, PC3 | 12 | 16,235,442 | 17,277,690 | 1,042,248 | MGST1, LMO3 |
| DU145, E006AA, LNCaP, PC82, VCaP | 13 | 72,316,041 | 74,078,996 | 1,762,955 | C13orf24, KLF5, KLF12 |
Homozygous Deletions Implicate New Genes Involved in Prostate Cancer
We used three different approaches in the identification of homozygous deletions, including the value of log2, SNP call (no call), and qPCR. Although, in general, it is not feasible to use default settings of CNAG2.0 to generate calls for a specific copy number alteration (other than homozygous deletion with a copy number equal to zero) in the presence of multiple levels of deletion in polyploid cells, there are obvious differences in log2 ratios between levels of deletion within a given genome when it is analyzed using the 500K SNP array. Taking the deletions on chromosome 5 in near-triploid cell lines PC3, for example (Figure 2a), we were able to clearly identify one-copy deletions (light blue arrows), two-copy deletions (black arrows), and complete or homozygous deletions (green arrows). In the regions of homozygous deletion, we found the no-call rate increased in comparison to the regions without homozygous deletion, consistent with loss of all copies of the specific genomic interval. To validate the homozygous deletion calls, we performed qPCR and confirmed the results as shown in Figure 2b.
Figure 2.
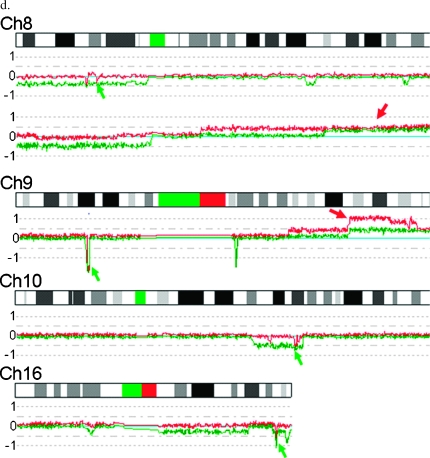
Homozygous deletions identified in PCa cell lines. (a) Log2 ratios (horizontal dot lines) are labeled for each of the chromosomes on the left as 1, 0, and -1, with 0 being the baseline (light blue). Dark blue curve indicates 10-SNP genomic smoothed log2 ratios of the 500K probes. Green arrows indicate homozygous deletion. Red arrow indicates amplifications with three or more extra copies. Light blue arrow indicates one-copy loss. Black arrow indicates two-copy loss. Brown arrow indicates one-extra-copy gain. Pink arrow indicates two-extra-copy gain. (b) Validation of homozygous deletions in PCa cell lines using qPCR. The raw signal intensities for each of the test loci were normalized against the signal intensity of control locus (GAPDH) with normal copy numbers for each of the cycles. The normalized relative signal intensities (Y-axis) are plotted for each of the PCR cycles (X-axis), with 083 from normal blood DNA used as a reference. No signals of PPP2R2A and BNIP3 detected in VCaP indicate complete loss of these two genes. (c) Physical locations and genes involved in homozygous deletions. Chr indicates chromosome; P tumor, clinical sample. (d) Examples of recurrent homozygous deletions (green arrows) and biallelic gains (red arrows) identified in different clinical samples by allele-specific analysis. Log2 ratios of the two alleles (red and green curves) are labeled for each of the chromosomes on the left as 1, 0, and -1, with 0 being the baseline (light blue). G6, G7, G8, and G9 represent Gleason scores of 6, 7, 8, and 9, respectively, for different subjects.
Using the 500K SNParray, we identified 12 homozygous deletions in five cell lines, with 7 apparently being novel (Figure 2, a and c). Half of these deletions occurred in a single cell line, PC3. The genes affected by these novel homozygous deletions include CDH18, AK130123, PPP2R2A, BNIP3L, MAT1A, LOC143241, AK125908, C10orf58, BC005871, TSPAN14, SH2D4B, MAP4K5, SPG3A, SAV1, USP10, CR593410, ZDHHC7, KIAA0513, FLJ44299, MGC22001, and PPAP2C. The cyclin-dependent kinase inhibitors CDKN2A and CDKN2B were found to be deleted in the E006AA line, the only line in this study derived from PCa arising in an African-American. Notably, each homozygous deletion was unique in that it was observed only once among the lines examined, although a subset of these deletions was observed additional times in the clinical specimens examined (see below). All of the homozygous deletions reside in larger regions of LOH (Figures W2, W5, W6, W8, W9, W10, W14, W16, W17, and W19). We confirmed five other regions that have been reported to be completely lost: on chromosome 2 in LNCaP; chromosomes 5, 10, and 17 in PC3; and on chromosome 6 in DU145, including the genes MSH2, KIAA0416, PTEN, SFTPA2, CTNNA1, STAT3, and STAT5B. Furthermore, using the 500K SNP array, we defined boundaries of previously reported homozygous deletions more closely and identified more affected genes, which include KCNK12, LRRTM2, C10orf59, LIPL1, LIPF, LGP2, GCN5L2, HSPB9, RAB5C, KCNH4, HCRT, AK075277, LGP1, STAT5B, STAT5A, PTRF, and ATP6V0A1 (Figure 2c).
Screening for these homozygous deletions in the clinical samples from 72 patients with PCa using allele-specific analysis, we found five recurrent biallelic losses on chromosomes 8, 9, 10, and 16, with examples shown in Figure 2d (green arrows). The most common complete loss of both alleles occurred at chromosome 10q23.31 and included PTEN, with a frequency of 21%, whereas the frequency of homozygous deletions at 9p21.3 and 16q24.1 was 4.2% (Figure 2c). The regions of complete loss at 9p21.3 in the clinical samples overlap with that observed in cell line E006AA, with an MCR affecting only CDKN2A and CDKN2B from 21,968 to 22,125 kb (Figure 2c). The MCR of complete loss at 16q24.1 in the VCaP cell line and clinical samples ranges from 83,300 to 83,541 kb, affecting the genes USP10 and CRISPLD2. We also observed single occurrences of homozygous deletions on 8p in the 72 clinical samples. The complete loss at 8p21.2 in a primary tumor affected AK130123, EBF2, PPP2R2A, and BNIP3L. We did not observe any complete losses at the other regions listed in Figure 2c in the clinical samples analyzed.
Amplifications with More Than Two Extra Copies
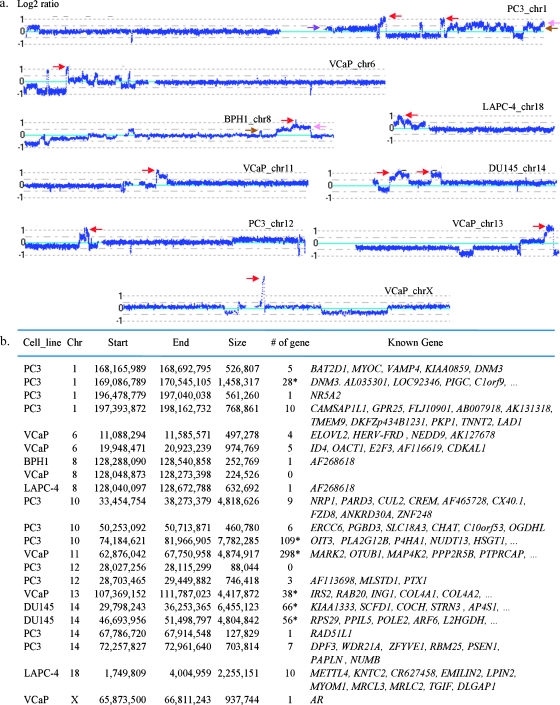
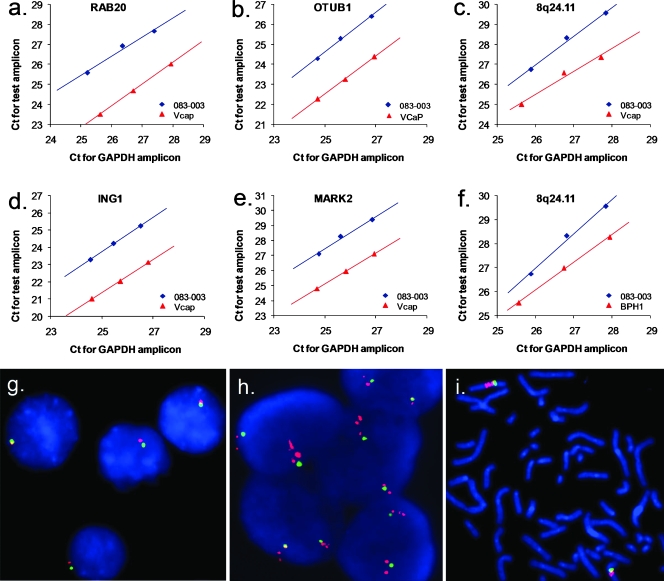
Using the same approach as described above for the analysis of multiple levels of deletion, we were able to define multiple levels of gain, as shown in Figure 3. Taking the gains on chromosomes 1 (Figure 3a) and 10 (Figure 2a) in PC3, for example, we identified gained regions with one, two, and three or more extra copies, marked by brown, pink, and red arrows, respectively, in this near-triploid genome. In comparison to homozygous deletions (Figure 2, a and c), the size of the amplifications, as defined by the presence of three or more extra copies, and ranging from approximately 88 kb to 7.8 Mb, was generally far larger than the homozygous deletions observed and affected a number of genes in various cell lines (Figure 3b). To confirm these amplifications, we selected five different regions on chromosomes 8, 11, and 13 and performed qPCR analysis of DNA copy number using GAPDH as the control locus. For example, in VCaP as shown Figure 4, the results of qPCR analysis revealed gains of multiple copies of RAB20, ING1, OTUB1, and MARK2. In addition, we also validated the amplification of an amplicon at 8q24.21 where several germ line PCa risk alleles have been identified in populations of both European and African descent. Whereas most of the regions with amplification were observed only once in this group of analyzed cell lines, the amplification at 8q24.21 was found in 3 of these 11 cell lines (Figure 3b).
Figure 3.
Examples of amplifications with three or more extra copies identified in PCa cell lines. (a) Log2 ratios (horizontal dot lines) are labeled for each of the chromosomes on the left as 1, 0, and -1, with 0 being the baseline (light blue). Red arrow indicates amplification with three or more extra copies. Brown arrow indicates one-extra-copy gain. Pink arrow indicates two-extra-copy gain. (b) Physical locations and genes involved in amplification. An asterisk (*) represents a known gene item count from the UCSC Table Browser utility (University of California, Santa Cruz, CA).
Figure 4.
Validation of amplifications in PCa cell lines using qPCR and FISH. Amplifications of RAB20 (a), OTUB1 (b), ING1 (d), MARK2 (e), and a locus at 8q24 (c and f) confirmed by qPCR. Cycle numbers (Ct) of the control (GAPDH; X-axis) with normal copy number and test amplicons (Y-axis) for the three dilutions of each DNA sample were plotted against each other and the offset between the reference sample (083-003; blue) and DNA from cell lines (red). Fluorescent in situ hybridization analysis of DNA copy number of AR in a normal male control cell line (g) and in the PCa cell line E006AA (h and i). Green indicates the X centromere probe. Red indicates the AR probe at Xq12. Metaphase analysis of E006AA indicated two copies of X centromere (green) and what seem to be multiple copies of the AR gene (red).
Analyzing the gains in the 72 clinical samples, we did not find the amplifications that we had observed in the cell lines shown in Figure 3b, except at chromosome 8q (Figure 2d). Among these clinical samples, approximately 38% harbor gains on 8q with most of them being hemizygous and affecting the entire q arm. In the allelespecific analysis, we identified three tumors harboring biallele gains, ranging from 98,928 kb to 8qter (Figure 2d, red arrows), which overlap with the amplifications at 8q24. However, the MCR ranging from 128,776 to 136,826 kb in the clinical samples is located 3′ from the MCR in the cell lines and affected a number of genes including MYC.
In addition, we detected an amplification event on chromosome X, ranging from 65,893 to 66,811 kb in VCaP. This alteration harbored only the AR gene and was present in at least five extra copies (Figure 3). Amplification of AR was also observed in the cell line E006AA, although the affected region was larger, ranging from 64,829 to 76,599 kb, covering a number of genes (Figure WX). The gain of AR in E006AA cells was confirmed by FISH where metaphase analysis indicated two copies of the X centromere (Figure 4i, green), consistent with the previously reported karyotype [18] and what seems to be two to three or more copies of the AR gene per X chromosome (Figure 4i, red). Interphase analysis demonstrated multiple red signals in relation to the number of green signals that could be more easily counted than in the metaphase. The number of green signals per nucleus ranged from 1 to 7 and the number of red signals per nucleus ranged from 3 to 18 (Figure 4h), indicating extensive genetic heterogeneity within these cells. On average, we observed 5.5 copies of AR with 2.2 copies of CEP X signals per nucleus, which is consistent with the result from the SNP array analysis (Figure WX). In comparison to E006AA, VCaP apparently harbors a higher level of AR amplification, according to the log2 ratios and metaphase analysis of FISH (Figure WX and data not shown).
Discussion
Alterations of DNA copy number in prostate cell lines, such as LNCaP, PC3, and DU145, have been systematically analyzed using cDNA microarray [5,8] and BAC tiling path array CGH [10]. Although the large-scale genomic alterations uncovered by various platforms are very similar, we find that the deletions and gains revealed by the 500K SNP array are more consistent with the results generated from the higher-resolution tiling path array than those from the cDNA arrays, which contain fewer and less evenly distributed probes. The varied results from study to study are most likely due to these factors, although variations in the cell lines themselves cannot be ruled out. Nevertheless, the comprehensive analysis and documentation of DNA copy number changes in these widely used in vitro model systems provides a detailed genetic background for studying PCa biology.
One of the hallmarks of cancers is genomic instability resulting in aneuploidy, which has been considered as a valuable predictor of clinical outcome [25]. To more specifically characterize the aneuploidy in PCa cell lines, we took advantage of the genotype data from the 500K SNP array and analyzed LOH across the entire genome using CNAG2.0.We found that some entire chromosomes or arms lost heterozygosity, although the DNA copy number either did not change or increased. This type of LOH apparently resulted from the loss of one chromosome or arm in combination with the amplification of the other chromosome or arm. It would be interesting to explore the biologic impact in processes, such as progression of PCa, of large LOH events resulting in the reduction to homozygosity of many hundreds to thousands of genes, with or without accompanying further changes through deletion and gain.
Complete losses of DNA at five chromosomal regions have been reported in PC3 and DU145 [5]. We have validated three of these regions on chromosomes of 5, 10, and 17. It is not surprising that the most frequent homozygous deletion in these 72 clinical samples analyzed in this study was found at 10q23.31, affecting PTEN. These results emphasize the significance of losing PTEN in the development of PCa. Somewhat surprisingly, occurrences of deletions between the TMPRSS2 and ERG genes were less common in these cancer cell lines (Figure W21), particularly considering the high frequency of these deletions in clinical samples of prostate cancer, although the number of the cancer cell lines analyzed is too small to be conclusive. This may suggest a counterselection for this genomic alteration in PCa cells, which can grow and can be established in culture. Furthermore, these results emphasize some of the different selection pressures that exist for PCa cells in patients versus in culture. Although these cell lines are valuable models to study human PCa in general, some critical differences should be borne in mind when extrapolating results from in vitro to in vivo settings.
Particularly interesting genes affected by complete losses identified in PCa cell lines and the clinical samples in this study include BNIP3L and PPP2R2A at 8p21.2, CDKN2A and CDKN2B at 9p21.3, USP10 and CRISPLD2 at 16q24.1, and MAP4K5 at 14q21.3. BNIP3L (BCL2/adenovirus E1B 19-kDa interacting protein) encodes a protein that is a functional homolog of BNIP3 and may play a role in tumor suppression [26]. Fei et al. [27] found that the expression of BNIP3L was induced by p53 under hypoxia, and its knockdown promoted tumor growth in vivo. Down-regulation of BNIP3L has been reported to be correlated with liver metastases and tumor invasion [28] and poor prognosis in hepatocellular carcinoma [29]. PPP2R2A belongs to the phosphatase 2 regulatory subunit B family. Protein phosphatase 2 is one of the four major Ser/Thr phosphatases and is implicated in the negative control of cell growth and division. Studying the fusion of CHEK2 and PPP2R2A in childhood teratoma, Jin et al. [30] reported that the deregulation of CHEK2 and/or PPP2R2A is of pathogenetic importance in at least a subset of germ cell tumors. CDKN2A and CDKN2B are cyclin-dependent kinase inhibitors. CDKN2A, also named as multiple tumor suppressor (MTS1), encodes p16INK4a and p14ARF and acts as a negative regulator of the proliferation of normal cells by interacting strongly with CDK4 and CDK6. Defect in CDKN2A is involved in tumor formation in a wide range of tissues and is the cause of familial atypical multiple mole melanoma-pancreatic carcinoma syndrome. Although hemizygous deletion of CDKN2A has been reported in prostate tumors [31–33], homozygous deletion of this gene in prostate tumors and cancer cell lines was not observed in several studies [33–35]. However, homozygous deletion of CDKN2A has been observed at low frequency in prostate tumors with a high Gleason score [36]. CDKN2B lies adjacent to the TSG CDKN2A. Using a systematic multiplex reverse transcription- polymerase chain reaction, Yamamoto et al. [37] found a decrease in expression of this gene in most breast and PCa cell lines. USP10 may function as a cofactor of the DNA-bound androgen receptor complex and is inhibited by a protein in the Ras-GTPase pathway, whereas CRISPLD2 seems to be highly expressed in prostate. MAP4K5 encodes a member of the serine/threonine protein kinase, which was shown to activate Jun kinase in mammalian cells, suggesting a role in stress response. This kinase is expressed at high levels in the ovary, testis, and prostate, whereas expression is decreased in PCa (ONCOMINE, http://www.oncomine.org/).
Most of the regions with amplification were large, affected a number of genes, and like homozygous deletions, were observed only once in this group of prostate cell lines. It is therefore hard to draw a connection between these amplifications and tumorigenesis because of the large number of genes affected and because most of the affected genes have little obvious relevance to the development of PCa on the basis the functional descriptions in public databases. However, the amplification at 8q24.21 was very interesting because it was found in two PCa cell lines and was located near MYC, an oncogene that is frequently up-regulated in a number of cancers. It is of particular interest that the gained region harbors a number of risk variants, including rs10086908, rs16901979, rs6983267, rs10505477, rs7017300, and rs1447295, which are associated with both prostate and colorectal cancer susceptibility in various human populations [38,39]. This amplification was also observed in BPH1, a nontumorigenic prostate epithelial cell line, indicating that the occurrence of this amplification event by itself is not sufficient for tumorigenesis. Further study is warranted to dissect the function of these DNA sequences in regulating the development of PCa.
The other recurring region of amplification was observed at Xq12, occurring in two of the PCa cell lines analyzed. In both cases, the AR gene was included in the amplified region, either alone or in combination with several other genes. It is well established that the androgen receptor plays a key role in the progression of PCa to hormone independence. A highly localized amplification of AR was apparently associated with the androgen-deprived selection of the PCa cells in the vertebral metastatic deposit from which the VCaP cell line was derived [40]. However, AR amplification was also observed in E006AA that was derived from a clinically localized primary prostate cancer from an African-American patient that apparently had not undergone hormone deprivation treatment. Whether the gain of AR sequences occurred before or after establishment of the E006AA cells in culture is unclear. In any case, these results emphasize that gain of AR sequences is a recurring but by no means a mandatory event associated with prostate cancer progression.
In summary, while documenting DNA copy number changes and LOH at >500,000 locations across the entire genome of 11 prostate cell lines, we have highlighted the following: 1) unreported LOH regions, including frequent monosomy with accompanying gain and loss, 2) novel homozygous deletions that implicate new genes involved in PCa, 3) gains of AR in two cell lines, and 4) recurrent amplification of regions that harbor a number of germ line risk SNPs that have been identified in both prostate and colorectal cancers. Further studies are needed to evaluate whether risk SNPs in the germ line play any role in the amplification of chromosome 8q including the oncogene MYC, and ultimately whether amplification of these SNPs in the somatic genome contributes to the formation of PCa.
Deletion and gain of a number of regions that affect numerous genes in the tumor genomes of PCa have been identified [41]. However, only a few these genes have been investigated in the functional analyses using in vitro and in vivo systems with limited knowledge on their genomic architectures. In this study, we provide the genomic architectures of 11 prostate cell lines at high resolution, which will facilitate further functional analysis of a number of candidate TSGs and oncogenes, including the novel ones identified in this study.
Supplementary Material
Footnotes
The study was partially supported by National Cancer Institute grants CA105055, CA106523, and CA95052 and by a Department of Defense grant PC051264 to J.X. and by a grant from the Dubie H. Holleman Trust for Cancer & Heart Research to W.L.
This article refers to supplementary materials, which are designated by Figures W1–W22 and WX and are available online at www.neoplasia.com.
References
- 1.Bernardino J, Bourgeois CA, Muleris M, Dutrillaux AM, Malfoy B, Dutrillaux B. Characterization of chromosome changes in two human prostatic carcinoma cell lines (PC-3 and DU145) using chromosome painting and comparative genomic hybridization. Cancer Genet Cytogenet. 1997;96:123–128. doi: 10.1016/s0165-4608(96)00258-0. [DOI] [PubMed] [Google Scholar]
- 2.Nupponen NN, Hyytinen ER, Kallioniemi AH, Visakorpi T. Genetic alterations in prostate cancer cell lines detected by comparative genomic hybridization. Cancer Genet Cytogenet. 1998;101:53–57. doi: 10.1016/s0165-4608(97)00060-5. [DOI] [PubMed] [Google Scholar]
- 3.Virgin JB, Hurley PM, Nahhas FA, Bebchuk KG, Mohamed AN, Sakr WA, Bright RK, Cher ML. Isochromosome 8q formation is associated with 8p loss of heterozygosity in a prostate cancer cell line. Prostate. 1999;41:49–57. doi: 10.1002/(sici)1097-0045(19990915)41:1<49::aid-pros7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Aurich-Costa J, Vannier A, Gregoire E, Nowak F, Cherif D. IPM-FISH, a new M-FISH approach using IRS-PCR painting probes: application to the analysis of seven human prostate cell lines. Genes Chromosomes Cancer. 2001;30:143–160. [PubMed] [Google Scholar]
- 5.Clark J, Edwards S, Feber A, Flohr P, John M, Giddings I, Crossland S, Stratton MR, Wooster R, Campbell C, et al. Genome-wide screening for complete genetic loss in prostate cancer by comparative hybridization onto cDNA microarrays. Oncogene. 2003;22:1247–1252. doi: 10.1038/sj.onc.1206247. [DOI] [PubMed] [Google Scholar]
- 6.van Bokhoven A, Caires A, Maria MD, Schulte AP, Lucia MS, Nordeen SK, Miller GJ, Varella-Garcia M. Spectral karyotype (SKY) analysis of human prostate carcinoma cell lines. Prostate. 2003;57:226–244. doi: 10.1002/pros.10291. [DOI] [PubMed] [Google Scholar]
- 7.Lieberfarb ME, Lin M, Lechpammer M, Li C, Tanenbaum DM, Febbo PG, Wright RL, Shim J, Kantoff PW, Loda M, et al. Genome-wide loss of heterozygosity analysis from laser capture microdissected prostate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res. 2003;63:4781–4785. [PubMed] [Google Scholar]
- 8.Zhao H, Kim Y, Wang P, Lapointe J, Tibshirani R, Pollack JR, Brooks JD. Genome-wide characterization of gene expression variations and DNA copy number changes in prostate cancer cell lines. Prostate. 2005;63:187–197. doi: 10.1002/pros.20158. [DOI] [PubMed] [Google Scholar]
- 9.Paris PL, Hofer MD, Albo G, Kuefer R, Gschwend JE, Hautmann RE, Fridyland J, Simko J, Carroll PR, Rubin MA, et al. Genomic profiling of hormone-naive lymph node metastases in patients with prostate cancer. Neoplasia. 2006;8:1083–1089. doi: 10.1593/neo.06421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson SK, deLeeuw RJ, Horsman DE, Squire JA, Lam WL. Cytogenetically balanced translocations are associated with focal copy number alterations. Hum Genet. 2007;120:795–805. doi: 10.1007/s00439-006-0251-9. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, Kumar-Sinha C, Cao X, Dash A, Wang L, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- 12.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 13.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Chang B, Sauvageot J, Dimitrov L, Gielzak M, Li T, Yan G, Sun J, Sun J, Adams TS, et al. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer. 2006;45:1018–1032. doi: 10.1002/gcc.20369. [DOI] [PubMed] [Google Scholar]
- 15.Liu W, Ewing CM, Chang BL, Li T, Sun J, Turner AR, Dimitrov L, Zhu Y, Sun J, Kim JW, et al. Multiple genomic alterations on 21q22 predict various TMPRSS2/ERG fusion transcripts in human prostate cancers. Genes Chromosomes Cancer. 2007;46:972–980. doi: 10.1002/gcc.20482. [DOI] [PubMed] [Google Scholar]
- 16.Hayward SW, Dahiya R, Cunha GR, Bartek J, Despande N, Narayan P. Establishment and characterization of an immortalized but non-tumorigenic human prostate epithelial cell line: BPH-1. In Vitro. 1995;31A:14–24. doi: 10.1007/BF02631333. [DOI] [PubMed] [Google Scholar]
- 17.Sramkoski R, Pretlow TG, Giaconia J, Pretlow TP, Schwartz S, Sy MS, Marengo S, Rhim J, Zhang D, Jacobberger J. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403–409. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 18.Koochekpour S, Maresh GA, Katner A, Parker-Johnson K, Lee TJ, Hebert FE, Kao YS, Skinner J, Rayford W. Establishment and characterization of a primary androgen-responsive African-American prostate cancer cell line, E006AA. Prostate. 2004;60:141–152. doi: 10.1002/pros.20053. [DOI] [PubMed] [Google Scholar]
- 19.Yasunaga Y, Nakamura K, Ewing MC, Isaacs WB, Hukku B, Rhim JS. A novel human cell culture model for the study of familial prostate cancer. Cancer Res. 2001;61:5969–5973. [PubMed] [Google Scholar]
- 20.Hoehn W, Schroeder FH, Reimann JF, Joebsis AC, Hermanek P. Human prostatic adenocarcinoma: some characteristics of a serially transplantable line in nude mice (PC 82) Prostate. 1980;1:95–104. doi: 10.1002/pros.2990010113. [DOI] [PubMed] [Google Scholar]
- 21.Romijn JC, Verkoelen CF, Schroeder FH. Determination of the growth rate of human prostatic cells in primary culture by a morphometric technique. Cell Biol Int Rep. 1984;8:363–371. doi: 10.1016/0309-1651(84)90138-3. [DOI] [PubMed] [Google Scholar]
- 22.Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, Hangaishi A, Kurokawa M, Chiba S, Bailey DK, Kennedy GC, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 23.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 24.Chang BL, Liu W, Sun J, Dimitrov L, Li T, Turner AR, Zheng SL, Isaacs WB, Xu J. Integration of somatic deletion analysis of prostate cancers and germline linkage analysis of prostate cancer families reveals two small consensus regions for prostate cancer genes at 8p. Cancer Res. 2007;67:4098–4103. doi: 10.1158/0008-5472.CAN-06-4570. [DOI] [PubMed] [Google Scholar]
- 25.Borre M, Hèyer M, Nerstrèm B, Overgaard J. DNA ploidy and survival of patients with clinically localized prostate cancer treated without intent to cure. Prostate. 1998;36:244–249. doi: 10.1002/(sici)1097-0045(19980901)36:4<244::aid-pros5>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 26.Mellor HR, Harris AL. The role of the hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer. Cancer Metastasis Rev. 2007;26:553–566. doi: 10.1007/s10555-007-9080-0. [DOI] [PubMed] [Google Scholar]
- 27.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Niedergethmann M, Alves F, Neff JK, Heidrich B, Aramin N, Li L, Pilarsky C, Grutzmann R, Allgayer H, Post S, et al. Gene expression profiling of liver metastases and tumour invasion in pancreatic cancer using an orthotopic SCID mouse model. Br J Cancer. 2007;97:1432–1440. doi: 10.1038/sj.bjc.6604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin Y, Mertens F, Kullendorff CM, Panagopoulos I. Fusion of the tumor-suppressor gene CHEK2 and the gene for the regulatory subunit B of protein phosphatase 2 PPP2R2A in childhood teratoma. Neoplasia. 2006;8:413–418. doi: 10.1593/neo.06139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarrard DF, Bova GS, Ewing CM, Pin SS, Nguyen SH, Baylin SB, Cairns P, Sidransky D, Herman JG, Isaacs WB. Deletional, mutational, and methylation analyses of CDKN2 (p16/MTS1) in primary and metastatic prostate cancer. Genes Chromosomes Cancer. 1997;19:90–96. [PubMed] [Google Scholar]
- 32.Mangold KA, Takahashi H, Brandigi C, Wada T, Wakui S, Furusato M, Boyd J, Chandler FW, Allsbrook WC., Jr p16 (CDKN2/MTS1) gene deletions are rare in prostatic carcinomas in the United States and Japan. J Urol. 1997;157:1117–1120. [PubMed] [Google Scholar]
- 33.Heidenreich B, Heidenreich A, Sesterhenn A, Srivastava S, Moul JW, Sesterhenn IA. Aneuploidy of chromosome 9 and the tumor suppressor genes p16(INK4) and p15(INK4B) detected by in situ hybridization in locally advanced prostate cancer. Eur Urol. 2000;38:475–482. doi: 10.1159/000020327. [DOI] [PubMed] [Google Scholar]
- 34.Komiya A, Suzuki H, Aida S, Yatani R, Shimazaki J. Mutational analysis of CDKN2 (CDK4I/MTS1) gene in tissues and cell lines of human prostate cancer. Jpn J Cancer Res. 1995;86:622–625. doi: 10.1111/j.1349-7006.1995.tb02443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W, Weghorst CM, Sabourin CL, Wang Y, Wang D, Bostwick DG, Stoner GD. Absence of p16/MTS1 gene mutations in human prostate cancer. Carcinogenesis. 1996;17:2603–2607. doi: 10.1093/carcin/17.12.2603. [DOI] [PubMed] [Google Scholar]
- 36.Konishi N, Nakamura M, Kishi M, Nishimine M, Ishida E, Shimada K. Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am J Pathol. 2002;160:1207–1214. doi: 10.1016/S0002-9440(10)62547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto M, Metoki R, Yamamoto F. Systematic multiplex polymerase chain reaction and reverse transcription-polymerase chain reaction analyses of changes in copy number and expression of proto-oncogenes and tumor suppressor genes in cancer tissues and cell lines. Electrophoresis. 2004;25:3349–3356. doi: 10.1002/elps.200406090. [DOI] [PubMed] [Google Scholar]
- 38.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 39.Zheng SL, Sun J, Cheng Y, Li G, Hsu FC, Zhu Y, Chang BL, Liu W, Kim JW, Turner AR, et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 40.Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, Lin DL, Pienta KJ. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 41.Sun J, Liu W, Adams TS, Sun J, Li X, Turner AR, Chang B, Kim JW, Zheng SL, Isaacs WB, et al. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.