Abstract
The twin-arginine translocation (Tat) pathway is a system used by some bacteria to export proteins out from the cytosol to the cell surface or extracellular environment. A functional Tat pathway exists in the important human pathogen Mycobacterium tuberculosis. Identification of the substrates exported by the Tat pathway can help define the role that this pathway plays in the physiology and pathogenesis of M. tuberculosis. Here we used a reporter of Tat export, a truncated β-lactamase, ′BlaC, to experimentally identify M. tuberculosis proteins with functional Tat signal sequences. Of the 13 proteins identified, one lacks the hallmark of a Tat-exported substrate, the twin-arginine dipeptide, and another is not predicted by in silico analysis of the annotated M. tuberculosis genome. Full-length versions of a subset of these proteins were tested to determine if the native proteins are Tat exported. For three proteins, expression in a Δtat mutant of Mycobacterium smegmatis revealed a defect in precursor processing compared to expression in the wild type, indicating Tat export of the full-length proteins. Conversely, two proteins showed no obvious Tat export in M. smegmatis. One of this latter group of proteins was the M. tuberculosis virulence factor phospholipase C (PlcB). Importantly, when tested in M. tuberculosis a different result was obtained and PlcB was exported in a twin-arginine-dependent manner. This suggests the existence of an M. tuberculosis-specific factor(s) for Tat export of a proven virulence protein. It also emphasizes the importance of domains beyond the Tat signal sequence and bacterium-specific factors in determining if a given protein is Tat exported.
In bacteria, protein export across the cytoplasmic membrane represents the first step in the delivery of proteins to the cell envelope or extracellular space. Two conserved systems are responsible for the majority of this protein export: the general secretion (Sec) and the twin-arginine translocation (Tat) pathways (for reviews, see references 31 and 35). Both systems export proteins that are synthesized as precursors with amino-terminal signal sequences. In both cases, the signal sequences are comprised of a tripartite structure: a charged amino-terminal region, a hydrophobic region, and a carboxy-terminal region containing a signal peptidase cleavage site (31, 62). With most exported proteins, the signal sequence is cleaved from the precursor during or immediately after translocation, which liberates the mature exported substrate.
A feature that distinguishes Tat signal sequences from Sec signal sequences is the presence of a consensus twin-arginine motif, which is defined as S/T-R-R-x-F-L-K (5). The arginine dipeptide, RR, in the motif is a major _targeting determinant of the signal sequence as shown by conservative replacement of “RR” with a lysine pair, KK, preventing Tat export of proteins (13, 14, 25, 54). Computational Tat signal sequence prediction programs, based on sequence and structural conservation, have been developed (4, 45, 52). These programs are valuable for identifying Tat substrates that adhere to the consensus motif; however, they cannot account for species-specific differences, unless modified to do so, and it remains to be established how useful they are for comprehensive identification of Tat-exported proteins. There are two Tat-exported proteins known to exist in nature that lack the twin arginines (26, 27). These exceptions may be members of a larger group of yet-to-be identified Tat proteins that rely on features other than the twin-arginine motif for Tat export.
Another distinguishing feature of the Tat export pathway is that Tat substrates are translocated across the membrane in a folded state, with folding being a prerequisite for Tat export (15). Some Tat substrates require cytoplasmic chaperones for export. These chaperones may be specific to one Tat substrate, or they can have a more general effect (23, 28, 38-40). In these cases, it is thought that chaperones function in folding substrates or _targeting them to the membrane-localized Tat translocase complex once folding is complete. The Tat translocase is composed of TatA, TatB, and TatC proteins, although not all bacteria with functional Tat export systems have TatB.
The Tat pathway is present in many, but not all, bacteria. In several bacterial pathogens, the Tat pathway plays an important role in exporting virulence factors (9, 10, 17, 30, 36, 42, 46, 60). Mycobacterium tuberculosis is the bacterial pathogen responsible for tuberculosis, which kills 1.8 million people a year (64). Mycobacteria have a functional Tat pathway. In the fast-growing nonpathogenic species Mycobacterium smegmatis, mutants lacking tatA, tatB, or tatC genes have multiple phenotypes including slow growth on agar and sensitivity to β-lactam antibiotics (34, 41). The latter phenotype is attributed to a failure to export the chromosomally encoded β-lactamase. In both M. smegmatis and M. tuberculosis the endogenous β-lactamases possess Tat signal sequences. β-Lactamases, which destroy cell wall-_targeting β-lactam antibiotics, must be exported to protect bacteria from the drugs. In M. tuberculosis it has not been possible to construct tat mutants (47). This indicates that, in pathogenic M. tuberculosis, the Tat pathway is essential under standard laboratory conditions. Without an M. tuberculosis tat mutant, there are fewer approaches available for identifying Tat-exported proteins and studying the significance of Tat export in this pathogen.
In this study, we used the M. tuberculosis β-lactamase (BlaC) as a reporter to identify M. tuberculosis proteins that possess functional Tat signal sequences. A truncated ′BlaC, lacking its endogenous signal sequence, is not exported and is unable to protect a mycobacterial β-lactam-sensitive mutant (M. smegmatis ΔblaS or M. tuberculosis ΔblaC strain) from the β-lactam antibiotic carbenicillin (34). When a signal sequence from a Tat-exported M. tuberculosis protein is fused to ′BlaC, the hybrid protein is exported and confers carbenicillin resistance on Δbla mutant mycobacteria. Exported ′BlaC fusion proteins can be identified by direct selection of drug-resistant colonies on agar containing carbenicillin. Importantly, the ′BlaC reporter is Tat specific. It works only when fused to Tat signal sequences and requires both the twin-arginine motif and a functional Tat pathway (34).
Using an M. tuberculosis genomic library constructed upstream of the ′blaC reporter, we identified signal sequences capable of exporting ′BlaC in a Tat-dependent manner. In addition to the demonstrated virulence factor phospholipase C (PlcB) (29, 43), we identified proteins with potential roles in carbohydrate and lipid metabolism, copper homeostasis, cell envelope maintenance, and nutrient import. The proteins identified included one lacking a twin-arginine dipeptide and one not predicted by in silico analysis. We also investigated full-length versions of a subset of the proteins identified. Importantly, full-length PlcB was exported and was twin arginine dependent when expressed in its native host, M. tuberculosis. However, when expressed in M. smegmatis this M. tuberculosis protein did not appear to be exported. This suggests the existence of an M. tuberculosis-specific factor(s) that is required for Tat export of a proven virulence protein.
MATERIALS AND METHODS
Bacterial strains and culture methods.
Bacterial strains used during this work are listed in Table 1. Luria-Bertani (LB) medium (Fisher) was used for culturing of Escherichia coli. Middlebrook 7H9 or 7H10 medium (Difco; BD Biosciences) was used for the culturing of M. smegmatis and M. tuberculosis. For M. smegmatis, Middlebrook medium was supplemented with 0.5% glycerol and 0.2% dextrose. For M. tuberculosis, Middlebrook medium was supplemented with 0.5% glycerol and 1× ADS (0.5% bovine serum albumin, fraction V [Roche]; 0.2% dextrose; and 0.85% NaCl). When necessary, medium was supplemented with 0.05 to 0.1% Tween 80 (Fisher). As required, antibiotics were added to Middlebrook media at the indicated concentrations: hygromycin B (Roche Applied Science), 50 μg/ml; carbenicillin (Sigma), 50 μg/ml; and kanamycin (Acros Chemicals), 20 μg/ml. l-Lysine at 40 and 80 μg/ml was added to agar and liquid media, respectively, for growth of M. smegmatis strains PM759 and JM578.
TABLE 1.
Strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| E. coli | ||
| K-12 DH5α | F− [φ80dΔlacZM15] Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 glnV44 thi-1 gyrA96 relA1 | Gibco-BRL |
| TOP10 | F−mcrA [φ80dΔlacZM15] Δ(mrr-hsdRMS-mcrBC) ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK rpsL nupG | Invitrogen |
| XL1-Blue | [F′ proAB lacIqZΔM15 Tn10(Tetr)] recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 λ−lac | Stratagene |
| M. smegmatis | ||
| mc2155 | ept-1 | 53 |
| MB692 | mc2155, ΔtatA | 34 |
| JM567 | mc2155, ΔtatC | 34 |
| PM759 | mc2155, ΔblaS1 ΔlysA4 rpsL6 | 18 |
| JM578 | PM759, ΔtatA | 34 |
| M. tuberculosis | ||
| H37Rv | Virulent | 1 |
| PM638 | H37Rv, ΔblaC1 | 18 |
Molecular biology procedures.
Standard molecular biology techniques were employed (48). The Expand High-Fidelity PCR system (Roche) was used in all PCRs, and 5.0% dimethyl sulfoxide was included in select reactions. DNA sequencing was performed either by the UNC-CH Automated DNA Sequencing Facility (Chapel Hill, NC) or by Eton Bioscience Inc. (San Diego, CA).
Construction of ′BlaC reporter libraries.
All plasmids used in this study are listed in Table 2. Two library plasmids were constructed with a truncated version of M. tuberculosis blaC (referred to as ′blaC), amplified from pJM106 by PCR using the primers Lib′blaCfor and Lib′blaCrev (see Table S1 in the supplemental material).
TABLE 2.
Plasmids used in this study
| Plasmid | Genotype | Description | Source or reference |
|---|---|---|---|
| pCR2.1 | bla aph ColE1 | TA cloning vector | Invitrogen |
| pMV206.hyg | hyg oriM ColE1 | Multicopy mycobacterial shuttle plasmid | 55 |
| pMV261.kan | aph Phsp60oriM ColE1 | Multicopy mycobacterial shuttle plasmid | 55 |
| pMV306.kan | aph int attP ColE1 | Single-copy mycobacterial shuttle plasmid | 55 |
| pJSC77 | aph Phsp60-HA oriM ColE1 | HA tag cloned into pMV261 | 21 |
| pJM106 | cat ′blaC (M. tuberculosis) oriV ori2 | Predicted M. tuberculosis ′blaC mature sequence cloned into pCC1 (Epicentre) | 34 |
| pJES112 | bla aph ColE1 ′blaC (M. tuberculosis) | M. tuberculosis ′blaC mature sequence cloned into pCR2.1 | This work |
| pJES113 | hyg oriM ColE1 ′blaC (M. tuberculosis) | M. tuberculosis ′blaC mature sequence cloned into pMV206.hyg | This work |
| pJM153 | bla aph ColE1Phsp60 (M. tuberculosis) | M. tuberculosis hsp60 promoter cloned into pCR2.1 | This work |
| pJM157 | hyg oriM ColE1 Phsp60 ′blaC (M. tuberculosis) | M. tuberculosis hsp60 promoter cloned into pJES113 | This work |
| Y49 pTrcHisB-BlaC | bla Ptrc-blaC (M. tuberculosis) ColE1 | E. coli expression vector for M. tuberculosis blaC | 59 |
| pJSS51 | bla aph ColE1 ′plcB (M. tuberculosis) | M. tuberculosis plcB (no signal sequence) cloned into pCR2.1 | This work |
| pJM171 | aph oriM ColE1 Phsp60 ′plcB-HA | 1.4-kbp MscI/HindIII fragment containing plcB (no signal sequence) from pJSS51 cloned into pJSC77 | This work |
| pJM210 | bla aph ColE1 Rv0774c (M. tuberculosis) | M. tuberculosis full-length Rv0774c cloned into pCR2.1 | This work |
| pJM211 | aph oriM ColE1 Phsp60 Rv0774c-HA | 913-bp MscI/HindIII fragment containing full-length Rv0774c from pJM210 cloned into pJSC77 | This work |
| pJES146 | bla aph ColE1 | M. tuberculosis Rv0774c (no signal sequence) cloned into pCR2.1 | This work |
| PJES147 | aph Phsp60 ′Rv0774c-HA oriM ColE1 | 797-bp MscI/HindIII fragment containing Rv0774c (no signal sequence) from pJES146 cloned into pJSC77 | This work |
| pJM212 | bla aph ColE1 Rv2843 (M. tuberculosis) | M. tuberculosis full-length Rv2843 cloned into pCR2.1 | This work |
| pJM214 | aph oriM ColE1 Phsp60 Rv2843-HA | 547-bp NruI/HindIII fragment containing full-length Rv2843 from pJM212 cloned into pJSC77 | This work |
| pJM196 | bla aph ColE1 PnativeplcB (M. tuberculosis) | M. tuberculosis full-length plcB and promoter cloned into pCR2.1 | This work |
| pJM199 | aph oriM ColE1 PnativeplcB-HA | 2.2-kbp XbaI/HindIII fragment containing full-length plcB and promoter from pJM196 cloned into pJSC77 | This work |
| pJM202 | bla aph ColE1 PnativeplcA plcB (M. tuberculosis) | M. tuberculosis full-length plcA, plcB, and promoter cloned into pCR2.1 | This work |
| pJM203 | aph oriM ColE1 PnativeplcA plcB-HA | 3.5-kbp XbaI/HindIII fragment containing full-length plcA, plcB, and promoter from pJM202 cloned into pJSC77 | This work |
| pJM197 | bla aph ColE1 Pnative Rv0315 (M. tuberculosis) | M. tuberculosis full-length Rv0315 and promoter cloned into pCR2.1 | This work |
| pJM206 | aph oriM ColE1 Pnative Rv0315-HA | 1.2-kbp XbaI/HindIII fragment containing full-length Rv0315 and promoter from pJM197 cloned into pJSC77 | This work |
| pJM216 | aph oriM ColEI PplcBplcB(KK)-HA | Overlapping, self-ligated PCR product amplified from pJM199; contains plcB-KK under the control of the native promoter | This work |
| pMP327 | aph PblaC-blaC (M. tuberculosis) oriM ColE1 | M. tuberculosis full-length blaC cloned into pMV261 | 34 |
| pJM113 | aph Phsp60-′blaC (M. tuberculosis) oriM ColE1 | ′blaC from pJM106 cloned into MscI-linearized pMV261 | 34 |
| pJM117 | aph PblaC-blaC(KK) (M. tuberculosis) oriM ColE1 | M. tuberculosis full-length blaC(KK) cloned into pMV261 | 34 |
(i) Library 1.
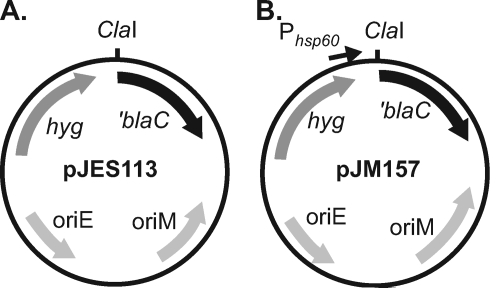
The resulting ′blaC amplicon was ligated into the multicopy mycobacterial shuttle vector pMV206.hyg, which had been digested with ClaI and NcoI. The final plasmid was pJES113 (Fig. 1A). Genomic DNA was isolated from M. tuberculosis strain H37Rv as previously described (6) and partially digested with AciI and HpaII, and digests with DNA fragments between 0.5 and 5.0 kbp were selected. The genomic digest was cloned into the unique ClaI site immediately upstream of ′blaC in pJES113. The resulting ligation reaction mixture was transformed into E. coli XL1-Blue (Stratagene). Approximately 1 × 106 hygromycin-resistant E. coli transformants were pooled for plasmid DNA isolation (Qiagen).
FIG. 1.
Plasmids used for construction of M. tuberculosis-′blaC fusion libraries. (A) Library 1 was constructed by ligating fragments of M. tuberculosis H37Rv genomic DNA into the unique ClaI site located upstream of ′blaC in pJES113. The truncated β-lactamase gene (′blaC) lacks a promoter, Shine-Dalgarno sequence, start codon, and signal sequence for export. (B) Library 2 was constructed with genomic DNA of M. tuberculosis PM638 (ΔblaC) ligated into the unique ClaI site located upstream of ′blaC in pJM157. The pJM157 plasmid additionally has the mycobacterial hsp60 promoter located upstream of the ClaI site but lacks a Shine-Dalgarno site and start codon for ′blaC. Both library plasmids carry a selectable hygromycin resistance gene (hyg) and have origins of replication for mycobacteria (oriM) and for E. coli (oriE).
(ii) Library 2.
The second library vector, pJM157, was constructed to carry a mycobacterial promoter upstream of the unique ClaI site. A fragment carrying the promoter and the +1 transcriptional start site from M. tuberculosis hsp60 was amplified by PCR from pMV261.hyg using the primers Hsp60for-BstBI and Hsp60rev2-ClaI (see Table S1 in the supplemental material). The resulting PCR product was ligated into pJES113, which had been linearized with ClaI, to produce pJM157 (Fig. 1B). For construction of library 2, genomic DNA was isolated from M. tuberculosis strain PM638 (ΔblaC). Digestion of genomic DNA and ligation into the single ClaI site upstream of ′blaC in pJM157 were conducted as described above. After electroporation into E. coli DH5α (Invitrogen), approximately 8 × 105 CFU were pooled and used to isolate plasmid DNA (Qiagen).
Selection of exported ′BlaC fusions.
Library DNA was electroporated into M. smegmatis ΔblaS strain PM759 (6, 18). The resulting transformants were plated onto 7H10 agar medium without Tween, containing 40 μg/ml lysine (Fischer Scientific), 50 μg/ml hygromycin, and carbenicillin at concentrations that ranged from 35 to 75 μg/ml, and incubated at 37°C for a minimum of 4 days. The drug resistance of colonies that grew up on 7H10 agar with hygromycin and carbenicillin was confirmed by spot test analysis on 7H10 agar plates containing (i) both 50 μg/ml hygromycin and 45 μg/ml carbenicillin and (ii) 50 μg/ml hygromycin only. Strains were further analyzed if spots revealed confluent growth on plates containing hygromycin and carbenicillin in comparison to the negative-control strain (ΔblaS M. smegmatis with plasmid pJM113, which carries a promoterless ′blaC gene) (34).
Recovery of ′blaC fusion plasmids.
Plasmid DNA was transferred from carbenicillin- and hygromycin-resistant ΔblaS M. smegmatis to E. coli DH5α by electroduction (2). A small amount of M. smegmatis was transferred from a colony into 20 μl of ice-cold 10% glycerol. The suspension was mixed by vortexing, incubated on ice for 10 min, and added to 40 μl of electrocompetent E. coli DH5α. The mixture was transferred to a chilled 0.2-cm-gap cuvette and pulsed using conditions typical for E. coli electroporation (25 μF, 200 Ω, and 2.5 kV). Immediately, 1 ml of LB broth was added to the cuvette, incubated at 37°C for 1 h, and then plated on LB with hygromycin. Plasmid DNA was purified from hygromycin-resistant E. coli (Qiagen). The M. tuberculosis genomic DNA insert upstream of ′blaC was identified by sequencing with the primer TnBlaCout (see Table S1 in the supplemental material).
Anti-BlaC antibody.
A six-histidine-tagged copy of M. tuberculosis BlaC was expressed from Y49 pTrcHisB-BlaC in E. coli DH5α (provided by Doug Kernodle) and purified by nickel affinity chromatography using HIS-Select nickel affinity gel (Sigma) as described previously (59). Purified BlaC was eluted from the nickel column with 300 mM imidazole at a concentration of 1.3 mg/ml and used to immunize rabbits together with TiterMax Gold adjuvant (Sigma). Rabbit immunizations and polyclonal antiserum collection were carried out by the BioSource Custom Immunology Department (Hopkinton, MA).
Immunoblot analysis.
Whole-cell lysates of M. smegmatis and whole-cell lysates of formalin-killed M. tuberculosis were prepared as described previously (7, 19, 33) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. Polyclonal BlaC antiserum was used in immunoblot analysis at a dilution of 1:10,000, and polyclonal 19-kDa antiserum (provided by Douglas Young) was used at a dilution of 1:40,000. Anti-rabbit peroxidase-conjugated antibodies (Bio-Rad) were used as secondary antibodies for both anti-BlaC and anti-19-kDa antiserum. Both monoclonal hemagglutinin (HA) antiserum (Covance) and monoclonal GroEL HAT5/IT-64 antiserum (World Health Organization collection) were used at a dilution of 1:20,000, and anti-mouse peroxidase-conjugated antibodies were used as secondary antibodies (Bio-Rad).
Construction of specific M. tuberculosis-′BlaC fusions.
The nucleotides encoding the signal sequences of the Rv2041c and Rv2525c genes were PCR amplified using the primers rv2041ssF and 2041ssR or 2525F and 2525R, respectively (see Table S1 in the supplemental material). Both the forward and reverse primers encoded a BstBI site, and the forward primers also included the Shine-Dalgarno sequence from M. tuberculosis hsp60. The amplified fragments were ligated into pCR2.1 (Invitrogen), digested with BstBI, and then ligated into ClaI-digested pJM157, upstream of ′blaC.
Construction of expression constructs for HA-tagged M. tuberculosis proteins. (i) hsp60 promoter-driven HA fusions: Rv0774c-HA, ΔssRv0774c-HA, Rv2843-HA, and ΔssPlcB-HA.
Oligonucleotide primers were designed to amplify full-length or truncated genes from M. tuberculosis genomic DNA. Forward and reverse primers were designed each with 5′ extension sequences carrying MscI and HindIII restriction sites, respectively (see Table S1 in the supplemental material). The resulting PCR product was first cloned into pCR2.1 (Invitrogen) and sequenced. The cloned gene was then digested with MscI and HindIII, and the appropriate fragment was isolated and ligated into the mycobacterial shuttle vector pJSC77 (21), which was digested with MscI and HindIII and which carries the hsp60 promoter and multiple cloning site upstream of a C-terminal HA tag (Table 2). Due to an MscI site within the Rv2843 gene, the strategy was revised in this instance and an NruI site was included instead of MscI on the forward primer (see Table S1 in the supplemental material).
(ii) Native promoter-driven HA fusions: PlcB-HA and Rv0315-HA.
Oligonucleotide primers were designed to amplify a fragment of M. tuberculosis genomic DNA which included the full-length gene of interest and upstream sequence containing the putative native promoter. Forward primers were designed each with 5′ extension sequences carrying XbaI and HindIII restriction sites, respectively (see Table S1 in the supplemental material). The resulting PCR product was first cloned into pCR2.1 (Invitrogen) and sequenced. The cloned gene and promoter were then digested with XbaI and HindIII, and the appropriate fragment was isolated and ligated into the mycobacterial shuttle vector pJSC77, which had been digested with XbaI and HindIII, which removes the hsp60 promoter (Table 2).
(iii) PlcB(KK)-HA.
The construct in which the codons for the twin-arginine pair of PlcB were mutated to encode lysines (KK) was generated as follows. The PCR primers plcBKKfor2 and plcBKKrev3 overlapped at the site of mutation and were used in inverse PCR to generate a product from pJM199. The resulting PCR product was then end repaired using the EndIt kit (Epicentre) following the manufacturer's instructions and self-ligated to create pJM216.
Subcellular fractionation.
Cell wall, membrane, and soluble fractions were prepared by differential ultracentrifugation as described previously (19, 44). Briefly, 100-ml cultures of M. tuberculosis were harvested by centrifugation at 3,000 × g. Cell pellets were sterilized by gamma irradiation in a JL Shephard Mark I 137Cs irradiator (Department of Radiobiology, University of North Carolina at Chapel Hill) with a dose of 2.4 megarads. All subsequent steps were performed at 4°C. Pellets were resuspended in 4 ml of breaking buffer (phosphate-buffered saline, 1 mM phenylmethylsulfonyl fluoride, 0.6 μg/ml each of DNase and RNase, and a cocktail of protease inhibitors [2 μg/ml each of aprotinin, E-64, leupeptin, and pepstatin A and 100 μg/ml Pefabloc SC]) and then lysed in a French pressure cell. Unbroken cells were pelleted at 3,000 × g for 20 min to generate a clarified whole-cell lysate, which was centrifuged at 27,000 × g for 30 min to pellet the cell wall. The supernatant was centrifuged at 100,000 × g for 2 h to separate the membrane fraction from the soluble fraction. The cell wall and membrane fractions were washed once and then resuspended in phosphate-buffered saline.
Bioinformatic identification of putative twin-arginine signal sequences.
Protein sequences corresponding to the 4,056 predicted open reading frames (ORFs) of M. tuberculosis H37Rv were obtained from TubercuList (Institut Pasteur [http://genolist.pasteur.fr/TubercuList/]) and were entered as query sequences in the TATFIND v.1.4 (45) (http://signalfind.org/tatfind.html) and TatP v.1.0 (4) (http://www.cbs.dtu.dk/services/TatP/) search algorithms. The output for TATFIND v.1.4 is either “true” or “false” for a given peptide. TatP v.1.0 has multiple outputs. We used the default search criteria RRx[FGAVML][LITMVF] and selected only those proteins that had a predicted tripartite signal peptide with a Tat motif according to the prediction program. We also reviewed the list of predicted Tat-exported proteins of M. tuberculosis defined by the TigrFAM motif (TIGR01409) (52). This list was obtained directly from the TIGR website (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?database=ntmt02).
RESULTS
Selection of exported M. tuberculosis ORF-′BlaC fusions.
A truncated M. tuberculosis β-lactamase ′BlaC, lacking its native signal sequence, can be used in mycobacteria as a reporter of export (34). A special feature of the ′BlaC reporter is that it works only when exported by the Tat pathway. In a β-lactam-sensitive background, such as the M. smegmatis ΔblaS strain, exported ′BlaC fusion proteins can be detected by their ability to promote growth in the presence of the β-lactam antibiotic carbenicillin.
To experimentally identify M. tuberculosis ORFs with functional Tat signal sequences, we constructed genomic M. tuberculosis libraries upstream of the truncated ′blaC reporter. Two libraries were constructed in multicopy vectors, pJES113 and pJM157, each of which carries truncated ′blaC immediately downstream of a unique ClaI cloning site (Fig. 1). The difference between the vectors is that pJM157 contains the mycobacterial hsp60 promoter upstream of the ClaI site to drive expression from genomic fragments that lack a promoter. The hsp60 sequence in pJM157, however, does not include a Shine-Dalgarno site or start codon; these elements must be provided by the genomic insert. For library 1, M. tuberculosis genomic DNA was prepared from wild-type strain H37Rv, and for library 2, the genomic DNA was prepared from the ΔblaC mutant of M. tuberculosis. In both cases the genomic DNA was cut with ClaI-compatible endonucleases for ligation into the vectors.
The libraries were electroporated into the ΔblaS mutant of M. smegmatis and directly plated on 7H10 medium containing carbenicillin. Plasmids expressing exported fusion proteins were selected by their ability to promote growth in the presence of carbenicillin. For library 1, 101 carbenicillin-resistant colonies were obtained from an estimated 1 × 106 M. smegmatis transformants. For library 2, 29 carbenicillin-resistant colonies were obtained from an estimated 9 × 105 transformants. Following confirmation of carbenicillin resistance of individual transformants, plasmid DNA was isolated and sequenced to determine the identity of the genomic DNA insert. With one notable exception, all plasmids sequenced revealed an in-frame fusion with ′blaC. The exception was plasmids in which the full-length M. tuberculosis blaC gene was cloned. In these cases, the blaC insert did not need to be cloned in frame with the reporter sequence on the plasmid. BlaC was identified in 50% of the carbenicillin-resistant clones from library 1. In library 2 this problem was avoided by using genomic DNA from ΔblaC M. tuberculosis.
From the two libraries, we identified amino-terminal sequences of 10 unique M. tuberculosis proteins that promote export of the ′BlaC reporter (Table 3). To confirm that the fusion proteins identified were exported in a Tat-dependent manner, we rescued the fusion plasmids and electroporated them into a double ΔblaS ΔtatA M. smegmatis mutant. This allowed us to test for export in the absence of a functional Tat pathway. All 10 fusions that conferred carbenicillin resistance in a ΔblaS mutant background failed to confer carbenicillin resistance in the ΔblaS ΔtatA strain. This indicated that all the fusion proteins identified require the Tat pathway to export functional ′BlaC.
TABLE 3.
Functional Tat signal sequences in M. tuberculosis proteins identified with the ′BlaC reporter
| Protein | Description | No. of times identified in libraries | In silico prediction
|
||
|---|---|---|---|---|---|
| TatP | TATFIND | TigrFAM | |||
| Rv0063 | Possible oxidoreductase | 1 | No | No | Yes |
| Rv0315 | Possible beta-1,3-glucanase precursor | 41 | Yes | Yes | Yes |
| Rv0483 (LprQ) | Possible conserved lipoprotein | 2 | No | No | No |
| Rv0519c | Possible lipase | 5 | Yes | No | Yes |
| Rv0774c | Possible lipase; short-chain alcohol dehydrogenase family | 12 | Yes | Yes | Yes |
| Rv0846c | Possible multicopper oxidase | 1 | Yes | Yes | Yes |
| Rv2068c (BlaC) | Class A β-lactamase | 63 | Yes | Yes | Yes |
| Rv2350c (PlcB) | Phospholipase C | 1 | Yes | Yes | Yes |
| Rv2833c (UgpB) | Possible glycerol-3-phosphate-binding lipoprotein | 3 | Yes | Yes | Yes |
| Rv2843 | Possible conserved transmembrane Ala-rich protein | 1 | No | Yes | Yes |
| Rv2041c | Possible sugar-binding lipoprotein | 0a | Yes | Yes | Yes |
| Rv2351c (PlcA) | Phospholipase C | 0a | Yes | No | Yes |
| Rv2525c | Conserved hypothetical protein | 0a | Yes | Yes | Yes |
Demonstrated by direct testing only.
Direct testing of candidate M. tuberculosis Tat signal sequences.
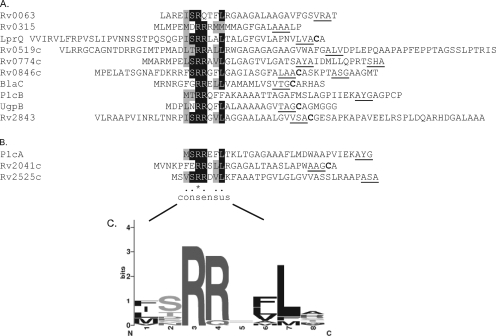
The M. tuberculosis sequences fused to ′BlaC in the 10 active fusions were all predicted to contain signal sequences, according to the Signal P 3.0 prediction algorithm (Fig. 2A) (3). Evaluation of the exported fusions revealed that the junction with ′BlaC always occurred close to the predicted signal sequence cleavage site of the M. tuberculosis protein. The greatest distance that we observed between a predicted cleavage site and the ′BlaC fusion junction was 34 amino acids. This revealed a requirement for an appropriately positioned restriction enzyme site near the cleavage site in order to identify a Tat signal sequence in our libraries. It also suggested that some proteins may have been missed for this reason. For example, in previous work we showed that the PlcA signal sequence promotes Tat export of the ′BlaC reporter (34), but PlcA was not identified in the libraries. From genome gazing and bioinformatic predictions (discussed below), we selected Rv2041c and Rv2525c as candidate Tat substrates that may have been missed. Construction and direct testing of ssRv2041c-′BlaC and ssRv2525c-′BlaC fusion proteins revealed that both confer resistance to β-lactam in a Tat-dependent manner. This provides experimental validation of these additional M. tuberculosis Tat signal sequences (Fig. 2B).
FIG. 2.
M. tuberculosis Tat signal sequences capable of driving the export of ′BlaC. Multiple sequence alignment (PROMALS [http://prodata.swmed.edu/promals/promals.php]) of M. tuberculosis sequences capable of directing the export of functional ′BlaC. (A) Alignment of sequences identified from the genomic libraries. For a given protein, the shortest stretch of amino acids up to the fusion junction with ′BlaC is shown. (B) Alignment of sequences from fusions directly tested. Identical amino acids are shaded in black, and similar amino acids are shaded in gray. Underlined are signal sequence cleavage sites predicted by Signal P (3). Cysteine residues in bold indicate conserved residues in lipobox motifs that are the predicted site of lipid modification for lipoproteins. (C) Graphical representation of the sequence alignment of the consensus region of the 13 twin-arginine signal sequences shown to export functional ′BlaC. The height of each stack represents the degree of sequence conservation, and the size of each letter is proportional to the frequency of the corresponding amino acid in that position (http://weblogo.berkeley.edu).
Alignment of functional Tat signal sequences identified with the ′BlaC reporter.
From studies largely conducted with E. coli, the generally accepted twin-arginine consensus motif is S/T-R-R-x-F-L-K (x is any polar amino acid). The twin arginines are nearly always invariant, and the frequency of occurrence of the other amino acids is reported to exceed 50% (5, 31, 39). Amino acid alignment of the M. tuberculosis signal sequences that we identified revealed that all but one contained the twin-arginine dipeptide (Fig. 2). The exception was the Rv0063 signal sequence, which has a glutamine in the position where the second arginine would be (R-Q-T-F-L). In terms of the other amino acids in the consensus, all were present in ≥40% of the sequences except for the final lysine (K). The alignment also revealed conservation of a hydrophobic residue just prior to the S/T amino acid in the consensus.
Comparison to in silico-predicted Tat signal sequences.
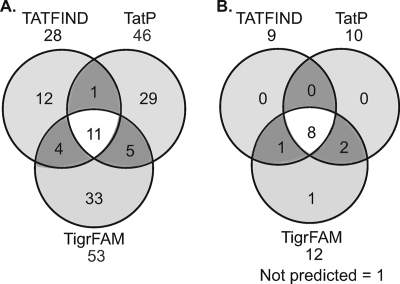
Multiple Tat signal prediction programs exist, but their ability to accurately and comprehensively identify Tat substrates within mycobacteria is unresolved. We applied two of these web-based programs, TatP v.1.0 (4) and TATFIND v1.4 (45), to the M. tuberculosis H37Rv genome sequence and compared the output to Tat signal sequences predicted by a third prediction program devised by The Institute for Genomic Research (TIGR) (TIGR01409) (52). Of the 4,056 ORFs in the M. tuberculosis H37Rv genome (11), 95 were predicted to encode proteins with Tat signal sequences by at least one of the prediction programs (see Table S2 in the supplemental material). There is surprisingly limited overlap between the algorithms, with only 11 proteins being predicted by all three programs (Fig. 3A).
FIG. 3.
Distribution of predicted and experimentally verified M. tuberculosis Tat signal sequences. (A) Venn diagram indicating the distribution of M. tuberculosis proteins predicted to have Tat signal sequences by TATFIND v.1.4, TatP v.1.0, and/or TigrFAM (TIGR01409) algorithms. (B) Venn diagram comparing the proteins predicted in silico with the M. tuberculosis Tat signal sequences experimentally identified with the ′BlaC reporter. Rv0063 was predicted by TigrFAM only. LprQ was not predicted by any of the programs.
We next compared the signal sequences that we identified experimentally (Table 3; Fig. 2) to the in silico predictions of the annotated M. tuberculosis genome. Eight of the proteins that we identified were predicted by all three programs, and three were predicted by two programs (Fig. 3B). Of the remaining two signal sequences, one was Rv0063, which lacks the twin arginine and was identified only by the TigrFAM prediction program, and the other was LprQ, which was not predicted by any program.
Assessment of Tat dependence for full-length proteins with functional Tat signal sequences.
In addition to there being a requirement for a Tat signal sequence, the mature domain also plays a role in whether a protein is a Tat substrate. For this reason, we tested full-length versions of a subset of the proteins that we identified to see if they were Tat dependent.
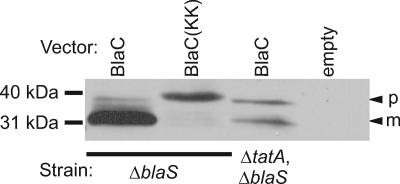
Signal sequence cleavage is commonly used as an indicator of protein export (37). To establish the utility of this approach for monitoring export of M. tuberculosis Tat substrates expressed in M. smegmatis, we first assayed signal sequence cleavage of full-length BlaC. Polyclonal antibodies were raised against BlaC and used to detect BlaC expressed in whole-cell lysates of ΔblaS M. smegmatis by immunoblotting ( Fig. 4). In the presence of a functional Tat apparatus, BlaC is observed as a predominant band running at 30 kDa, which is the predicted size of the exported and processed mature species. Expression of BlaC(KK), which has a KK substitution for the RR dipeptide, produced a slower-migrating species, which is consistent with a lack of export and accumulation of full-length unprocessed BlaC precursor (Fig. 4). In the absence of a functional Tat pathway (in a ΔblaS ΔtatA mutant), a larger presumptive precursor species was observed, although some smaller mature protein was detected as well. These experiments indicated that the protein species observed for each of these strains was a good indicator of Tat- and twin-arginine-dependent export.
FIG. 4.
Tat-dependent processing of BlaC in M. smegmatis. Full-length BlaC was assessed for signal sequence processing by immunoblot analysis of whole-cell lysates. Full-length BlaC was expressed in a tat+ strain (ΔblaS) or a tat deletion strain (ΔblaS ΔtatA) of M. smegmatis. BlaC[KK] was expressed in a tat+ strain. Whole-cell lysates of these strains were separated by SDS-PAGE and compared to a whole-cell lysate of ΔblaS M. smegmatis carrying an empty vector (empty). Bands were detected by immunoblot analysis using anti-BlaC antibodies. The presumed precursor (p) and mature (m) forms of select proteins are indicated by arrowheads.
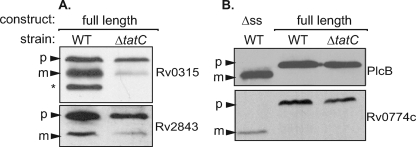
We then tested full-length versions of four additional proteins identified with the ′BlaC reporter in wild-type and ΔtatC M. smegmatis. Proteins were expressed as a C-terminal fusion to the HA epitope tag, and immunoblot analysis was performed on whole-cell lysates. For Rv0315 and Rv2843, Tat-dependent processing was observed in M. smegmatis, which is indicative of the full-length proteins being Tat exported. A lower-molecular-weight and presumably processed form of the protein was seen when expressed in the wild type, but this species was significantly reduced in abundance in the ΔtatC mutant (Fig. 5A). Three bands were observed for Rv0315-HA expressed in wild-type M. smegmatis. The highest-molecular-mass band is similar in size (∼34 kDa) to that of the predicted Rv0315 precursor and is present in both wild-type and ΔtatC strains. The two smaller protein species are absent or greatly reduced in the ΔtatC mutant background. This suggests that Rv0315 is subject to multiple processing events.
FIG. 5.
Assessment of signal sequence processing of M. tuberculosis full-length candidate Tat substrates. (A) Full-length constructs expressing HA-tagged Rv0315 and Rv2843 were assessed for signal sequence processing in wild-type (WT) or ΔtatC M. smegmatis. Whole-cell lysates were prepared, and proteins were separated by SDS-PAGE. Bands were detected by immunoblot analysis using anti-HA antibodies. The presumed precursor (p) and mature (m) forms of select proteins are indicated by arrowheads. The asterisk indicates a second proteolytic cleavage product for Rv0315. (B) Constructs of HA-tagged PlcB and Rv0774c lacking their presumed signal sequences (Δss) were run alongside full-length versions of PlcB-HA and Rv0774-HA expressed in wild-type or ΔtatC strains of M. smegmatis.
For the other two full-length proteins tested, no obvious Tat dependence was observed in immunoblots of M. smegmatis whole-cell lysates (Fig. 5B). One of these proteins was the virulence factor PlcB-HA. The protein species seen in the wild-type and ΔtatC M. smegmatis strains migrated at the predicted precursor size of 57 kDa. Moreover, compared to a truncated form of PlcB-HA lacking the predicted signal sequence (ΔssPlcB-HA), the full-length product that we expressed migrated slower than the expected mature product did (Fig. 5B). Similar results were obtained with Rv0774c-HA and a truncated ΔssRv0774c-HA protein expressed in M. smegmatis. This suggested that these two predicted Tat substrates were not being processed or exported when expressed in M. smegmatis.
Full-length PlcB is exported by the Tat pathway when expressed in its native host, M. tuberculosis.
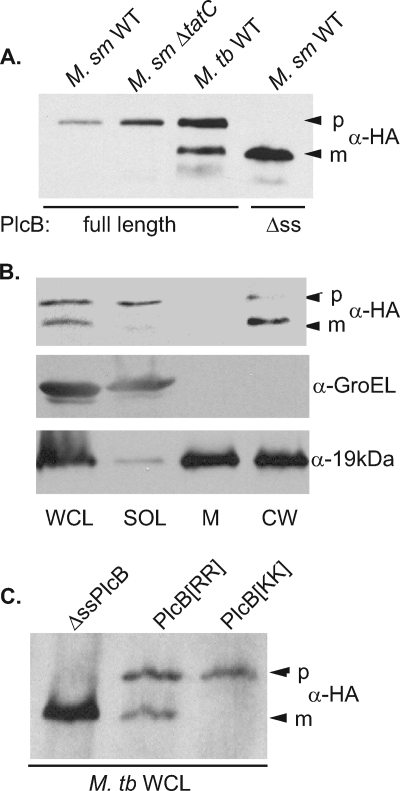
Since M. smegmatis does not have phospholipase C homologs, we considered the possibility that PlcB-HA can be exported only by its native host, M. tuberculosis. To test this idea, we expressed full-length PlcB-HA in M. tuberculosis H37Rv and assayed whole-cell lysates by immunoblot analysis. Unlike what was observed in M. smegmatis, immunoblot assays for PlcB-HA in M. tuberculosis whole-cell lysates yielded two products: a larger species that migrated like the full-length precursor seen in M. smegmatis and a smaller species that migrated like the expected mature ΔssPlcB-HA product (Fig. 6A). This suggested that in M. tuberculosis, PlcB-HA is exported and processed. Subcellular fractions prepared from the M. tuberculosis strain were used to show that the observed faster-migrating product was exported. Soluble, membrane, and cell wall fractions were prepared from whole-cell lysates of H37Rv expressing PlcB-HA and analyzed by immunoblotting (Fig. 6B). Of the two protein species seen in the whole-cell lysate, the larger product, the presumptive nonexported precursor, was in the soluble cytosolic fraction and the smaller product was primarily in the cell wall. Thus, when expressed in M. tuberculosis the PlcB-HA protein was processed and exported to the cell wall. As controls for the fractionation, we showed that the GroEL protein and the 19-kDa lipoprotein were enriched in the soluble and cell envelope (cell wall and membrane) fractions, respectively.
FIG. 6.
Full-length PlcB is processed and exported in M. tuberculosis but not in M. smegmatis. (A) Full-length PlcB-HA and PlcB-HA lacking its signal sequence (Δss) were expressed in wild-type (M. sm WT) and tat deletion (M. sm ΔtatC) strains of M. smegmatis or a wild-type strain of M. tuberculosis (M. tb WT). Bands were detected by immunoblot analysis using anti-HA antibodies. The presumed precursor (p) and mature (m) forms of PlcB are indicated by arrowheads. (B) Irradiated cultures of M. tuberculosis expressing HA-tagged full-length PlcB-HA were fractionated from whole-cell lysates (WCL) into soluble (SOL), membrane (M), and cell wall (CW) fractions. Immunoblot analysis was performed to localize HA-tagged PlcB-HA and GroEL and 19-kDa lipoprotein controls. (C) HA-tagged truncated ΔssPlcB, full-length PlcB[RR] (wild type), and PlcB[KK] were expressed in M. tuberculosis. Whole-cell lysates of each strain were analyzed by immunoblot analysis.
The essential nature of the Tat pathway in M. tuberculosis precludes testing full-length PlcB-HA in an M. tuberculosis Δtat mutant. To address whether full-length M. tuberculosis PlcB is a Tat substrate, the RR pair in the PlcB signal sequence was replaced with KK. When the PlcB(KK)-HA protein was expressed in M. tuberculosis, a single precursor-sized species was observed (Fig. 6C). Together, these experiments demonstrated that PlcB is a Tat substrate exported to the cell wall in M. tuberculosis but not in M. smegmatis.
DISCUSSION
The Tat pathway is important to the virulence and physiology of several bacterial pathogens. Because it is absent in mammals, it has been considered a _target for development of new antibacterial agents (8, 31). Here we used ′BlaC as a reporter to identify proteins with functional Tat signal sequences and to begin understanding the role that Tat export plays in M. tuberculosis. This approach allows for the experimental investigation of Tat export in a system where obtaining a Δtat mutant is not feasible. ′BlaC is not the only recognized Tat-specific reporter (14, 63), but it is the only such reporter that works in a direct selection, as opposed to a screen. This property of ′BlaC allowed us to exploit it on a genome-wide level. Previously, Tat reporters have been used only to validate preselected candidates (49, 57, 58, 63).
All active ′BlaC fusions obtained from our libraries were in frame with an ORF, had predicted signal sequences, and were confirmed to be exported in a Tat-dependent manner. This attests to the power of the ′BlaC reporter. Our objective was to comprehensively identify the Tat signal sequences of M. tuberculosis. Our two libraries were composed of ∼2 × 106 plasmids combined, which is sufficiently complex to have >99% probability of every M. tuberculosis ORF being represented as a single in-frame ′blaC fusion (48). However, the unanticipated requirement for a fusion junction to be positioned just beyond the signal sequence cleavage site means that, despite this level of complexity, some proteins were not picked up. This restriction most likely reflects extended sequences of a mature protein preventing ′BlaC folding into an active conformation compatible with Tat export. Future libraries could be improved by increasing the complexity and using smaller randomly sheared genomic DNA inserts to overcome limitations that include the lack of available restriction enzyme cleavage sites.
Of the 13 Tat signal sequences identified with the reporter, many are in proteins (Rv0063, Rv0315, Rv0774c, Rv2525c, BlaC, and PlcB) previously shown to be secreted or cell wall associated by proteomics (22, 24, 32, 43, 47, 51). Six of them (Rv0846c, Rv2041c, Rv2843, BlaC, LprQ, and UgpB) are predicted lipoproteins with a lipobox motif in their signal sequence (Fig. 2A), which predicts cell envelope localization (56). Recently, in the archaeon Haloferax volcanii it was shown that lipoproteins can be Tat substrates (20). BlaC, PlcA, and PlcB are the only proteins that we identified that have demonstrated functions (β-lactamase and phospholipase C activities, respectively) (29, 43, 59). The Plc proteins are also shown to function in M. tuberculosis pathogenesis. Simultaneous deletion of multiple plc genes results in attenuated virulence of M. tuberculosis in a mouse model of infection (43).
Much less is known about the remaining Tat signal sequence-containing proteins that we identified (Table 3). Sequence analysis suggests that these proteins have diverse functions as lipases (Rv0519c and Rv0774c), a copper oxidase (Rv0846c), a glycosyl hydrolase (Rv0315), an oxidoreductase (Rv0063), and substrate-binding proteins of sugar uptake systems (UgpB and Rv2041c). Rv2525c is one of the proteins identified with no predicted function. On the basis of bioinformatic predictions, this protein was previously hypothesized to be an M. tuberculosis Tat substrate and, because of coregulation with other genes, was suggested to have a role in cell wall biogenesis (47). Both UgpB and Rv2041c are predicted by transposon saturation mutagenesis to be essential to M. tuberculosis (50), providing a potential clue as to why the Tat pathway cannot be inactivated in M. tuberculosis.
An advantage of using a genetic reporter to directly identify functional Tat signal sequences is that there is no imposed requirement that conserved sequence elements, as defined by other studies, need be present. In this regard, the functional Tat signal sequences that we identified should be useful for refining Tat prediction algorithms. Most of the sequences that we experimentally identified were predicted by at least two of three Tat signal sequence prediction programs consulted. This suggests that the common core elements of these programs are the best predictors. The signal sequence of Rv0063, which possesses RQ in the Tat motif, was the only one of our experimentally identified sequences lacking a twin-arginine dipeptide. Our identification of Rv0063 is consistent with the recent demonstration that its signal sequence can direct Tat export of an agarase reporter when expressed in Streptomyces lividans (63). Although no natural Tat substrate is known to have an RQ pair, there are two examples of bacterial Tat signal sequences lacking an RR dipeptide (26, 27). In addition, an RQ substitution in the Tat signal sequence of the E. coli TorA protein is able to promote Tat export of a green fluorescent protein reporter (14). Interestingly, the TigrFAM program predicted Rv0063 to have a Tat signal sequence (52). We also identified one protein, LprQ, which was not identified by any of the three prediction programs. LprQ was likely missed due to its extended N terminus upstream of the twin-arginine motif (Fig. 2A). However, it is also possible that the true start codon of LprQ is incorrectly annotated, as there are additional GTG codons between the annotated start and the twin-arginine motif.
In addition to having an amino-terminal Tat signal sequence, there are features of the mature domain of a protein, which must be folded, that determine whether it can be translocated by the Tat pathway (15). Recently, some putative Tat signal sequences of E. coli were shown to be able to promote export of a fused reporter through Sec or Tat pathways, depending on the unfolded or folded nature of the reporter (58). These apparently promiscuous signal sequences highlight the importance played by the mature domain of a protein. The basic question of how often it is that a functional Tat signal sequence is present on a bona fide Tat substrate has only begun to be investigated (58). For this reason, we examined full-length versions of a subset of proteins that we identified in wild-type and Δtat strains of M. smegmatis. Three proteins (BlaC, Rv0315, and Rv2843) showed a Tat-dependent effect, which indicates that the native proteins are subject to Tat export. For Rv0315-HA, the immunoblot analysis revealed three protein species in whole-cell lysate of wild-type M. smegmatis and two species in lysate of tat mutant M. smegmatis (Fig. 5A). A homologous β-glucanase of Bacillus circulans is subject to progressive proteolytic processing postexport (61). Similar proteolytic processing of Rv0315 would explain the multiple species seen by immunoblotting.
For two other M. tuberculosis proteins tested, there was no difference in the protein species seen in whole-cell lysates from wild-type and Δtat strains of M. smegmatis (Fig. 5B). In these cases, the full-length protein species observed was larger than the predicted mature protein. Thus, no signal sequence cleavage or obvious export occurred when full-length versions of these proteins were expressed in M. smegmatis.
The fact that PlcB-HA was unaffected in an M. smegmatis tat mutant was surprising since the Plc proteins are demonstrated to be exported to cell wall fractions of M. tuberculosis (43). Moreover, virtually all phospholipase C orthologs in a wide variety of organisms have predicted Tat signal sequences (16), and there are bacterial Plc enzymes proven to be exported by the Tat pathway (9, 36, 46, 60). In contrast to what was seen in M. smegmatis, when expressed in M. tuberculosis two species of PlcB-HA were evident by immunoblot analysis. The faster of the two species was exported to the cell wall and migrated at the same molecular weight as did ΔssPlcB-HA, the expected size of processed mature PlcB. A KK substitution for the twin-arginine motif of full-length PlcB prevented its processing in M. tuberculosis. These results demonstrate that in its native host PlcB is an authentic twin-arginine-dependent protein. These data provide an important link between the Tat pathway and virulence factor export in M. tuberculosis.
The above result is also interesting in suggesting that Tat export of an M. tuberculosis virulence factor requires a pathogen-specific component. There are a small number of Tat substrates shown to require dedicated chaperones for export (28, 38, 39). There are also examples of chaperones, such as DnaK, that work with a broader collection of Tat substrates (23, 40). Perhaps the pathogen-specific factor is a chaperone for PlcB that is present in M. tuberculosis but absent in M. smegmatis. Although the exact function of these chaperones remains to be discerned, proposals for how they work include promoting proper folding, protecting the Tat substrate from degradation or delivery to the translocon before folding is complete, and delivering the substrate to the translocase (39). Some of these functions relate to the mature domain of the protein, which could account for why an ssPlcB-′BlaC protein was exported by M. smegmatis but the full-length PlcB-HA was not. Notably, PlcH of P. aeruginosa requires PlcR chaperones for its export (12). However, no obvious PlcR homologs exist in M. tuberculosis. Another possibility is that a conserved component of the M. smegmatis and M. tuberculosis Tat systems differs in its substrate recognition abilities.
The work presented here demonstrates the power of using a Tat-specific reporter to identify functional Tat signal sequences without any preconceived bias regarding the features that define them. By examining full-length versions of proteins in wild-type and Δtat strains of M. smegmatis, we showed three of the proteins identified to be true Tat substrates. The results with the other two proteins tested emphasize the importance of domains beyond the Tat signal sequence and bacterium-specific factors in defining a true Tat substrate. In our quest to prove that the virulence factor PlcB is a true Tat substrate, we discovered that this protein is exported by the Tat pathway only its native host, indicating the existence and requirement for pathogen-specific host factors in its Tat export.
Supplementary Material
Acknowledgments
This research was funded by a grant from NIAID (AI54540 to M.B.) and a developmental grant from the University of North Carolina at Chapel Hill Center for AIDS Research (NIH #9P30 AI50410-04). J.A.M. was also supported by the Training in Sexually Transmitted Diseases and AIDS NIH NIAID training grant 5T32 AI07001-28 and a Society of Fellows dissertation completion fellowship from the Graduate School at the University of North Carolina at Chapel Hill. J.R.M. was also supported by an NIH Cell and Molecular Biology training grant (NIH 5-T32-GM008581). J.S.S. was supported by NSF REU grant DBI-0453235.
We thank Doug Young for 19-kDa lipoprotein antibodies, Doug Kernodle for providing us with the His-tagged BlaC expression vector, Adrienne Cox and Henry Gibbons for assistance with irradiation and fractionation of M. tuberculosis cultures, and Ruth Silversmith for advice on purification of His-tagged BlaC. We also thank Martin Pavelka and members of the Braunstein laboratory for critical review of the manuscript.
Footnotes
Published ahead of print on 25 July 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bange, F. C., F. M. Collins, and W. R. Jacobs, Jr. 1999. Survival of mice infected with Mycobacterium smegmatis containing large DNA fragments from Mycobacterium tuberculosis. Tuber. Lung Dis. 79171-180. [DOI] [PubMed] [Google Scholar]
- 2.Baulard, A., C. Jourdan, A. Mercenier, and C. Locht. 1992. Rapid mycobacterial plasmid analysis by electroduction between Mycobacterium spp. and Escherichia coli. Nucleic Acids Res. 204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, D. Widdick, T. Palmer, and S. Brunak. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22393-404. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein, M., S. S. Bardarov, and W. R. J. Jacobs. 2002. Genetic methods for deciphering virulence determinants of Mycobacterium tuberculosis. Methods Enzymol. 35867-99. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein, M., A. M. Brown, S. Kurtz, and W. R. Jacobs, Jr. 2001. Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 1836979-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronstein, P., M. Marrichi, and M. P. DeLisa. 2004. Dissecting the twin-arginine translocation pathway using genome-wide analysis. Res. Microbiol. 155803-810. [DOI] [PubMed] [Google Scholar]
- 9.Bronstein, P. A., M. Marrichi, S. Cartinhour, D. J. Schneider, and M. P. DeLisa. 2005. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J. Bacteriol. 1878450-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldelari, I., S. Mann, C. Crooks, and T. Palmer. 2006. The Tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol. Plant-Microbe Interact. 19200-212. [DOI] [PubMed] [Google Scholar]
- 11.Camus, J. C., M. J. Pryor, C. Medigue, and S. T. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 1482967-2973. [DOI] [PubMed] [Google Scholar]
- 12.Cota-Gomez, A., A. I. Vasil, J. Kadurugamuwa, T. J. Beveridge, H. P. Schweizer, and M. L. Vasil. 1997. PlcR1 and PlcR2 are putative calcium-binding proteins required for secretion of the hemolytic phospholipase C of Pseudomonas aeruginosa. Infect. Immun. 652904-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cristobal, S., J. W. de Gier, H. Nielsen, and G. von Heijne. 1999. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 182982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLisa, M. P., P. Samuelson, T. Palmer, and G. Georgiou. 2002. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 27729825-29831. [DOI] [PubMed] [Google Scholar]
- 15.DeLisa, M. P., D. Tullman, and G. Georgiou. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. USA 1006115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 1851478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding, Z., and P. J. Christie. 2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores, A. R., L. M. Parsons, and M. S. Pavelka, Jr. 2005. Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology 151521-532. [DOI] [PubMed] [Google Scholar]
- 19.Gibbons, H. S., F. Wolschendorf, M. Abshire, M. Niederweis, and M. Braunstein. 2007. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J. Bacteriol. 1895090-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gimenez, M. I., K. Dilks, and M. Pohlschroder. 2007. Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol. Microbiol. 661597-1606. [DOI] [PubMed] [Google Scholar]
- 21.Glickman, M. S., J. S. Cox, and W. R. Jacobs, Jr. 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5717-727. [DOI] [PubMed] [Google Scholar]
- 22.Gomez, M., S. Johnson, and M. L. Gennaro. 2000. Identification of secreted proteins of Mycobacterium tuberculosis by a bioinformatic approach. Infect. Immun. 682323-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graubner, W., A. Schierhorn, and T. Bruser. 2007. DnaK plays a pivotal role in Tat _targeting of CueO and functions beside SlyD as a general Tat signal binding chaperone. J. Biol. Chem. 2827116-7124. [DOI] [PubMed] [Google Scholar]
- 24.Gu, S., J. Chen, K. M. Dobos, E. M. Bradbury, J. T. Belisle, and X. Chen. 2003. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics 21284-1296. [DOI] [PubMed] [Google Scholar]
- 25.Halbig, D., T. Wiegert, N. Blaudeck, R. Freudl, and G. A. Sprenger. 1999. The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur. J. Biochem. 263543-551. [DOI] [PubMed] [Google Scholar]
- 26.Hinsley, A. P., N. R. Stanley, T. Palmer, and B. C. Berks. 2001. A naturally occurring bacterial Tat signal peptide lacking one of the ‘invariant’ arginine residues of the consensus _targeting motif. FEBS Lett. 49745-49. [DOI] [PubMed] [Google Scholar]
- 27.Ignatova, Z., C. Hornle, A. Nurk, and V. Kasche. 2002. Unusual signal peptide directs penicillin amidase from Escherichia coli to the Tat translocation machinery. Biochem. Biophys. Res. Commun. 291146-149. [DOI] [PubMed] [Google Scholar]
- 28.Jack, R. L., G. Buchanan, A. Dubini, K. Hatzixanthis, T. Palmer, and F. Sargent. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J. 233962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansen, K. A., R. E. Gill, and M. L. Vasil. 1996. Biochemical and molecular analysis of phospholipase C and phospholipase D activity in mycobacteria. Infect. Immun. 643259-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavander, M., S. K. Ericsson, J. E. Broms, and A. Forsberg. 2006. The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect. Immun. 741768-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, P. A., D. Tullman-Ercek, and G. Georgiou. 2006. The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malen, H., F. S. Berven, K. E. Fladmark, and H. G. Wiker. 2007. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 71702-1718. [DOI] [PubMed] [Google Scholar]
- 33.McCann, J. R., J. A. McDonough, M. S. Pavelka, and M. Braunstein. 2007. Beta-lactamase can function as a reporter of bacterial protein export during Mycobacterium tuberculosis infection of host cells. Microbiology 1533350-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonough, J. A., K. E. Hacker, A. R. Flores, M. S. Pavelka, Jr., and M. Braunstein. 2005. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 1877667-7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9494-500. [DOI] [PubMed] [Google Scholar]
- 36.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 998312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver, D. B., and J. Beckwith. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25765-772. [DOI] [PubMed] [Google Scholar]
- 38.Oresnik, I. J., C. L. Ladner, and R. J. Turner. 2001. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 40323-331. [DOI] [PubMed] [Google Scholar]
- 39.Palmer, T., F. Sargent, and B. C. Berks. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13175-180. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Rodriguez, R., A. C. Fisher, J. D. Perlmutter, M. G. Hicks, A. Chanal, C. L. Santini, L. F. Wu, T. Palmer, and M. P. Delisa. 2007. An essential role for the DnaK molecular chaperone in stabilizing over-expressed substrate proteins of the bacterial twin-arginine translocation pathway. J. Mol. Biol. 367715-730. [DOI] [PubMed] [Google Scholar]
- 41.Posey, J. E., T. M. Shinnick, and F. D. Quinn. 2006. Characterization of the twin-arginine translocase secretion system of Mycobacterium smegmatis. J. Bacteriol. 1881332-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pradel, N., C. Ye, V. Livrelli, J. Xu, B. Joly, and L. F. Wu. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 714908-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raynaud, C., C. Guilhot, J. Rauzier, Y. Bordat, V. Pelicic, R. Manganelli, I. Smith, B. Gicquel, and M. Jackson. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 45203-217. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, A. D., J. T. Belisle, A. M. Cooper, and I. M. Orme. 1998. Specific models: murine model of tuberculosis. Methods Microbiol. 25389-417. [Google Scholar]
- 45.Rose, R. W., T. Bruser, J. C. Kissinger, and M. Pohlschroder. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45943-950. [DOI] [PubMed] [Google Scholar]
- 46.Rossier, O., and N. P. Cianciotto. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 732020-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saint-Joanis, B., C. Demangel, M. Jackson, P. Brodin, L. Marsollier, H. Boshoff, and S. T. Cole. 2006. Inactivation of Rv2525c, a substrate of the twin arginine translocation (Tat) system of Mycobacterium tuberculosis, increases β-lactam susceptibility and virulence. J. Bacteriol. 1886669-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Santini, C. L., A. Bernadac, M. Zhang, A. Chanal, B. Ize, C. Blanco, and L. F. Wu. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 2768159-8164. [DOI] [PubMed] [Google Scholar]
- 50.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 9812712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt, F., S. Donahoe, K. Hagens, J. Mattow, U. E. Schaible, S. H. Kaufmann, R. Aebersold, and P. R. Jungblut. 2004. Complementary analysis of the Mycobacterium tuberculosis proteome by two-dimensional electrophoresis and isotope-coded affinity tag technology. Mol. Cell. Proteomics 324-42. [DOI] [PubMed] [Google Scholar]
- 52.Selengut, J. D., D. H. Haft, T. Davidsen, A. Ganapathy, M. Gwinn-Giglio, W. C. Nelson, A. R. Richter, and O. White. 2007. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 35D260-D264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 54.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein _targeting in Escherichia coli. J. Biol. Chem. 27511591-11596. [DOI] [PubMed] [Google Scholar]
- 55.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, and G. F. Hatfull. 1991. New use of BCG for recombinant vaccines. Nature 351456-460. [DOI] [PubMed] [Google Scholar]
- 56.Sutcliffe, I. C., and D. J. Harrington. 2004. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol. Rev. 28645-659. [DOI] [PubMed] [Google Scholar]
- 57.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 3947-53. [DOI] [PubMed] [Google Scholar]
- 58.Tullman-Ercek, D., M. P. Delisa, Y. Kawarasaki, P. Iranpour, B. Ribnicky, T. Palmer, and G. Georgiou. 2007. Export pathway selectivity of Escherichia coli twin-arginine translocation signal peptides. J. Biol. Chem. 2828309-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voladri, R. K., D. L. Lakey, S. H. Hennigan, B. E. Menzies, K. M. Edwards, and D. S. Kernodle. 1998. Recombinant expression and characterization of the major beta-lactamase of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 421375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voulhoux, R., G. Ball, B. Ize, M. L. Vasil, A. Lazdunski, L. F. Wu, and A. Filloux. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 206735-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe, T., N. Kasahara, K. Aida, and H. Tanaka. 1992. Three N-terminal domains of beta-1,3-glucanase A1 are involved in binding to insoluble beta-1,3-glucan. J. Bacteriol. 174186-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickner, W., and R. Schekman. 2005. Protein translocation across biological membranes. Science 3101452-1456. [DOI] [PubMed] [Google Scholar]
- 63.Widdick, D. A., R. T. Eijlander, J. M. van Dijl, O. P. Kuipers, and T. Palmer. 2008. A facile reporter system for the experimental identification of twin-arginine translocation (Tat) signal peptides from all kingdoms of life. J. Mol. Biol. 375595-603. [DOI] [PubMed] [Google Scholar]
- 64.World Health Organization. 2007, posting date. WHO information tuberculosis fact sheet. http://www.who.int/mediacentre/factsheets/fs104/en/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.