Abstract
Purpose
Bif-1 protein is a member of the endophilin B family that plays a critical role in apoptosis, autophagy and mitochondrial morphology. Loss of Bif-1 suppresses programmed cell death and promotes tumorigenesis. The connection of Bif-1 to colorectal cancer remains to be evaluated.
Experimental Design
To determine Bif-1 expression in human colorectal adenocarcinoma (CRC), we performed immunohistochemistry using stage oriented cancer tissue microarrays containing 102 CRC of different stage, and 38 samples of normal colorectal mucosa (NR). Formalin-fixed, paraffin-embedded core sections on the tissue array were immunostained using the avidin-biotin-peroxidase method and the anti-Bif-1 murine monoclonal antibody. Bif-1 staining was scored by two independent observers. To examine Bif-1 mRNA levels, we performed DNA microarray analysis of 205 CRC and 10 NR samples.
Results
Bif-1 expression was negative in 22.5% (23/102) of CRC. Moderate to strong Bif-1 staining was identified in 36.3% (37/102) of the tumors, and weak stain was seen in 41.2% (42/102) of them. Twenty-six of 38 (68.4%) NR samples exhibited moderate to strong Bif-1 immunoreactivity, and none of them was negative. In 12 cases (31.6%) NR showed weak Bif-1 stain. The mean (median) scores for CRCs and NR differed significantly [3.2 (3.0) and 5.2 (6.0) respectively, p=0.0003], and the percent of cases with negative expression also differed significantly between NR and CRC (p=0.002). Decreased Bif-1 expression in CRCs was confirmed at mRNA level by microarray analysis.
Conclusions
We report the down regulation of Bif-1 during the transition from NR to CRC, a novel finding in agreement with the tumor suppressor function of Bif-1.
Condensed abstract
Using gene expression profiling and immunohistochemistry we demonstrated the down regulation of Bif-1during the transition from normal colonic mucosa to adenocarcinoma.
Keywords: Bif-1, colon adenocarcinoma, microarray, immunohistochemistry
Introduction
Colorectal adenocarcinoma (CRC) is one of the most common malignancies, accounting for approximately 15% of all cancer-related deaths in the US (1). The prevalence of CRC increases with age, the largest number of tumors occurring during the sixth decade. The expected annual incidence of this tumor has risen over the last decade and 153,760 new cases are estimated in 2007 (1, 2). If not diagnosed and treated early, this tumor spreads through the entire bowel wall, extends to adjacent organs, and eventually metastasizes to regional lymph nodes and distant sites. The majority of deaths from CRC occur in patients with late stage tumors, which are usually incurable (3).
It has been shown that inhibition of apoptosis is critical to colorectal tumorigenesis (4). For example, it has been proposed that overexpression of Bcl-XL in cancer may suppress the activity of the proapoptotic molecules Bax and Bak, contributing to cancer progression (5, 6). It seems that, also in CRC, the dissociation of Bax and Bcl-XL promotes Bax multimerization and mitochondrial translocation, triggering apoptosis (7). Similarly, dysregulation of autophagy has also been proposed to play a role in the pathogenesis of cancer. As of an example, the autophagy activator Beclin 1 is found to be monoallelically deleted in a high percentage of ovarian, breast, and prostate cancers, and overexpression of Beclin 1 in MCF7 cells promotes autophagy and inhibits tumor formation in nude mice (8, 9). Moreover, the Beclin 1 binding protein UVRAG has been shown to promote autophagy and suppress the tumorigenesis of colon cancer cells in nude mice (10).
Bif-1 (Bax-Interacting Factor-1) has been shown to interact with Bax and induce its conformational change in mammalian cells during apoptosis (11). Knockout of Bif-1 suppresses Bax/Bak conformational change, cytochrome c release, caspase activation and cell death (12). Interestingly, we have recently discovered that Bif-1 also regulates autophagy by forming a multi-protein complex with PI3KC3-Beclin1 through UVRAG and loss of Bif-1 suppresses autophagic cell death and promotes tumorigenesis (13). Along this line, a recent study has reported the decrease of Bif-1 expression in malignant gastric epithelial cells as compared to the normal gastric mucosal cells (14). To date the expression of Bif-1 in CRC has not been reported.
In this study we focused on the evaluation of Bif-1 expression and significance in CRC. Bif-1 expression levels in CRC were determined using semi-quantitative immunohistochemistry and microarray analysis of archival specimens. The results of this study may help in allowing evolution of improved therapies for CRC, based on the better understanding of the underlying biology of this disease process.
Materials and methods
Selection of human tissues
Using stage oriented human colorectal cancer tissue microarrays (prepared in the Histology laboratory of the Moffitt Cancer Center Tissue Core Facility), 140 tissue samples (102 CRC and 38 samples of normal colonic mucosa (NR)) were analyzed for Bif-1 expression by immunohistochemistry. All of the tumors used for the tissue array construction were CRC identified from the Moffitt Cancer Center Anatomic Pathology Division’s database, CoPath®, and representing surgical resection specimens obtained between 1990 and 2002. All of the specimens were preserved in 10% buffered formalin prior to embedding in paraffin. The patients had a median age of 65 years (range 24-92), 61 were male and 41 were female. The tumors ranged in size between 1.4 cm and 14.5 cm. The tumors were staged according to the TNM system, following the recommendations of the American Joint Committee on Cancer, 1988. The stage of the invasive tumors was as follows: 10 patients had stage I (Duke’s stage A), 33 stage II (Duke’s stage B), 38 stage III (Duke’s stage C), and 21 stage IV (Duke’s stage D). All tumors occurred in the absence of genetic cancer syndromes such as human non polyposis colon cancer syndrome (HNPCC), familial adenomatous polyposis syndrome (FAP), etc.; also cancers arising in the background of ulcerative colitis or Crohn’s disease were excluded from the study. The NR samples were taken near the resected colorectal margin, away from the tumor site, from CRC colon resection specimens included in this study.
Immunohistochemistry
The tissues were stained for Bif-1, using a mouse monoclonal antibody (Imgenex, San Diego, CA). The slides were dewaxed by heating at 55° C for 30 minutes and by three washes, five minutes each, with xylene. Tissues were rehydrated by a series of five-minute washes in 100%, 95%, and 80% ethanol, and distilled water. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 20 minutes. After blocking with universal blocking serum (Ventana Medical Systems, Inc., Tucson, Arizona) for 30 minutes, the samples were incubated with anti-Bif-1 mouse monoclonal antibody (Imgenex, dilution 1:2500) at 4° C overnight. The samples were then incubated with biotin-labeled secondary antibody and streptavidin-horseradish peroxidase for 30 minutes each (Ventana Medical Systems). The slides were developed with 3,3′-diaminobenzidine tetrahydrochloride substrate (Ventana Medical Systems Inc.) and counterstained with hematoxylin (Ventana Medical Systems Inc. Tucson, Arizona). The tissue samples were dehydrated and coversliped. Standard cell conditioning (following the Ventana proprietarian recommendations) was used for antigen retrieval. The specificity of the anti-Bif-1 monoclonal antibody was confirmed by immunostains of Bif-1 overexpression and knockout cell lines (12, 13). Negative control was included by using non immune mouse sera and omitting the monoclonal Bif-1 antibody during the primary antibody incubation step.
Immunohistochemical data analysis
The Bif-1 stained tissue cores were examined by two independent observers (FK, DC) simultaneously and a consensus score was reached for each specimen. The positive reaction of Bif-1 was scored into four grades, according to the intensity of the staining: 0, 1+, 2+, and 3+. The percentages of Bif-1 positive cells were also scored into four categories: 0 (0%), 1 (1-33%), 2 (34-66%), and 3 (67-100%). The product of the intensity by percentage scores was used as the final score. The final scores were classified as: 0 negative; 1-3, weak; 4-6, moderate; and 7-9, strong. The specimens were also classified by the types of tissue staining positive: normal colonic mucosa and adenocarcinoma.
Statistical analysis
Descriptive statistics for the scores were generated and reported for each tissue group. The initial method used to compare Bif-1 expression in CRC and NR was the Wilcoxon Rank sum test. In addition, Fisher’s exact test was used to compare Bif-1 negativity between CRC and NR. For CRC the exact Cochran-Armitage trend test was used to compare Bif-1 negativity across stage. The Holm step down method was used to adjust for multiple testing. Age and gender differences between cohorts were examined using the Wilcoxon rank sum test and the Chi-square test, respectively. Spearman’s correlation was used to examine the correlation between age and Bif-1 expression, and the Wilcoxon rank sum test was used to compare Bif-1 expression differences between genders.
mRNA microarray analysis
To evaluate whether the variation in Bif-1 protein expression between NR and CRC reflected a corresponding modulation of Bif-1 mRNA, we resorted to the Moffitt Cancer Center gene profiling data base. Two hundred and five (205) colorectal adenocarcinoma tumor specimens and 10 samples of normal colorectal mucosa, from patients treated at the Moffitt Cancer Center under a University of South Florida IRB-approved protocol were arrayed on Affymetrix HG-U133+ GeneChip microarrays. The tumors, used for the mRNA microarray analysis, included mirror image samples from all of the CRC used to construct the colon tissue microarray (TMA) utilized in this study.
The data was processed using MAS5.0 and scaled to a mean intensity of 500. Three probe sets were identified by Affymetrix NetAffx as detecting Bif-1: 209090_s_at, 209091_s_at and 210101_x_at. The R statistical software was used for expression analysis (freely available open source statistical package: www.r-project.org). The Anderson-Darling test for normality was used to verify the distribution of gene expression for each probe set across samples and a t test was used to compare differences between groups (normal vs. tumor and normal vs. each stage). Expression is graphed using the mean and standard error for each probe set across the different groups.
Results
Clinical pathologic findings
The patients had a median age of 65 years (range, 24-92). Sixty-one were male and 41 were female. The tumors ranged in size between 1.4 cm and 14.5 cm, mostly polypoid and ulcerated. Twenty tumors involved the cecum, 26 the ascending colon, 4 the transverse colon, 9 the descending colon, 24 the sigmoid, 10 the rectosigmoid junction, and 9 the rectum. Fourteen tumors were well differentiated, 74 moderately differentiated, and 14 poorly differentiated. Ten tumors were Duke’s stage A, 33 Duke’s stage B, 38 Duke’s stage C, and 21 Duke’s stage D. Only 2 patients, both with rectal cancer, received preoperative radiation, to decrease the size of their tumors..
Immunohistochemical observations
All of the positively stained cases had cytoplasmic staining, which was diffusely granular with variation in intensity seen within the same lesion of some cases. Cases with variable staining were graded based on the predominant staining intensity and the percentage of positive stain was determined based on the amount of the lesion demonstrating the predominant intensity. In CRC specimens, approximately 41.2% (42/102) had weak Bif-1 staining (Fig. 1A), 36.3% (37/102) exhibited moderate to strong Bif-1 staining (Fig. 1B, 1C), and 22.5% (23/102) were Bif-1 negative (Fig. 1D). In contrast, in NR samples, 68.4% (26/38) exhibited moderate to strong Bif-1 immunoreactivity (Fig. 1 E, 1F), 31.6% (12/38) showed weak Bif-1 staining and none was Bif-1 negative. The specificity of the anti-Bif-1 antibody was confirmed by immunostaining of Bif-1 overexpressing cells (G) as compared to the Bif-1 knockout cell line (H).
Fig. 1.
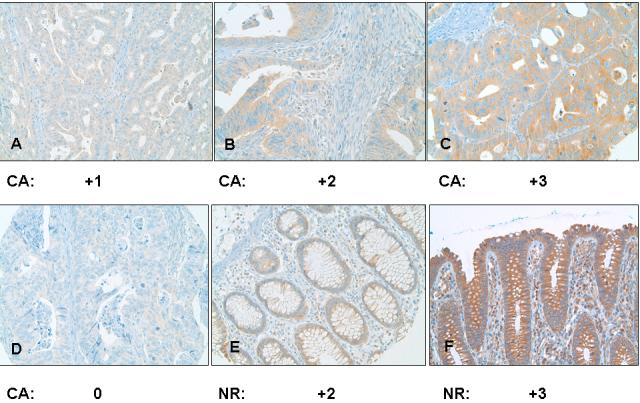
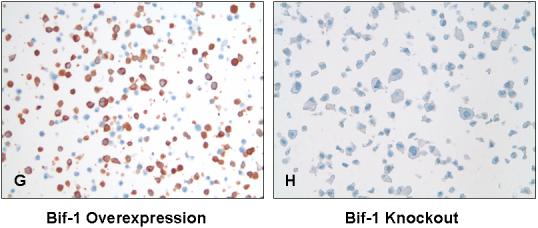
Analysis of Bif-1 in human colorectal adenocarcinoma (A-D) and normal colonic mucosa (E and F) by immunohistochemistry. Representative light photomicrographs are shown for strong (C, X400; and F, X200), moderate (B, X400; and E, X200), weak (A, X200), and negative (D, X200) Bif-1 immunoreactivity in the cytoplasm. The specificity of the anti-Bif-1 antibody was confirmed using Bif-1 overexpression (G, X200) and knockout (H, X200) cell lines.
Statistical analysis
There was a statistically significant difference found in Bif-1 staining score between NR and CRC, using the Wilcoxon rank sum test (p=0.0003), and there was also a significant difference in Bif-1 negativity (p=0.002) when comparing the percentage negative (as dichotomous groups) between the two tissue types. Using the exact Cochran-Armitage trend test, there was no statistically significant increasing trend between Bif-1 negativity score and tumor stage (p-value = 0.29). When considering the expression of Bif-1 in the NR versus the CRC it became evident that while 22.5 % of CRC were negative, none of the NR was negative. This difference was statistically significant (p=0.002). There were no statistically significant differences found in age or gender between the NR and CRC cohorts (p=0.39 and p=0.63, respectively: Table 1), and age and gender were not found to be significantly correlated with Bif-1 staining score (p=0.68 and p=0.26, respectively).
TABLE 1. Patients’ Demographics.
| Normal (n=38) | CA (n=102) | p-value | |
|---|---|---|---|
| Gender Male Female |
21 (55.3%) 17 (44.7%) |
61 (59.8%) 41 (40.2%) |
0.63* |
| Age Median Range |
68 years (34 - 91 years) |
65 years (24 - 92 years) |
0.39** |
Chi-square test.
Wilcoxon rank sum test
Gene profiling
To determine whether Bif-1 changes in protein level during human colorectal cancer development reflect changes in gene expression, we compared the Bif-1 mRNA levels by DNA microarray in 10 normal human colon tissues and 205 colorectal tumors grouped by Dukes’ staging system. The samples included 32 stage A, 66 stage B, 65 stage C, and 42 stage D colorectal adenocarcinomas. The expression of Bif-1 mRNA decreased significantly between NR and stage A CRCs and kept about the same levels during tumor progression (Fig. 2), suggesting that loss of Bif-1 expression may play a role at an early stage of colorectal tumorigenesis. The mRNA levels of Bif-1 in the 10 NR were about 3 fold higher than the Bif-1 mRNA levels in the tumors.
Fig. 2. Bif-1 mRNA expression is downregulated in human colorectal tumors.
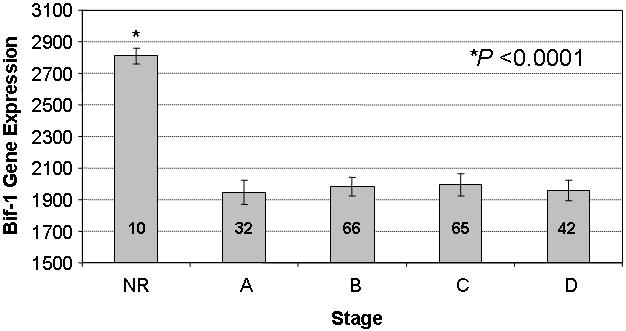
The mRNA levels of Bif-1 were analyzed by DNA microarray in 10 NR and 205 CRC (32 stage A, 66 stage B, 65 stage C, and 42 stage D) samples. The data shown are means ± standard deviations.
Discussion
Programmed cell death (PCD) is defined as a physiological process that plays a critical role in normal development, cellular differentiation, and tissue homeostasis of multicellular organisms (15-17). Dysregulation of this physiological cell death process contributes to the pathogenesis of human diseases including cancer (18). In addition to apoptosis (type I cell death), which has long been used as a synonym for PCD, accumulating evidence suggests that autophagy (type II cell death) also belongs to PCD (19). Autophagy is a highly orchestrated self-digestion process that involves multiple steps from the formation of autophagic vesicles to lysosomal degradation of the vesicles and their contents (19-21). As with apoptosis, autophagy also contributes to proper morphogenesis during development and tissue homeostasis in mature organisms (21).
Bif-1, also known as endophilin B1 and SH3GLB1 (SH3 domain GRB2-like endophilin B1), was originally identified as a Bax-binding protein by yeast two-hybrid screens using Bax as the bait (11, 22). The human Bif-1 gene encodes a 365 amino-acid polypeptide that contains an N-terminal BAR (Bin/Amphiphysin/Rvs) domain, a central coiled-coil domain, and a C-terminal SH3 domain. The N-terminal part (1-27 amino acids) of Bif-1 is required for its binding to Bax (11, 22). Moreover, the interaction between Bif-1 and Bax is enhanced in mammalian cells during apoptosis, which is accompanied by a conformational change in the Bax protein (11, 12, 23).
Overexpression of Bif-1 promotes Bax activation and apoptosis (11), whereas inhibition of Bif-1 expression suppresses Bax/Bak conformational activation, cytochrome c release, caspase activation, and cell death in response to intrinsic apoptosis signals (12). It has also been demonstrated that Bif-1 regulates apoptosis by mediating the mitochondrial fission process (24). This suggests that Bif-1 may represent a new type of Bax activator controlling the mitochondrial pathway of apoptosis.
Besides its role as a Bax activator, Bif-1 has recently been found to be involved in autophagosome formation and autophagic cell death (13). Bif-1 interacts with Beclin 1 through UVRAG to positively regulate the class III PI3-kinase (PI3KC3) lipid kinase during autophagy. Although the C-terminal SH3 domain of Bif-1 is sufficient for binding to UVRAG, the N-terminal BAR domain of Bif-1 is also required for Bif-1 to activate PI3KC3 lipid kinase and induce autophagosome formation. Suppression of Bif-1 expression inhibits autophagy and prolongs cell survival under nutrient starvation. Moreover, Bif-1 ablation promotes the development of spontaneous tumors in mice, consistent with the notion that both apoptosis and autophagy play crucial roles in tumor suppression (19, 25-27).
It has been shown that the Bif-1 mRNA levels are downregulated in lung carcinomas (28), and that approximately 60% gastric carcinomas express undetectable levels of Bif-1 protein (14). In addition, loss of heterozygosity (LOH) on 1p22, where the bif-1 gene is localized, is frequently observed in many types of tumor (29-37). In CRC 1p22 deletions were identified in >70% of advanced stage and metastatic tumors (36). These results are in agreement with other studies showing that 1p deletion was significantly more common in metastatic as compared to primary CRC (37). Others have described a 38% incidence of 1p deletions in 34 sporadic colorectal adenomas, using a centromeric probe for chromosome 1 and a simultaneous telomeric probe mapping to 1p36. These authors concluded that 1p deletion is an early event in colorectal tumorigenesis (38). These data and the observation that inhibition of Bif-1 expression promotes tumor development in mice (12, 13), propose Bif-1 as a candidate tumor suppressor gene.
In this study we found that the expression of Bif-1 was absent in 22.5% of CRC but all of the NR samples were Bif-1 positive. This difference was statistically significant (p=0.002). This finding is in agreement with the tumor suppressor function of Bif-1. Remarkably, this trend of Bif-1 protein expression down regulation in CRC was mirrored by significant decrease in the mRNA levels of Bif-1 at an early stage of colorectal cancer development. Since loss of Bif-1 not only suppresses Bax/Bak activation and apoptosis (12) but also inhibits PI3KC3 activation and autophagy (13), it remains to be determined whether the tumor suppressor activity of Bif-1 is due to its pro-apoptotic activity, pro-autophagic activity, or both.
A previous study has reported allelic loss in 1p36 and 1p32 regions of chromosome 1 as independent predictor of poor prognosis in patients with CRC (39). Here, complete follow up information was available only for a subset of the patients studied. The correlation of Bif-1 protein expression and patient survival in CRC is warrant further evaluation.
Acknowledgments
This study was supported by grants from the National Cancer Institute and the American Cancer Society.
We thank the Histology Section of the Tissue Core at the Moffitt Cancer Center and Research Institute for the support in performing the immunohistochemical stains. The DNA microarray analysis was performed in the Moffitt Microarray Core facility. This work was supported in part by grants from ACS (RSG-05-244-01-CCG) and NIH (CA82197) to HGW.
Footnotes
The authors have no conflict of interests.
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA: a cancer journal for clinicians. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Nishisho I, Nakamura Y, Miyoshi Y, et al. Science (New York, NY. Vol. 253. 1991. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients; pp. 665–9. [DOI] [PubMed] [Google Scholar]
- 4.Rupnarain C, Dlamini Z, Naicker S, Bhoola K. Colon cancer: genomics and apoptotic events. Biological chemistry. 2004;385:449–64. doi: 10.1515/BC.2004.053. [DOI] [PubMed] [Google Scholar]
- 5.Nechushtan A, Smith CL, Lamensdorf I, Yoon SH, Youle RJ. Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J Cell Biol. 2001;153:1265–76. doi: 10.1083/jcb.153.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei MC, Zong WX, Cheng EH, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science (New York, NY. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ming L, Wang P, Bank A, Yu J, Zhang L. PUMA Dissociates Bax and Bcl-X(L) to induce apoptosis in colon cancer cells. J Biol Chem. 2006;281:16034–42. doi: 10.1074/jbc.M513587200. [DOI] [PubMed] [Google Scholar]
- 8.Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 9.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 10.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 11.Cuddeback SM, Yamaguchi H, Komatsu K, et al. Molecular cloning and characterization of Bif-1: A novel SH3 domain-containing protein that associates with Bax. J Biol Chem. 2001;276:20559–65. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y, Karbowski M, Yamaguchi H, et al. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Molecular and cellular biology. 2005;25:9369–82. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, Jeong EG, Soung YH, et al. Decreased expression of tumour suppressor Bax-interacting factor-1 (Bif-1), a Bax activator, in gastric carcinomas. Pathology. 2006;38:312–5. doi: 10.1080/00313020600820880. [DOI] [PubMed] [Google Scholar]
- 15.Raff MC. Social controls on cell survival and cell death. Nature. 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 16.Green DR. Apoptotic pathways: the roads to ruin. Cell. 1998;94:695–8. doi: 10.1016/s0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- 17.Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–54. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 18.Reed JC. In: Mechanisms of Bcl-2 family protein function and dysfunction in health and disease. Krammer, Nagata, editors. Behring Institute Mitteilungen: Behring Institute Mitteilungen; 1996. pp. 72–100. [PubMed] [Google Scholar]
- 19.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 20.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nature reviews. 2001;2:211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 21.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 22.Pierrat B, Simonen M, Cueto M, Mestan J, Ferrigno P, Heim J. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics. 2001;71:222–34. doi: 10.1006/geno.2000.6378. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi H, Paranawithana SR, Lee MW, Huang Z, Bhalla KN, Wang H-G. Epothilone B analogue (BMS-247550)-mediated cytotoxicity through induction of Bax conformational change in human breast cancer cells. Cancer Res. 2002;62:466–71. [PubMed] [Google Scholar]
- 24.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166:1027–39. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science (New York, NY. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathew R, White E. Why sick cells produce tumors: the protective role of autophagy. Autophagy. 2007;3:502–5. doi: 10.4161/auto.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 28.Bonner AE, Lemon WJ, Devereux TR, Lubet RA, You M. Molecular profiling of mouse lung tumors: association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene. 2004;23:1166–76. doi: 10.1038/sj.onc.1207234. [DOI] [PubMed] [Google Scholar]
- 29.Balakrishnan A, von Neuhoff N, Rudolph C, et al. Quantitative microsatellite analysis to delineate the commonly deleted region 1p22.3 in mantle cell lymphomas. Genes Chromosomes Cancer. 2006;45:883–92. doi: 10.1002/gcc.20352. [DOI] [PubMed] [Google Scholar]
- 30.Walker GJ, Indsto JO, Sood R, et al. Deletion mapping suggests that the 1p22 melanoma susceptibility gene is a tumor suppressor localized to a 9-Mb interval. Genes Chromosomes Cancer. 2004;41:56–64. doi: 10.1002/gcc.20056. [DOI] [PubMed] [Google Scholar]
- 31.Lee WC, Balsara B, Liu Z, Jhanwar SC, Testa JR. Loss of heterozygosity analysis defines a critical region in chromosome 1p22 commonly deleted in human malignant mesothelioma. Cancer Res. 1996;56:4297–301. [PubMed] [Google Scholar]
- 32.Roberts T, Chernova O, Cowell JK. Molecular characterization of the 1p22 breakpoint region spanning the constitutional translocation breakpoint in a neuroblastoma patient with a t(1;10)(p22;q21) Cancer Genet Cytogenet. 1998;100:10–20. doi: 10.1016/s0165-4608(97)00013-7. [DOI] [PubMed] [Google Scholar]
- 33.Kakinuma H, Habuchi T, Ito T, et al. BCL10 is not a major _target for frequent loss of 1p in testicular germ cell tumors. Cancer Genet Cytogenet. 2001;126:134–8. doi: 10.1016/s0165-4608(00)00405-2. [DOI] [PubMed] [Google Scholar]
- 34.Mora J, Cheung NK, Kushner BH, et al. Clinical categories of neuroblastoma are associated with different patterns of loss of heterozygosity on chromosome arm 1p. J Mol Diagn. 2000;2:37–46. doi: 10.1016/S1525-1578(10)60613-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emi M, Matsumoto S, Iida A, et al. Correlation of Allelic Losses and Clinicopathological Factors in Primary Breast Cancers. Breast Cancer. 1997;4:243–6. doi: 10.1007/BF02966514. [DOI] [PubMed] [Google Scholar]
- 36.Bello MJ, de Campos JM, Vaquero J, Kusak ME, Sarasa JL, Rey JA. High-resolution analysis of chromosome arm 1p alterations in meningioma. Cancer Genet Cytogenet. 2000;120:30–6. doi: 10.1016/s0165-4608(99)00249-6. [DOI] [PubMed] [Google Scholar]
- 37.Knosel T, Petersen S, Schwabe H, et al. Incidence of chromosomal imbalances in advanced colorectal carcinomas and their metastases. Virchows Arch. 2002;440:187–94. doi: 10.1007/s004280100493. [DOI] [PubMed] [Google Scholar]


