Abstract
Mutant p53 gain of function contributes to cancer progression, increased invasion and metastasis potentials, and resistance to anticancer therapy. The ability of mutant p53 to acquire its gain of function is shown to correlate with increased expression of progrowth genes, such as c-MYC, MDR1, and NF-κB2. However, most of the published studies to identify mutant p53 _target genes were performed in a cell system that artificially overexpresses mutant p53. Thus, it remains unclear whether such mutant p53 _targets can be regulated by endogenous physiological levels of mutant p53. Here, we utilized SW480 and MIA-PaCa-2 cells, in which endogenous mutant p53 can be inducibly knocked down, to identify mutant p53 _target genes that potentially mediate mutant p53 gain of function. We found that knockdown of mutant p53 inhibits GRO1 expression, whereas ectopic expression of mutant R175H in p53-null HCT116 cells increases GRO1 expression. In addition, we found that endogenous mutant p53 is capable of binding to and activating the GRO1 promoter. Interestingly, ectopic expression of GRO1 can rescue the proliferative defect in SW480 and MIA-PaCa-2 cells induced by knockdown of mutant p53. Conversely, knockdown of endogenous GRO1 inhibits cell proliferation and thus abrogates mutant p53 gain of function in SW480 cells. Taken together, our findings define a novel mechanism by which mutant p53 acquires its gain of function via transactivating the GRO1 gene in cancer cells. Thus, _targeting GRO1 for cancer therapy would be applicable to a large portion of human tumors with mutant p53, but the exploration of GRO1 as a potential _target should take the mutation status of p53 into consideration.
It is known that mutation of p53 and/or inactivation of the p53 pathway are a hallmark of ∼50% human tumors. p53 is a tumor suppressor, and mutation of p53 leads to loss of its tumor-suppressing functions. Furthermore, a growing number of studies have provided compelling evidence that some p53 mutants have acquired additional functions that promote cancer development and progression, called gain-of-function (1, 2). Accordingly, gain of function was found to include the ability of mutant p53 to modulate specific _target genes, such as MDR1 and c-MYC (3, 4), which mediate tumor cells to acquire various tumorigenic properties. However, most of the published studies to identify mutant p53 _target genes were performed in a cell system that artificially overexpresses exogenous mutant p53 proteins. Thus, it remains unclear whether these mutant p53 _targets can be regulated by endogenous physiological levels of mutant p53.
Growth-regulated oncogene 1 (GRO1 or CXCL1) is found as an inflammatory factor and necessary for wound healing by modulating cell migration and angiogenesis (5, 6). GRO1, a 73-amino acid secreted protein, contains a 3-amino acid motif (ELR; glutamate, leucine, and arginine) between the N terminus and the first cysteine (7). GRO1 is structurally and functionally related to GRO2 (CXCL2), GRO3 (CXCL3), and interleukin-8. These four ligands constitute the CXC chemokine subfamily. All four ligands bind to the seven-transmembrane G-protein-coupled receptor CXCR2, with GRO1 having the highest affinity (8). Upon ligand-receptor interaction, various signaling cascades are activated, including ERK1/2, JAK2-STAT3, phosphatidylinositol 3-kinase, and tyrosine kinases, to promote cell proliferation, angiogenesis, and cell invasion and migration (9–11).
GRO1 is also found to be an oncogene and has pleiotropic effects on cell proliferation, tumor angiogenesis, invasion, and metastasis (9, 12). GRO1 is frequently overexpressed in many cancers, such as squamous cell carcinoma (12), breast cancer (13), ovarian cancer (14), colorectal cancer (15), and prostate cancer (16). Consistent with this, ectopic expression of GRO1 promotes low GRO1-producing cells to proliferate and spontaneous metastasis in BALB/c mice (17). Conversely, abolition of GRO1 via anti-GRO1-neutralizing antibody leads to reduction of tumor growth with decreased microvessel formation (15). Furthermore, GRO1 was found to be a potent mediator of tumor-associated angiogenesis in colorectal and breast cancers (15, 18). However, how GRO1 expression is deregulated in malignant cells is still poorly understood. Transcriptional analysis revealed that the GRO1 promoter contains several cis-acting elements, including the ones recognized by nuclear factor κB (NF-κB), Sp1, and human CUT homeodomain protein/CCAAT displacement protein (19). Indeed, NF-κB activity is found to be necessary for GRO1 expression in microvascular endothelial cells (20) and breast cancer cells (13). Thus, GRO1 is regarded as one of NF-κB _target genes (21).
In the present study, we demonstrated that GRO1 is transactivated by endogenous mutant p53 and serves as a crucial determinant for mutant p53 gain of function. We showed that endogenous mutant p53 directly binds to the GRO1 promoter and that knockdown of mutant p53 inhibits GRO1 expression. In addition, overexpression of mutant R175H in p53-null HCT116 cells increases GRO1 expression. Interestingly, ectopic expression of GRO1 can rescue the proliferative defect in SW480 and MIA-PaCa-2 cells induced by knockdown of mutant p53. Conversely, knockdown of endogenous GRO1 inhibits cell proliferation and thus abrogates mutant p53 gain of function in SW480 cells.
EXPERIMENTAL PROCEDURES
Small Hairpin RNA Oligonucleotides and Plasmids—To transiently knock down wild-type p53, a 21-bp p53 short hairpin RNA (shRNA),2 5′-GACUCCAGUGGUAAUCUACdTdT-3′, and a randomly selected scrambled shRNA, 5′-GCAGUGUCUCCACGUACUAdTdT-3′, were purchased from Dharmacon RNA Technologies. Transfection of p53 or scrambled shRNA was performed by using SilentFect™ lipid reagent (Bio-Rad).
To generate a construct that expresses p53 shRNA under the control of tetracycline, one pair of oligonucleotides was cloned into pBabe-H1 at HindIII and BglII sites, and the resulting construct was designated as pBabe-H1-p53shRNA. pBabe-H1 is a PolIII promoter-driven plasmid with a tetracycline operator sequence inserted before the transcriptional starting site (22). The shRNA oligonucleotides cloned into pBabe-H1-p53shRNA are sense (5′-GAT CCC C GA CTC CAG TGG TAA TCT ACT TCA AGA GAG TAG ATT ACC ACT GGA GTC TTT TTG GAA A-3′) and antisense (5′-AGC TTT TCC AAA AA G ACT CCA GTG GTA ATC TAC TCT CTT GAA GTA GAT TAC CAC TGG AGT CGG G-3′), with the shRNA _targeting region shown in boldface type.
To generate a construct that expresses GRO1 shRNA, one pair of oligonucleotides (sense, 5′-GAT CCC CGC TCA CTG GTG GCT GTT CCT TCA AGA GAG GA A CAG CCA CCA GTG AGC TTT TTG GAA A-3′; antisense, 5′-AGC TTT TCC AAA AA G CTC ACT GGT GGC TGT TCC TCT CTT GAA GGA ACA GCC ACC AGT GAG CGG G-3′; shRNA _targeting region shown in boldface type) was synthesized and cloned into pBabe-H1 at HindIII and BglII sites, and the resulting construct was designated as pBabe-H1-GRO1shRNA.
For stable or inducible overexpression of GRO1 in cancer cells, a cDNA fragment containing the GRO1 open reading frame was amplified from an expressed sequence tag clone (GenBank™ accession code NM_001511) with the forward primer (5′-GGA TCC ACC ATG GCC CGC GCT GCT CTC TCC G-3′) and the reverse primer (5′-CTC GAG TCA GTT GGA TTT GTC ACT GTT CAG-3′), confirmed by DNA sequencing, and then cloned into pcDNA3 and pcDNA4 tetracycline-inducible expression vector (Invitrogen). The resulting plasmids were designated as pcDNA3-GRO1 and pcDNA4-GRO1, respectively.
To generate a construct that expresses mutant p53(R175H), a DNA fragment containing the entire open reading frame of mutant p53(R175H) was amplified with a forward primer (5′-AAG CTT ACC ATG GGC TAC CCA TAC GAT GTT CCA GAT TAC GCT GAG GAG CCG CAG TCA GAT CC-3′) and a reverse primer (5′-CTC GAG TCA GTC TGA GTC AGG CCC TTC-3′) and confirmed by sequencing. The fragment was cloned into pcDNA4 and the resulting plasmid designated as pcDNA4-HA-p53(R175H). To generate a construct that expresses wild-type p53, a DNA fragment containing the entire open reading frame of wild-type p53 was amplified with a forward primer (5′-AAG CTT ACC ATG GAG GAG CCG CAG TCA GAT CC-3′) and a reverse primer (5′-CTC GAG TCA GTC TGA GTC AGG CCC TTC-3′) and confirmed by sequencing. The fragment was cloned into pcDNA3, and the resulting plasmid was designated as pcDNA3-wtp53.
The luciferase reporter under the control of the p21 promoter, pGL2-p21A, was as previously described (23). To generate a luciferase reporter under the control of the GRO1 promoter, a 100-bp DNA fragment containing the GRO1 promoter from nucleotide (nt) -50 to +50 was amplified using genomic DNA from SW480 cells with forward primer 5′-CTC GAG TTT CCA GCC CCA ACC ATG C-3′ and reverse primer 5′-AAG CTT GAG AGG AGC GGA AGA GCT G-3′. The PCR product, GRO1-50, was cloned into pGEM-T-Easy vector and confirmed by DNA sequencing. After digesting with XhoI and HindIII, the fragment was cloned into pGL2-Basic vector. The resulting luciferase reporter was designated as pGL2-GRO1-50.
Cell Culture—Human colon adenocarcinoma cell line SW480 (containing R273H/P309S), pancreatic cancer cell line MIA-PaCa-2 (containing R248W), and colon carcinoma cell lines HCT116 and HCT116 (p53-/-) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with ∼10% fetal bovine serum (Hyclone). SW480-p53-KD and MIA-PaCa-2-p53-KD cell lines, in which shRNA _targeting p53 is inducibly expressed under the control of the tetracycline-regulated promoter, were generated as described (24). To generate cell lines in which mutant p53 is inducibly knocked down and GRO1 is stably overexpressed, pBabe-H1-p53shRNA was cotransfected with pcDNA3-GRO1 into SW480 and MIA-PaCa-2 cells, which express a tetracycline repressor by pcDNA6. The resulting p53 knockdown and GRO1-stable overexpression cell lines were selected with puromycin and G418. The efficiency of p53 knockdown and GRO1-stable overexpression was confirmed by Western blot analysis with anti-p53 and anti-GRO1 antibodies.
To generate cell lines that inducibly overexpress GRO1, pcDNA4-GRO1 was transfected into SW480 cells, which express a tetracycline repressor by pcDNA6. The resulting GRO1 overexpression cell lines were selected with Zeocin, and inducible GRO1 overexpression was then confirmed by Western blot analysis with anti-GRO1 antibody.
To generate cell lines that inducibly overexpress mutant p53(R175H), pcDNA4-HA-p53(R175H) was transfected into HCT116(p53-/-) cells, which express a tetracycline repressor by pcDNA6. The resulting p53(R175H) overexpression cell lines were selected with Zeocin, and inducible mutant p53 overexpression was then confirmed by Western blot analysis with anti-p53 antibody.
Affymetrix GeneChip, Northern Blot, and Real-time PCR Analyses—Total RNA was isolated from cells using TRIzol reagent (Invitrogen). U133 Plus 2.0 arrays, which contain probe sets for analysis of over 47,000 human transcripts, were purchased from Affymetrix. GeneChip analysis was performed according to the manufacturer's instructions. Northern blot analysis was performed as described previously (24). The GRO1 probe was prepared from an expressed sequence tag clone (GenBank™ accession code NM_001511). The mRNA level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was examined as a loading control. Real-time PCR was conducted using a Realplex2 system (Eppendorf). cDNA was synthesized using a Iscript™ cDNA Synthesis kit (Bio-Rad). To quantify the GRO1 mRNA, real-time PCR was done with the forward primer 5′-AGG GAA TTC ACC CCA AGA AC-3′ and the reverse primer 5′-ACT ATG GGG GAT GCA GGA TT-3′.To quantify the p65 mRNA, quantitative real-time PCR was done with the forward primer 5′-CTG CCG GGA TGG CTT CTA T-3′ and the reverse primer 5′-CCG CTT CTT CAC ACA CTG GAT-3′. To quantify the p52 mRNA, real-time PCR was done with the forward primer 5′-GGG GCA TCA AAC CTG AAG ATT TCT-3′ and the reverse primer 5′-TCC GGA ACA CAA TGG CAT ACT GT-3′. GAPDH was amplified as internal control using the forward primer 5′-AGC CTC AAG ATC ATC AGC AAT G-3′ and the reverse primer 5′-ATG GAC TGT GGT CAT GAG TCC TT-3′. The changes in the levels of GRO1, p65, and p52 mRNA relative to that of GAPDH mRNA were determined using a threshold cycle method to calculate changes in threshold cycle and then folds. An average value for each gene was obtained from triplicate reactions.
Luciferase Assay—The assay was performed in triplicate according to the manufacturer's instructions (Promega). Briefly, 0.25 μg of a luciferase reporter, 0.25 μg of pcDNA3 or pcDNA3 that expresses mutant p53 or wild-type p53, and 5 ng of Renilla luciferase assay vector pRL-CMV (Promega) were co-transfected into p53-null HCT116 cells. The -fold increase in relative luciferase activity is a product of the luciferase activity induced by mutant p53 divided by that induced by an empty pcDNA3 vector. The pGL2-p21A luciferase reporter under the control of the two p53-responsive elements in the p21 promoter was used as a control (19).
Chromatin Immunoprecipitation (ChIP) Assay—The assay was performed as previously described (17, 25). SW480-p53-shRNA cells, which were uninduced (-) or induced (+) to knock down mutant p53 for 3 days, were cross-linked by 1% formaldehyde for 10 min at room temperature. Nuclear extracts were prepared, and chromatin was sonicated to generate 200–1000-bp DNA fragments. Protein-DNA complexes were immunoprecipitated with various antibodies. The DNA-protein cross-links were reversed by heating at 65 °C for 4 h. After phenol and chloroform extraction, DNA was purified by ethanol precipitation. To detect mutant p53 binding site in the GRO1 promoter, three regions, including GRO1-C1 (from nt -219 to +6), GRO1-C2 (from nt -1938 to -1719), and GRO1-C3 (from nt -3806 to -3611), were amplified by PCR. Three pairs of PCR primers were used to amplify: GRO1-C1 with forward 5′-TCA GAG TCC ACA GGA GTT ACT-3′ and reverse 5′-TCT GTG GCT CTC CGA GAT CC-3′; GRO1-C2 with forward 5′-AGA AAC AAC ATT CTA GCA CAC C-3′ and reverse 5′-TAT ATC AGT TAC TGC TAC CCA AC-3′; and GRO1-C3 with forward 5′-TGA CAC ATT ACT TTT AAC TGA AC-3′ and reverse 5′-CTC TTC AAA ATT GTC AAT GTC ATG-3′.
To amplify the region from nt -2312 to -2131 in the p21 promoter, PCR was performed with forward primer 5′-CAG GCT GTG GCT CTG ATT GG-3′ and reverse primer 5′-TTC AGA GTA ACA GGC TAA GG-3′. A region in the GAPDH promoter was amplified to serve as a nonspecific binding control with forward primer 5′-AAA AGC GGG GAG AAA GTA GG-3′ and reverse primer 5′-AAG AAG ATG CGG CTG ACT GT-3′.
DNA Pull-down Assay—The pull-down assay was performed as described (25, 26). The biotin-labeled promoter probe (nt -219 to +6 in the GRO1 gene) was synthesized by PCR with 5′-biotinated forward primer 5′-TCA GAG TCC ACA GGA GTT ACT and reverse primer 5′-TCT GTG GCT CTC CGA GAT CC. The biotin-labeled probe (nt -205 to +16 in the GAPDH gene), which does not contain a known NF-κB binding site, was also synthesized by PCR with 5′-biotinated forward primer 5′-CTA CGT CGG GGC CCA CAC and reverse primer 5′-GCT GCG GGC TCA ATT TAT AG and used as a negative binding control. The biotinated probes were then bound to streptavidin-agarose beads and used to precipitate a protein of interest from nuclear protein extracts. All steps of nuclear extract isolation were done at 4 °C. Cells were resuspended in two package cell volumes of a buffer containing 10 mm HEPES, pH 7.9, 1.5 mm MgC12, 10 mm KCl, 300 mm sucrose, 0.5% Nonidet P-40, 1 mm Na3VO4, 0.5 mm dithiothreitol, 0.5 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 25 mm β-glycerophosphate, and 10 mm NaF. After centrifugation, the cells were resuspended in two-thirds package cell volume of a buffer containing 20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, 2.5% glycerol, and various protein inhibitors as above. After sonication and centrifugation, the supernatant was isovolumetrically diluted with a buffer containing 20 mm HEPES, pH 7.9, 100 mm KC1, 0.2 mm EDTA, 8% glycerol, and inhibitors. The binding assay was performed by mixing 200 μg of nuclear protein extracts, 2 μg of a biotin-labeled probe, 20 μl of 50% streptavidin-agarose beads (Prozyme), and 500 μl of phosphate-buffered saline with various protein inhibitors (PBSI). The mixture was incubated at room temperature for 2 h with rocking. Beads were then pelleted down by centrifugation, washed with PBSI three times, and resuspended in 2× Laemmli sample buffer. After centrifugation, the supernatant was collected for Western blot assay.
Immunofluorescence Staining—Cells were grown on a 4-well chamber slide and treated as indicated. After being washed with phosphate-buffered saline, cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100 for 10 min, blocked with 10% fetal bovine serum for 30 min, and then incubated with mouse anti-p53 and rabbit anti-p65 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4 °C overnight, followed by incubation with fluorescein isothiocyanate- and Texas Red-conjugated secondary antibodies (Jackson Immuno-Research and Molecular Probes). Cells were also stained with 4′,6′-diamidino-2-phenylindole (Sigma) to visualize nuclei and then mounted with a solution containing 0.1% of purified protein derivative (Sigma) and 80% glycerol in phosphate-buffered saline. Intracellular distribution of proteins was analyzed by immunofluorescence microscopy.
Antibodies—Mouse anti-GRO1 and rabbit anti-p65 were purchased from Abcam Inc. and Santa Cruz Biotechnology, respectively. Antibodies against p53, hemagglutinin epitope, and actin were described previously (26). The level of actin protein was measured as a loading control.
Colony Formation Assay—SW480 or MIA-PaCa-2 cells (1000 cells/well) in a 6-well plate were cultured in the absence or presence of tetracycline (1.0 μg/ml) for 72 h and then untreated or treated with camptothecin as indicated for 4 h, followed by one wash with Dulbecco's modified Eagle's medium to remove the drug. The cells were maintained in fresh medium for the next 15–17 days and then fixed with methanol/glacial acetic acid (7:1) and stained with 0.1% crystal violet.
Statistics—All experiments were performed in triplicates. Two-group comparisons were analyzed by two-sided Student's t test. p values were calculated, and p < 0.05 was considered significant.
RESULTS
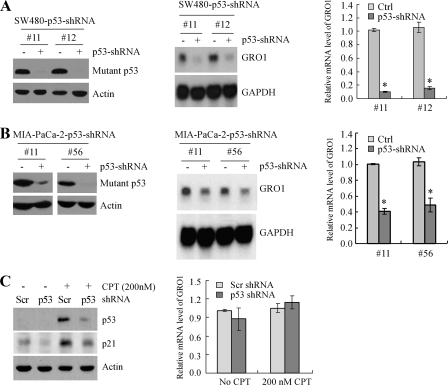
Knockdown of Mutant p53 Inhibits GRO1 Expression in SW480 and MIA-PaCa-2 Cells—Both in vitro and in vivo studies demonstrated that in addition to loss of tumor-suppressing functions, mutant p53 contributes to the malignant process by acquiring additional activities, including resistance to anticancer therapy and increased invasion and metastasis potential (2, 27). Through overexpression, some mutant p53 is capable of transactivating genes involved in cell growth and survival, such as c-MYC, MDR1, and NF-κB2 (4, 28, 29). However, it is not certain whether these genes can be regulated by physiologically relevant levels of mutant p53 in tumor cells. To address this, we utilized SW480 and MIA-PaCa-2 cell lines in which endogenous mutant p53 can be inducibly knocked down by shRNA under the control of the tetracycline-regulated promoter. Several representative p53 knockdown SW480 (Fig. 1A, left) and MIA-PaCa-2 (Fig. 1B, left) cell lines were shown. Upon induction of shRNA against p53, the levels of mutant p53 protein were obviously reduced. Furthermore, to determine whether endogenous mutant p53 has a transcriptional activity, microarray analyses were used to examine the pattern of gene expression in both SW480 and MIA-PaCa-2 cells, which were uninduced or induced to knock down endogenous mutant p53. We found that GRO1 expression was significantly down-regulated upon mutant p53 knockdown. Next, Northern blot analyses and quantitative real-time PCRs were performed and showed that the levels of GRO1 mRNA were markedly decreased upon knockdown of mutant p53 in SW480 (Fig. 1A, middle and right) and MIA-PaCa-2 (Fig. 1B, middle and right) cells. The levels of GAPDH mRNA were examined as a control in both assays. To examine whether down-regulation of GRO1 in these cells is specifically due to mutant p53 knockdown, wild-type p53 in HCT116 cells was transiently knocked down by p53 shRNA. We found that the protein levels of wild-type p53 and its _target p21 under both the basal and stress (treated with camptothecin) conditions were reduced by shRNA against p53 but not scrambled mock shRNA (Fig. 1C, left). However, knockdown of wild-type p53 had little if any effect on the relative mRNA level of GRO1 (Fig. 1C, right). These data suggest that GRO1 is likely to be a common _target of mutant R273H/P309S in SW480 cells and mutant R248W in MIA-PaCa-2 cells.
FIGURE 1.
Knockdown of mutant p53 inhibits GRO1 expression in SW480 and MIA-PaCa-2 cells. A, left, generation of SW480 cell lines in which mutant p53 can be inducibly knocked down. Western blots were prepared with extracts from SW480 cells uninduced (-) or induced (+) to knock down mutant p53 for 3 days and then probed with antibodies against p53 and actin, respectively. Middle, knockdown of mutant p53 in SW480 cells down-regulates GRO1 expression. Northern blots were prepared with total RNAs isolated from SW480 cells treated as above. The blots were probed with cDNAs derived from the GRO1 and GAPDH genes, respectively. The level of GAPDH mRNA was measured as a loading control. Right, the relative levels of GRO1 transcript were measured by quantitative reverse transcription-PCR and normalized by levels of GAPDH mRNA. B, left, generation of MIA-PaCa-2 cell lines in which mutant p53 can be inducibly knocked down. Western blot analyses were performed as in A. Middle and right, knockdown of mutant p53 in MIA-PaCa-2 cells down-regulates GRO1 expression. Northern blot and quantitative reverse transcription-PCR analyses were performed as in A. C, left, transient knockdown of wild-type p53 has no effect on Gro1 expression in HCT116 cells. Western blots were prepared with extracts from HCT116 cells with transient knockdown of wild-type p53 for 3 days and then probed with antibodies against p53, p21, and actin, respectively. A scrambled shRNA was used as a negative control. Right, the relative levels of GRO1 transcript in HCT116 cells with or without wild-type p53 knockdown were quantified by real-time PCR.
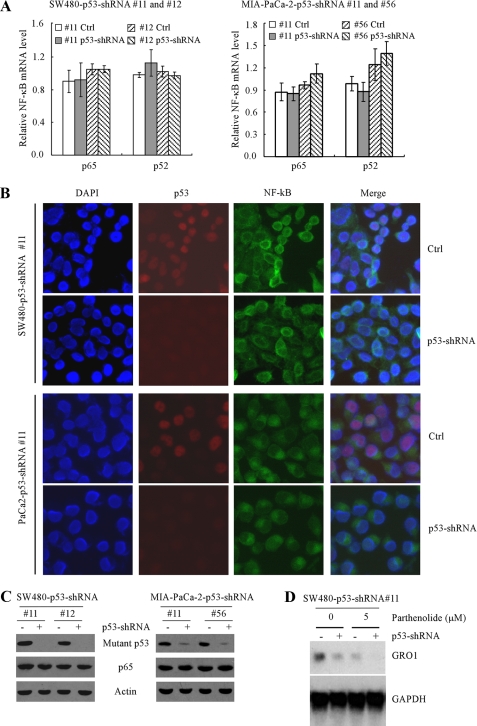
The NF-κB Pathway Is Not Required for Mutant p53 to Regulate GRO1 Expression—Previous studies showed that GRO1 is an NF-κB _target gene (21), and down-regulation of the NF-κB activity inhibits GRO1 expression (13, 20). Interestingly, wild-type p53 was found to inhibit, whereas mutant p53 was found to increase, the expression and/or activity of the NF-κB pathway (28, 30, 31). Thus, we assessed whether knockdown of mutant p53 leads to reduced expression or activity of NF-κB, which then down-regulates GRO1 expression. To test this, real-time PCR was used to quantify the relative levels of p65 (RelA) and p52 (NF-κB2) transcripts in two SW480 cell lines (Fig. 2A, left) and two MIA-PaCa-2 cell lines (Fig. 2A, right) that were uninduced or induced to knock down mutant p53. We found that knockdown of mutant p53 had little if any effect on the level of p65 and p52 mRNAs in SW480 cells (Fig. 2A, left) as well as in MIA-PaCa-2 cells (Fig. 2A, right). Consistent with this, we found that the localization (Fig. 2B) and level (Fig. 2C) of p65 protein were not obviously altered in SW480 and MIA-PaCa-2 cells with knockdown of mutant p53. To further address the contribution of mutant p53 and NF-κB pathways to GRO1 expression, SW480-p53-shRNA cells (clone 11) were treated with 5 μm parthenolide, a specific inhibitor of the NF-κB pathway, along with inducible knockdown of mutant p53. Northern blot analysis was performed and showed that although parthenolide reduced the overall level of GRO1 expression, knockdown of mutant p53 significantly inhibited GRO1 expression regardless of treatment with parthenolide (Fig. 2D). Thus, we conclude that mutant p53 regulates GRO1 independent of the NF-κB pathway.
FIGURE 2.
Down-regulation of GRO1 induced by mutant p53 knockdown is independent of the NF-κB pathway in SW480 and MIA-PaCa-2 cells. A, knockdown of mutant p53 had no obvious effect on the levels of p65 and p52 mRNA in SW480 cells (left) and MIA-PaCa-2 cells (right). Quantitative reverse transcription-PCR was performed with total RNAs isolated from SW480 and MIA-PaCa-2 cells that were uninduced or induced to knock down mutant p53 for 3 days. B, knockdown of mutant p53 had no obvious effect on the localization of p65 protein in SW480 cells (top two panels) and MIA-PaCa-2 cells (bottom two panels). Immunofluorescence staining was performed to detect the cellular location of p65 and p53 as described under “Experimental Procedures.” C, knockdown of mutant p53 had no obvious effect on the levels of p65 protein in SW480 cells (left) and MIA-PaCa-2 cells (right). Western blots were prepared with extracts from SW480 and MIA-PaCa-2 cells treated as in A and then probed with antibodies against p53, p65, and actin, respectively. D, knockdown of mutant p53 decreased the level of GRO1 mRNA regardless of inhibition of the NF-κB pathway. SW480-p53-shRNA cells (clone 11) were uninduced (-) or induced (+) to knock down mutant p53 along with treatment of 0 or 5 μm parthenolide, a specific inhibitor of the NF-κB pathway. The levels of GRO1 mRNA were measured by Northern blot analysis as in Fig. 1.
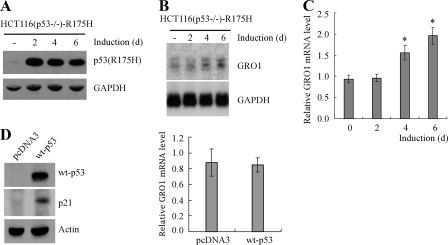
Ectopic Expression of Mutant p53 (R175H) in p53-null HCT116 Cells Increases GRO1 Expression—To further confirm that GRO1 is a _target gene of mutant p53, GRO1 expression was examined in p53-null HCT116 cells that inducibly expressed tumor-derived mutant R175H under the tetracycline-regulated promoter. As seen in Fig. 3A, mutant R175H protein was induced over a 6-day testing period. To examine the effect of mutant p53 on GRO1 expression, Northern blot analysis was performed and showed that the level of GRO1 transcripts was progressively increased upon expression of mutant R175H over the 6-day testing period (Fig. 3B). Alternatively, quantitative real-time PCR was performed and showed that the relative level of GRO1 transcripts was significantly increased upon ectopic expression of mutant p53 at days 4 and 6 after normalization by the level of GAPDH mRNA (Fig. 3C). As a control, we examined the effect of wild-type p53 on GRO1 expression in p53-null HCT116 cells. We found that wild-type p53 was expressed and transcriptionally active as evidenced by the induction of p21 (Fig. 3D, left). However, overexpression of wild-type p53 had little if any effect on the relative level of GRO1 mRNA (Fig. 3D, right). We note here that the ability of exogenous mutant p53 to regulate GRO1 in p53-null HCT116 cells is less potent with delayed kinetics compared with that of endogenous mutant p53 in SW480 and MIA-PaCa-2 cell lines. This is not surprising, since endogenous and exogenous p53 can be differentially modified and thus have different transcriptional activity (32). Together, we concluded that GRO1 is likely to be a mutant p53 _target gene.
FIGURE 3.
GRO1 expression was increased by ectopic expression of mutant p53 (R175H) in p53-null HCT116 cells. A, Western blots were prepared with extracts from p53-null HCT116 cells uninduced (-) or induced to express mutant R175H for 2, 4, or 6 days and then probed with antibodies against p53 and actin, respectively. B, endogenous GRO1 expression was increased by ectopic expression of mutant R175H. Northern blots were prepared with total RNAs purified from p53-null HCT116 cells uninduced (-) or induced to express mutant p53 for 2, 4, or 6 days and probed with cDNAs derived from the GRO1 and GAPDH genes, respectively. C, the relative levels of GRO1 transcript in p53-null HCT116 cells induced to express R175H for 0–6 days were quantified by real-time reverse transcription-PCR. D, left, Western blots were prepared with extracts from p53-null HCT116 cells with or without transient overexpression of wild-type p53 for 4 days. Right, the relative levels of GRO1 transcript in p53-null HCT116 cells in the presence or absence of wild-type p53 for 4 days were quantified by real-time PCR.
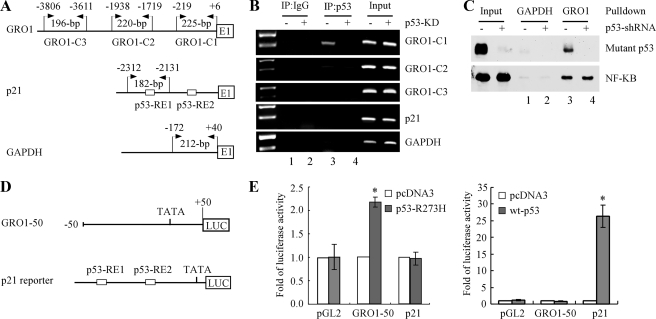
Mutant p53 Binds to and Transactivates the GRO1 Promoter—To determine whether GRO1 is a bona fide _target gene of mutant p53 in vivo, we attempted to identify a potential binding site responsible for activation by mutant p53. Although a consensus p53-responsive element has been defined for wild-type p53, such a responsive element for mutant p53 has not been established. Thus, a ChIP assay was performed with three pairs of primers to scan a large section throughout the GRO1 promoter (nt -3806 to -1) (Fig. 4A). Chromatins were isolated from SW480 cells, which were uninduced or induced to knock down mutant p53, and then immunoprecipitated with anti-p53 antibody or an IgG as a control. In addition, potential binding of mutant p53 to the p21 and GAPDH promoters was also analyzed (Fig. 4A). We showed that a 225-bp fragment located in the proximal GRO1 promoter (nt -219 to +6) was detected in mutant p53-containing chromatins but not mutant p53-knockdown chromatins (Fig. 4B, GRO1-C1 panel, compare lanes 3 and 4). The fragment between nt -1938 and -1719 in the GRO1 promoter was weakly amplified from mutant p53-containing chromatins, whereas the distal upstream region between nt -3806 and -3611 was not detected (Fig. 4B, GRO1-C2 and -C3 panels). For IgG antibody control, no GRO1 promoter fragment was detected (Fig. 4B). In addition, the p53-responsive element in the p21 gene and the GAPDH promoter were not found to interact with mutant p53 (Fig. 4B, p21 and GAPDH panels).
FIGURE 4.
Mutant p53 can bind to and transactivate the GRO1 promoter. A, schematic presentation of the GRO1 (top), p21 (middle), and GAPDH (bottom) promoters and the location of PCR primers used for ChIP assay. B, the binding of mutant p53 to the GRO1 promoter in SW480 cells was measured by ChIP. Mutant p53-DNA complexes were captured with anti-p53 antibody along with rabbit IgG as a control. The binding of mutant p53 protein to the p21 and GAPDH promoters was also measured as negative controls. C, pull-down assay of mutant p53 (top) and NF-κB (p65) (bottom) binding to the GRO1 promoter in vitro. The biotinated GRO1 promoter probe bound to streptavidin-agarose beads was used to precipitate p65 and mutant p53 from nuclear protein extracts from SW480 cells uninduced (-) or induced (+) to knock down mutant p53 for 3 days. The biotinated GAPDH promoter probe was used as a negative binding control. The nuclear proteins pulled down by biotinated promoter probes were analyzed by Western blot assay with antibodies against p53 and p65, respectively. D, schematic presentation of the luciferase reporter constructs under the control of the GRO1 (top) and p21 (bottom) promoters, respectively. E, the GRO1 promoter is responsive to mutant p53-R273H (left) but not wild-type p53 (right). The response of the p21 promoter to wild-type p53 was measured as a positive control. The luciferase activity for the promoterless pGL2 plasmid in the presence of wild-type and mutant p53 was measured as a negative control. Luciferase assay was performed as described under “Experimental Procedures.”
In parallel, we performed a DNA pull-down assay to confirm the physical interaction between mutant p53 and the GRO1 promoter in vitro using the biotinated GRO1 promoter probe (nt -219 to +6) immobilized on streptavidin-agarose beads and nuclear protein extracts from SW480 cells uninduced or induced to knock down mutant p53. The biotin-labeled GRO1 promoter probe also contains a NF-κB binding site (nt -77 to -67) (11). Therefore, a pull-down assay was conducted to detect p65 as a positive control. Indeed, we found that p65 protein was pulled down in SW480 cells regardless of mutant p53 knockdown (Fig. 4C, bottom, lanes 3 and 4). In addition, we found that mutant p53 was pulled down by the biotinated GRO1 promoter probe in SW480 cells with mutant p53 (Fig. 4C, top, lane 3). However, upon induction of shRNA against p53, mutant p53 was undetectable (Fig. 4C, top, lane 4). Furthermore, mutant p53 and p65 proteins were not pulled down when the biotinated GAPDH promoter probe was used (Fig. 4C, lanes 1 and 2).
Next, a luciferase reporter assay was performed to determine whether the GRO1 promoter is responsive to mutant p53. To test this, a proximal GRO1 promoter region (nt -50 to +50), with which mutant p53 was found to interact by ChIP assay, was cloned into the pGL2 promoterless luciferase reporter vector. The resulting vector was designated GRO1-50 (Fig. 4D). The luciferase reporter was cotransfected into p53-null HCT116 cells with either a pcDNA3 control vector or a vector that expresses mutant p53(R273H). We found that the luciferase activity for GRO1-50 was significantly increased by mutant p53(R273H) (Fig. 4E, left) but not wild-type p53 (Fig. 4E, right). However, the luciferase reporter under the control of the p21 promoter was responsive to wild-type p53 (Fig. 4E, right) but not p53(R273H) (Fig. 4E, left). Furthermore, the luciferase activity from empty pGL2 plasmid was not responsive to either mutant or wild-type p53 (Fig. 4E, left and right). These are consistent with the observations by ChIP and DNA pull-down assays. However, the precise responsive element for mutant p53 in the GRO1 promoter needs to be further characterized, which is beyond the scope of this study.
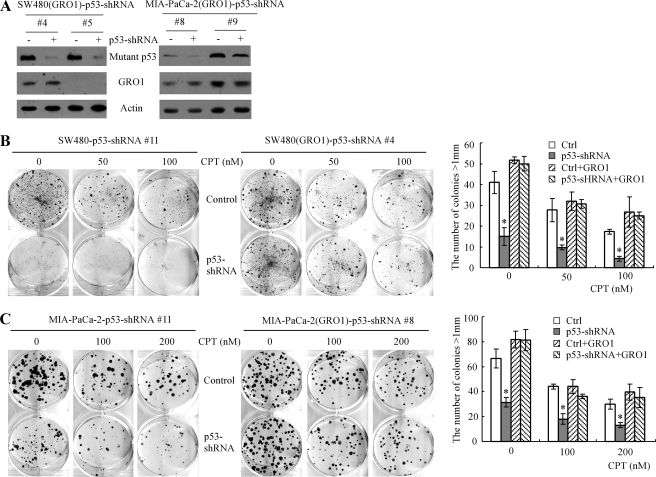
Ectopic Expression of GRO1 Can Rescue the Proliferative Defect Induced by Knockdown of Mutant p53 in SW480 and MIA-PaCa-2 Cell Lines—To assess whether GRO1 is sufficient to mediate mutant p53 gain of function, we generated SW480 and MIA-PaCa-2 cell lines in which GRO1 is stably overexpressed along with inducible knockdown of mutant p53. Two representative SW480 cell lines (clones 4 and 5) and two representative MIA-PaCa-2 cell lines (clones 8 and 9) were chosen for further study. Upon induction of shRNA against p53, the levels of p53 protein were obviously reduced (Fig. 5A, top). In addition, GRO1 was found to be stably expressed in one SW480 cell line (clone 4) and in both MIA-PaCa-2 cell lines (clones 8 and 9) (Fig. 5A, GRO1 panel). Next, a colony formation assay was performed to measure the effect of GRO1 on mutant p53 gain of function. We showed that upon inducible knockdown of mutant p53 in SW480 cells (Fig. 5B, left and right) and in MIA-PaCa-2 cells (Fig. 5C, left and right), the colony formation ability was markedly inhibited regardless of treatment with camptothecin. In contrast, upon stable expression of GRO1 in SW480 (Fig. 5B, middle and right) and in MIA-PaCa-2 (Fig. 5C, middle and right), inducible knockdown of mutant p53 had little if any effect on colony formation both under mock treatment and treatment with two different doses of camptothecin. This finding suggests that mutant p53 is required for cell proliferation of SW480 and MIA-PaCa-2 cells, and ectopic expression of GRO1, as a _target of mutant p53, is sufficient to compensate for knockdown of mutant p53.
FIGURE 5.
Ectopic expression of GRO1 can rescue the proliferative defect induced by mutant p53 knockdown. A, generation of SW480 and MIA-PaCa-2 cell lines in which GRO1 was stably expressed and endogenous mutant p53 can be inducibly knocked down. Western blots were prepared with extracts from SW480 and MIA-PaCa-2 cells uninduced (-) or induced (+) to knock down mutant p53 for 3 days and then probed with antibodies against p53 and actin, respectively. B and C, left, knockdown of mutant p53 inhibits colony formation in SW480 cells (B) and MIA-PaCa2 cells (C) that were mock-treated or treated with 50–200 nm camptothecin (CPT). Middle, ectopic expression of GRO1 rescues the proliferative defect induced by mutant p53 knockdown in SW480 cells (B) and MIA-PaCa2 cells (C) that were mock-treated or treated with 50–200 nm camptothecin. Right, quantification of the number of colonies with a diameter of >1 mm. *, p < 0.05.
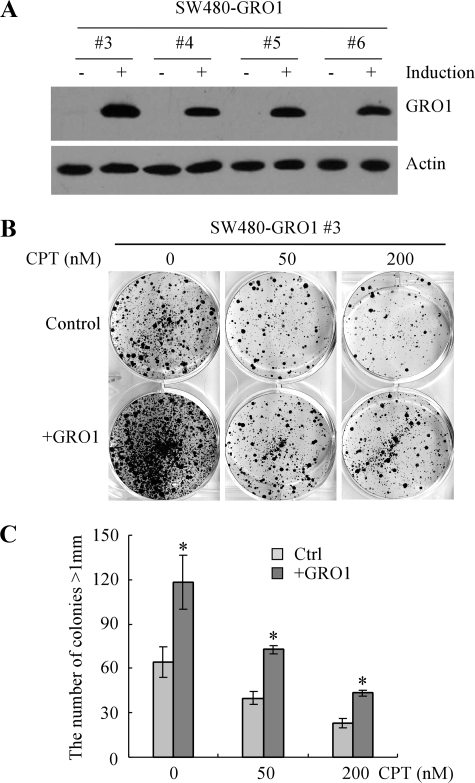
Inducible Expression of GRO1 Promotes Cell Proliferation and Confers Cell Resistance to DNA Damage—Since mutant p53 has been implicated in promoting cell survival and conferring cell resistance to DNA damage by inducing GRO1 expression, we tested whether ectopic expression of GRO1 is capable of mimicking mutant p53 activity. For this purpose, we generated SW480 cell lines in which GRO1 can be inducibly expressed under the control of the tetracycline-regulated promoter. Upon induction with tetracycline, GRO1 protein was inducibly expressed (Fig. 6A). A colony formation assay showed that upon inducible expression of GRO1, the colony-forming ability of SW480 cells was increased (Fig. 6, B and C). In addition, upon inducible expression of GRO1, the ability of SW480 cells to proliferate following treatment with camptothecin was also markedly increased (Fig. 6, B and C). These data suggest that overexpression of GRO1 in SW480 cells is able to enhance cell proliferation and desensitize cancer cells to anticancer drugs.
FIGURE 6.
Ectopic expression of GRO1 promotes cell proliferation in SW480 cells. A, generation of SW480 cell lines in which GRO1 can be inducibly expressed under the control of the teteracycline-regulated promoter. Western blots were prepared with extracts from SW480 cells uninduced (-) or induced (+) to express GRO1 for 24 h and then probed with antibodies against GRO1 and actin, respectively. B, ectopic expression of GRO1 alone is capable of promoting cell proliferation in SW480 cells. SW480-GRO1 cells were uninduced or induced to express GRO1 along with treatment of 0, 50, or 200 nm CPT, cultured for ∼3 weeks, and then fixed and stained to measure the number and size of colonies formed. C, quantification of the number of colonies with a diameter of >1 mm shown in B.*, p < 0.05.
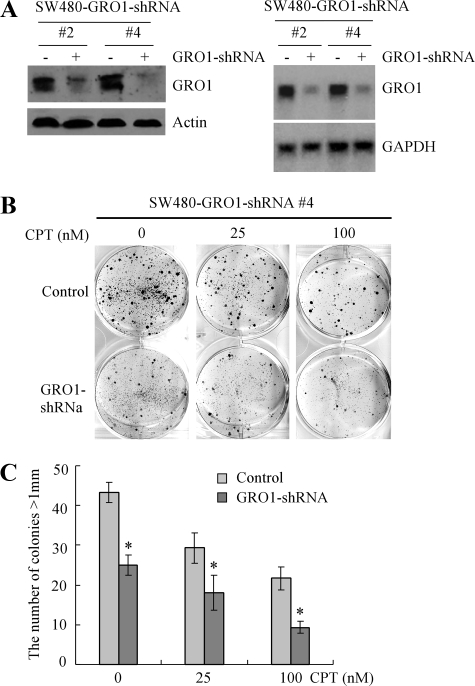
GRO1 Is Required for Mutant p53 Gain of Function—To determine whether GRO1 is required for mutant p53 gain of function, we generated SW480 cell lines in which endogenous GRO1 can be inducibly knocked down by GRO1-specific shRNA. Two representative cell lines were chosen for further study. We showed that upon induction of shRNA against GRO1, the levels of GRO1 protein (Fig. 7A, left) and transcript (Fig. 7A, right) were obviously reduced. Next, we examined the colony-forming ability of SW480 cells upon GRO1 knockdown. We showed that cell proliferation was inhibited by knockdown of GRO1 (Fig. 7, B (left) and C). In addition, the susceptibility of SW480 cells to camptothecin was also increased as the number and especially the size of colonies were decreased upon GRO1 knockdown (Fig. 7, B (middle and right) and C). Together, our data indicate that one mechanism by which mutant p53 promotes cell proliferation and chemoresistance in cancer cells is by up-regulation of GRO1 expression.
FIGURE 7.
Knockdown of endogenous GRO1 inhibits cell proliferation in SW480 cells. A, generation of SW480 cell lines in which endogenous GRO1 can be inducibly knocked down. Left, Western blots were prepared with extracts from SW480 cells uninduced (-) and induced (+) to knock down GRO1 for 3 days and then probed with antibodies against GRO1 and actin, respectively. Right, Northern blots were prepared with total RNAs purified from SW480 cells uninduced (-) and induced (+) to knock down GRO1 for 3 days and probed with cDNAs derived from the GRO1 and GAPDH genes, respectively. B, knockdown of GRO1 inhibits colony formation in SW480 cells. SW480 cells were uninduced or induced to knock down GRO1 along with treatment of 0, 25, or 100 nm CPT, cultured for ∼3 weeks, and then fixed and stained to measure the number and size of colonies formed. C, quantification of the number of colonies with a diameter of >1 mm shown in B.*, p < 0.05.
DISCUSSION
Both GRO1 and wild-type p53 are known to regulate a network associated with cell growth (2, 33, 34). Deregulation of GRO1 and p53, which occurs frequently in cancer, leads to abnormal cell growth and subsequently cancer development and progression (2, 34). Until recently, GRO1 and mutant p53 are still regarded as two independent oncogenic factors. However, recent evidence points to a multilevel and complex cooperation between the CXC chemokine family and wild-type p53. For example, wild-type p53 represses CXCR4 and CXCL12 expression (35) and thus suppresses cell migration and invasion. Here, we showed that knockdown of mutant p53 inhibits, whereas ectopic expression of mutant p53 increases, GRO1 expression. In addition, the GRO1 promoter contains a region that interacts with and is responsive to mutant p53. Interestingly, ectopic expression of GRO1 can rescue the proliferative defect induced by knockdown of mutant p53 in SW480 and MIA-PaCa-2 cells. Conversely, knockdown of endogenous GRO1 inhibits cell proliferation and abrogates mutant p53 gain of function in SW480 cells. Thus, _targeting GRO1 for cancer therapy would be applicable to a large portion of human tumors with mutant p53, but exploration of GRO1 as a potential _target should take into consideration the mutation status of p53.
Due to its potent oncogenic properties, GRO1 has been subject to extensive investigations, especially its transcriptional regulation by NF-κB (36). NF-κB was found to be one of the major regulators for GRO1 expression (37, 38). The proximal GRO1 promoter was found to contain an NF-κB binding site along with a TATA box and an immediate upstream region. Since mutant p53 was found to increase NF-kB2 expression for its gain of function (28), it is possible that mutant p53 regulation of GRO1 may be mediated by NF-κB. However, we demonstrated that knockdown of mutant p53 has little if any effect on expression levels of NF-κB subunits, p65 (RelA) and p52 (NF-κB2), and localization of p65. Considering the fact that the mutant p53 was found to interact with and activate the GRO1 promoter, it is reasonable to believe that GRO1 is a direct _target of mutant p53. Consistent with this observation, a recent report showed that in young adult murine colon cells that overexpress mutant p53(R175H) and activated H-RasV12, the level of GRO1 was increased (39), although it is not clear whether the increased expression is due to mutant p53, H-RasV12, or both.
In conclusion, our present study provides evidence that GRO1, which is known to play a major role in tumorigenesis, mediates mutant p53 gain of function. Thus, _targeting GRO1 may have therapeutic effects on clinical trials of tumors with mutant p53.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA121137.
Footnotes
The abbreviations used are: shRNA, short hairpin RNA; nt, nucleotide(s); GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ChIP, chromatin immunoprecipitation.
References
- 1.Kalo, E., Buganim, Y., Shapira, K. E., Besserglick, H., Goldfinger, N., Weisz, L., Stambolsky, P., Henis, Y. I., and Rotter, V. (2007) Mol. Cell. Biol. 27 8228-8242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossi, G., Lapi, E., Strano, S., Rinaldo, C., Blandino, G., and Sacchi, A. (2006) Oncogene 25 304-309 [DOI] [PubMed] [Google Scholar]
- 3.Atema, A., and Chene, P. (2002) Cancer Lett. 185 103-109 [DOI] [PubMed] [Google Scholar]
- 4.Frazier, M. W., He, X., Wang, J., Gu, Z., Cleveland, J. L., and Zambetti, G. P. (1998) Mol. Cell. Biol. 18 3735-3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane, B. R., Liu, J., Bock, P. J., Schols, D., Coffey, M. J., Strieter, R. M., Polverini, P. J., and Markovitz, D. M. (2002) J. Virol. 76 11570-11583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rennekampff, H. O., Hansbrough, J. F., Woods, V., Jr., Dore, C., Kiessig, V., and Schroder, J. M. (1997) Arch. Dermatol. Res. 289 204-212 [DOI] [PubMed] [Google Scholar]
- 7.Bechara, C., Chai, H., Lin, P. H., Yao, Q., and Chen, C. (2007) Med. Sci. Monit. 13 RA87-RA90 [PubMed] [Google Scholar]
- 8.Hammond, M. E., Shyamala, V., Siani, M. A., Gallegos, C. A., Feucht, P. H., Abbott, J., Lapointe, G. R., Moghadam, M., Khoja, H., Zakel, J., and Tekamp-Olson, P. (1996) J. Biol. Chem. 271 8228-8235 [DOI] [PubMed] [Google Scholar]
- 9.Wang, B., Hendricks, D. T., Wamunyokoli, F., and Parker, M. I. (2006) Cancer Res. 66 3071-3077 [DOI] [PubMed] [Google Scholar]
- 10.Burger, M., Hartmann, T., Burger, J. A., and Schraufstatter, I. (2005) Oncogene 24 2067-2075 [DOI] [PubMed] [Google Scholar]
- 11.Dhawan, P., and Richmond, A. (2002) J. Leukocyte Biol. 72 9-18 [PMC free article] [PubMed] [Google Scholar]
- 12.Warner, K. A., Miyazawa, M., Cordeiro, M. M., Love, W. J., Pinsky, M. S., Neiva, K. G., Spalding, A. C., and Nor, J. E. (2008) Neoplasia 10 131-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachmeier, B. E., Mohrenz, I. V., Mirisola, V., Schleicher, E., Romeo, F., Hohneke, C., Jochum, M., Nerlich, A. G., and Pfeffer, U. (2008) Carcinogenesis 29 779-789 [DOI] [PubMed] [Google Scholar]
- 14.Yang, S., Lim, M., Pham, L. K., Kendall, S. E., Reddi, A. H., Altieri, D. C., and Roy-Burman, P. (2006) Cancer Res. 66 4285-4290 [DOI] [PubMed] [Google Scholar]
- 15.Wang, D., Wang, H., Brown, J., Daikoku, T., Ning, W., Shi, Q., Richmond, A., Strieter, R., Dey, S. K., and DuBois, R. N. (2006) J. Exp. Med. 203 941-951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engl, T., Relja, B., Blumenberg, C., Muller, I., Ringel, E. M., Beecken, W. D., Jonas, D., and Blaheta, R. A. (2006) Life Sci. 78 1784-1793 [DOI] [PubMed] [Google Scholar]
- 17.Loukinova, E., Dong, G., Enamorado-Ayalya, I., Thomas, G. R., Chen, Z., Schreiber, H., and Van Waes, C. (2000) Oncogene 19 3477-3486 [DOI] [PubMed] [Google Scholar]
- 18.Caunt, M., Hu, L., Tang, T., Brooks, P. C., Ibrahim, S., and Karpatkin, S. (2006) Cancer Res. 66 4125-4132 [DOI] [PubMed] [Google Scholar]
- 19.Nirodi, C., Hart, J., Dhawan, P., Moon, N. S., Nepveu, A., and Richmond, A. (2001) J. Biol. Chem. 276 26122-26131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karl, E., Warner, K., Zeitlin, B., Kaneko, T., Wurtzel, L., Jin, T., Chang, J., Wang, S., Wang, C. Y., Strieter, R. M., Nunez, G., Polverini, P. J., and Nor, J. E. (2005) Cancer Res. 65 5063-5069 [DOI] [PubMed] [Google Scholar]
- 21.Wood, L. D., and Richmond, A. (1995) J. Biol. Chem. 270 30619-30626 [DOI] [PubMed] [Google Scholar]
- 22.Brummelkamp, T. R., Bernards, R., and Agami, R. (2002) Science 296 550-553 [DOI] [PubMed] [Google Scholar]
- 23.Nozell, S., and Chen, X. (2002) Oncogene 21 1285-1294 [DOI] [PubMed] [Google Scholar]
- 24.Yan, W., and Chen, X. (2007) Cancer Res. 67 9117-9124 [DOI] [PubMed] [Google Scholar]
- 25.Zhu, Y., Saunders, M. A., Yeh, H., Deng, W. G., and Wu, K. K. (2002) J. Biol. Chem. 277 6923-6928 [DOI] [PubMed] [Google Scholar]
- 26.Wu, K. K. (2006) Methods Mol. Biol. 338 281-290 [DOI] [PubMed] [Google Scholar]
- 27.Lang, G. A., Iwakuma, T., Suh, Y. A., Liu, G., Rao, V. A., Parant, J. M., Valentin-Vega, Y. A., Terzian, T., Caldwell, L. C., Strong, L. C., El-Naggar, A. K., and Lozano, G. (2004) Cell 119 861-872 [DOI] [PubMed] [Google Scholar]
- 28.Scian, M. J., Stagliano, K. E., Anderson, M. A., Hassan, S., Bowman, M., Miles, M. F., Deb, S. P., and Deb, S. (2005) Mol. Cell. Biol. 25 10097-10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisz, L., Oren, M., and Rotter, V. (2007) Oncogene 26 2202-2211 [DOI] [PubMed] [Google Scholar]
- 30.Phillips, A. C., Ernst, M. K., Bates, S., Rice, N. R., and Vousden, K. H. (1999) Mol. Cell 4 771-781 [DOI] [PubMed] [Google Scholar]
- 31.Weisz, L., Damalas, A., Liontos, M., Karakaidos, P., Fontemaggi, G., Maor-Aloni, R., Kalis, M., Levrero, M., Strano, S., Gorgoulis, V. G., Rotter, V., Blandino, G., and Oren, M. (2007) Cancer Res. 67 2396-2401 [DOI] [PubMed] [Google Scholar]
- 32.Ko, L. J., and Prives, C. (1996) Genes Dev. 10 1054-1072 [DOI] [PubMed] [Google Scholar]
- 33.Singh, S., Sadanandam, A., and Singh, R. K. (2007) Cancer Metastasis Rev. 26 453-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou, Y., Zhang, J., Liu, Q., Bell, R., Muruve, D. A., Forsyth, P., Arcellana-Panlilio, M., Robbins, S., and Yong, V. W. (2005) Carcinogenesis 26 2058-2068 [DOI] [PubMed] [Google Scholar]
- 35.Mehta, S. A., Christopherson, K. W., Bhat-Nakshatri, P., Goulet, R. J., Jr., Broxmeyer, H. E., Kopelovich, L., and Nakshatri, H. (2007) Oncogene 26 3329-3337 [DOI] [PubMed] [Google Scholar]
- 36.Anisowicz, A., Messineo, M., Lee, S. W., and Sager, R. (1991) J. Immunol. 147 520-527 [PubMed] [Google Scholar]
- 37.Amiri, K. I., Ha, H. C., Smulson, M. E., and Richmond, A. (2006) Oncogene 25 7714-7722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood, L. D., Farmer, A. A., and Richmond, A. (1995) Nucleic Acids Res. 23 4210-4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McMurray, H. R., Sampson, E. R., Compitello, G., Kinsey, C., Newman, L., Smith, B., Chen, S.-R., Klebanov, L., Salzman, P., Yakovlev, A., and Land, H. (2008) Nature 453 1112-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]