Abstract
Background
Recent studies have reported widespread copy number alterations and p53 mutations arising in cancer associated stromal cells. The aim of this study was to determine if pancreatic cancer associated fibroblasts display similar genetic alterations.
Design
Cancer-associated fibroblast cultures were established from 7 primary pancreatic adenocarcinomas. These fibroblasts and corresponding normal tissues when available were analyzed for genome-wide copy number changes using Affymetrix 250K SNP microarrays. Evidence of p53 protein expression, an indicator of p53 mutation was determined by immunohistochemical labeling of tissue microarrays containing 117 pancreatic ductal adenocarcinomas.
Results
Pancreatic cancer associated fibroblasts did not show any evidence of somatic copy number gains or losses. p53 protein expression was confined to invasive pancreatic adenocarcinoma cells and was not expressed in cancer-associated fibroblasts.
Conclusions
We find no evidence that pancreatic cancer associated fibroblasts harbor somatic copy number changes or immunohistochemical evidence of p53 mutations.
Keywords: pancreatic cancer, fibroblast, LOH, p53, SNP, microarray
Introduction
Increasing evidence suggests that tumor-stromal cell interactions within the cellular microenvironment of a variety of cancers are critical to tumor development and progression. The stromal component, consisting primarily of cancer-associated fibroblasts, inflammatory and endothelial cells as well as extracellular matrix, interacts with and undergo changes in gene expression in response to the invasive cancer to contribute to tumor progression of pancreatic1,2–5 and other cancers.6–7,8 While the importance of the stromal response to tumor development is recognized, the mechanisms underlying these molecular changes are only beginning to be understood.6–7,9
Recently, several investigators have reported genome-wide loss of heterozygosity (LOH) or allelic imbalance (AI) in cancer-associated stromal cells in many patients with breast cancer.10–17 These studies used archived tissue samples and laser capture microdissection to isolate stromal cells for genotyping by PCR amplifying microsatellite markers using limited amounts of formalin-fixed paraffin embedded DNA. However, PCR analysis of poor quality formalin fixed DNA can induce spurious genetic alterations.18–23 In addition, investigators have reported evidence of p53 mutations in breast cancer stromal cells.13 These authors conclude that the stromal compartment undergoes genetic alterations which contribute to carcinogenesis and tumor progression and that these stromal fibroblasts have similar chromosomal loss patterns to their epithelial cancer cells. Such somatic genetic alterations in cancer associated stromal cells are perhaps surprising given that cancer associated fibroblasts do not display morphological and metastatic features of cancer cells. Indeed, fibroblasts can be transformed into cancers in vitro using a limited number of genetic alterations.7 Furthermore, patients with inherited cancer syndromes due to defects in genome stability genes such as Li-Fraumeni syndrome carry a genetic deficiency in every cell and have normal tissue fibroblasts that are prone to genetic instability and can develop aneuploidy and immortalize in vitro.24,25 In contrast, we have found that cancer associated fibroblasts undergo senescence typical of normal tissue fibroblasts (unpublished observations).To determine if genetic alterations are found in pancreatic cancer associated fibroblasts (pancreatic CAFs), we performed a genome-wide analysis of pancreatic CAFs cultured directly from primary pancreatic ductal adenocarcinomas. We also analyzed stromal expression of the tumor suppressor protein TP53 for evidence of expression indicative of p53 mutation.13
Results
Generation of cancer associated fibroblast cultures
We established cancer associated fibroblast cultures from resection specimens of seven primary pancreatic adenocarcinomas. Initially, these cultures contain cancer cells as well as fibroblasts until fibroblast outgrowth is achieved. Once established, these cultures were shown by immunohistochemistry to express alpha smooth muscle actin, a marker of activated fibroblasts typically expressed in cancer associated fibroblasts but not normal quiescent fibroblasts (data not shown). They lacked expression of the pancreatic ductal epithelial-specific protein cytokeratin19, indicating that the cultures were free of contaminating epithelial cells (data not shown).
Analysis of DNA copy number in cancer associated fibroblasts
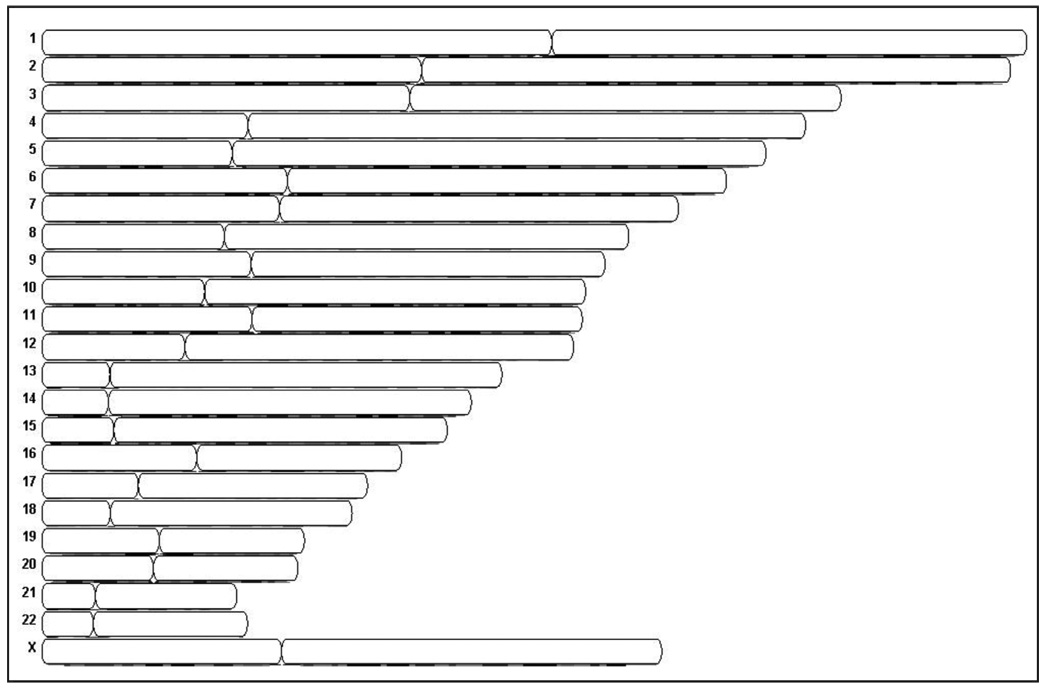
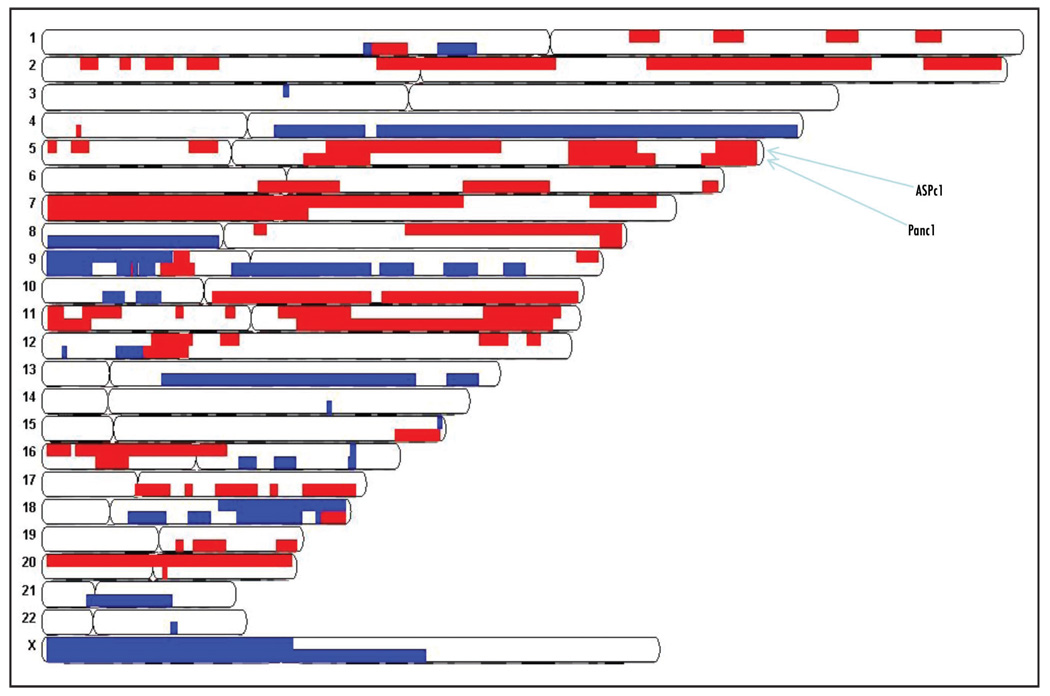
We employed the Affymetrix GeneChip Human Mapping 250K Sty I Array to analyze copy number and to detect regions of LOH using matched normal lymphocyte DNA and data from the HapMap collection (available online) as a reference. We determined CAF genotypes using the Affymetrix BRLMM algorithm, which generates homozygous or heterozygous genotype calls at any given SNP. The average call rate for all of the CAFs and matched normal lymphocytes was 99.2%. All of the samples had call rates of ~99% apart from one CAF sample that had a genotype call rate of 95.4%. Heterozygous SNP calls were located throughout the genome of all pancreatic CAFs (Fig. 1). Genotype analysis using a Hidden Markov model (Partek Genomics Suite Software) predicted no LOH events in the CAF samples (Fig. 1). We observed small (<1 megabase) copy number changes typical of normal germline copy number variation in each of the seven CAFs compared to the HapMap collection, but no large copy number gains or losses typical of the chromosomal instability found in cancer cells were detected. We did not detect any somatic copy number gains or losses in four of the CAF samples compared to their normal counterparts (Fig. 1). For comparison, we also examined the genome copy number of two pancreatic cancer cell lines using the 270 controls from the HapMap collection as a reference (Fig. 2). These two pancreatic cancers showed widespread megabase copy number gains and losses typical of those found with pancreatic ductal adenocarcinoma.27 For example, both cancer cell lines harbored losses at chromosomes 9p and 18q, and gains at chromosomes 7p and 11q as well as numerous other gains and losses in chromosomal regions (Fig. 3).
Figure 1.
Copy number analysis of four cancer associated fibroblast cultures and matching normal genomis DNA hybridized o SNP arrays. No copy number gains or losses were predicted by Hidden Markov’s model.
Figure 2.
Copy number gains (red) and losses (blue) in pancreatic cancer cell lines ASPc1 and Panc1 relative to the 270 HapMap data collection. For each chromosome, ASPc1 is represented in the top panel and Panc1 is represented in the bottom panel.
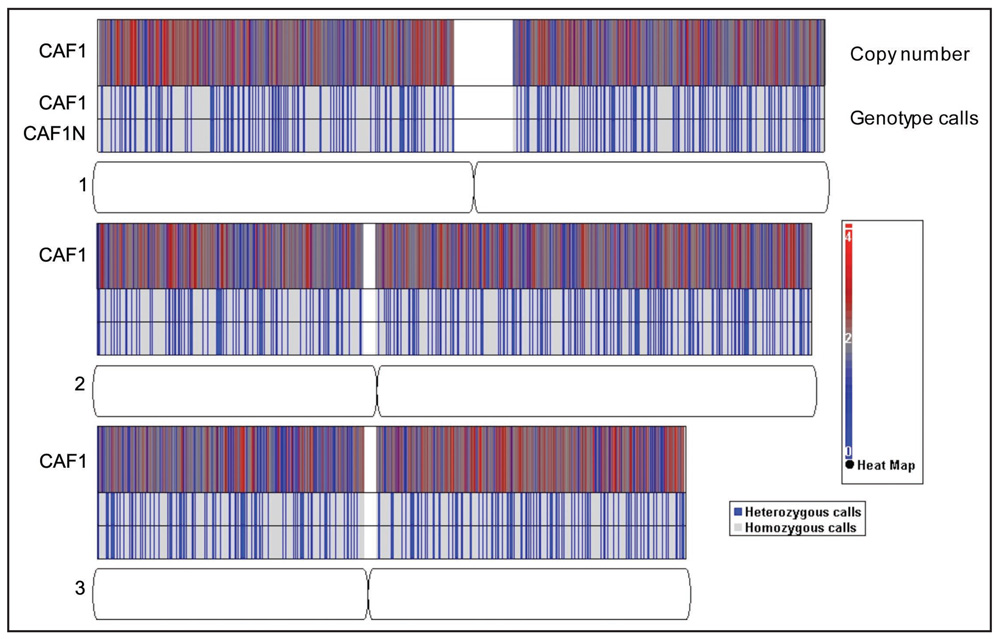
Figure 3.
Representative heat map of copy number values (top panel) and plot of genotype calls (bottom panel) on chromosomes 1, 2, amd 3 for one CAF sample relative to its match normal DNA. The heat map plots copy number at each SNP. The paired row of genotype calls represents one CAF and its match normal DNA
Analysis of p53 expression pattern in of pancreatic cancers associated stromal tissue
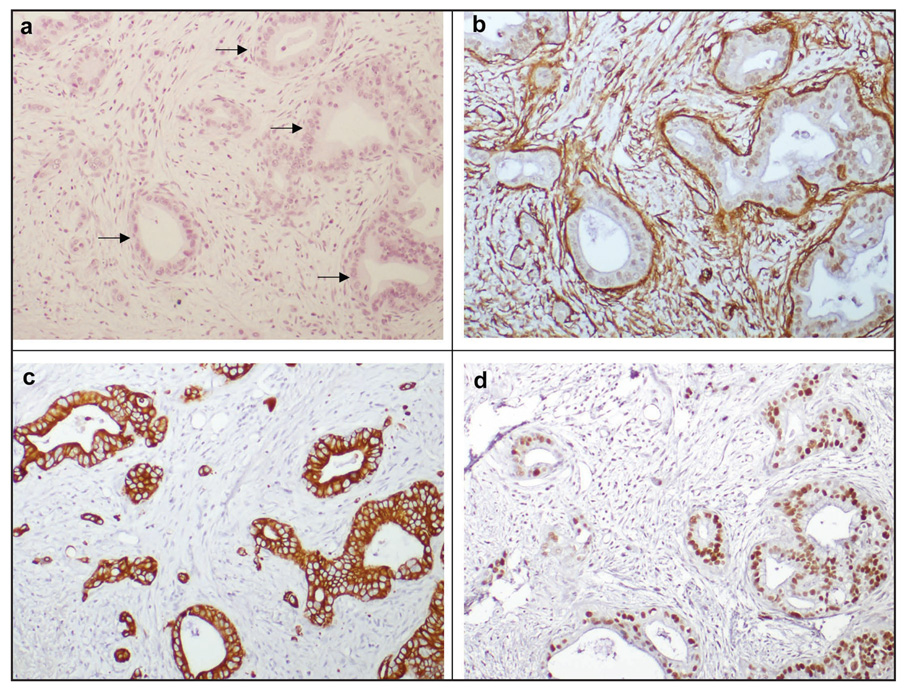
We analyzed the infiltrating cancer and stroma of 117 pancreatic cancers for evidence of p53 mutation using immunohistochemical detection of p53 expression. While 71 of 117 pancreatic cancers (61%) demonstrated strong labeling of p53 in cancer epithelial cells, none of the cancers expressed p53 in the associated stromal tissue (Fig. 4). For comparison, α-SMA expression of stromal fibroblasts and cytoplasmic cytokeratin 19 expression of malignant epithelial cells is also shown. Cytokeratin-19 immunostaining highlights isolated epithelial cells present in a sea of stroma (Fig. 4c). Immunohistochemical staining of p53 protein shows overexpression in the malignant neoplastic epithelial cells with no detectable expression in the surrounding stromal fibroblasts.
Figure 4.
Localizatin of p53 protien in ductal adenocarcinomaof the pancreas. (A) Hematoxylin and eosin staining demonstrates scattered malignant epithelial glands (arrows) and surrounding stromal fibroblasts and infiltration of inflammatory cells. (B) *-SMA expressiom is observed in stromal fibroblasts and (C) expression of cytoplasmic cytokeratin 19 is exclusively observed in malignant epithelial cells. (D) Immunohistochemical staining of p53 protien show overexpression in the malignant neoplastic epithelial cells with no detectable expression in the surrounding stromal fibroblasts.
Discussion
The integral role of the stromal component of neoplasms in their development and progression has been increasingly recognized. Molecular analysis of stromal cells can no doubt increase our understanding of their role in tumor progression. Isolating pure populations of stromal cells is often necessary for molecular analysis and can be achieved using laser-capture microdissection (LCM) of individual cells from formalin-fixed, paraffin-embedded tissue. Twenty-eight LCM enables the analysis of specifically defined cell populations, but the quality and quantity of DNA that can be obtained for downstream applications is often limited. Formalin fixation in particular cross links nucleic acids and results in highly fragmented DNA and RNA molecules; thus, material extracted from formalin-fixed, paraffin-embedded tissues is not optimal for large scale genetic analysis and the risk of artifacts during PCR amplification is high, particularly if picogram amounts of DNA are used in analysis.18–23 In contrast, one can obtain sufficient amounts of high-quality DNA from specific cell populations by isolating and growing the cells of interest in culture. This method has been widely used to study pure populations of cancer-associated fibroblasts (CAFs), which can be harvested from primary cultures and do not require degradative processes such as freezing or formalin fixation.7,29
We find no evidence of widespread copy number alterations indicative of genomic instability in stromal fibroblasts derived from primary pancreatic cancers. Previous reports suggest that genomic alterations occur in both the epithelium and stroma of cancers. In these studies, LOH was defined by genotyping of microsatellite markers, using genomic DNA from archival tissue. DNA from these tissues is typically degraded and of suboptimal quality, and at low DNA concentrations the allelic dropout rate due to stochastic error of PCR can be greater than 50%.19–21,30,31 Allelic dropout at a heterozygous locus results in a false positive prediction of LOH due to apparent homozygosity at that locus, leading to overestimation of LOH rate. Therefore, the high rate of LOH previously reported in tumor associated stroma could well be the result of stochastic PCR errors leading to erroneous LOH calls generated from poor quality DNA.10–17
Patocs et al. recently found p53 mutations in the stromal compartment of a subset of breast cancers analyzed. The majority of p53 gene mutations in human cancers are missense mutations which result in overexpression of mutant p53 protein in tumor cells.32 They also found p53 overexpression in tumor associated stromal cells, although the authors did not describe a close correlation between cases with stromal expression of p53 and the presence of stromal p53 mutations. In contrast, although we find that ~60% of the 117 pancreatic cancers analyzed displayed p53 overexpression, only the cancer epithelial cells displayed such overexpression, with no evidence of p53 overexpression in the stromal cells of any of the pancreatic cancers examined. Since the use of formalin-fixed tissue can also create artifacts in cycle sequencing of p53 and other genes33, it is possible that the somatic p53 mutations reported in tumor associated stromal cells by Patocs et al are the result of PCR artifacts. Another possibility is that microdissections of stroma could contain isolated infiltrating cancer cells.
Several mouse genetic models highlight the important role of stromal fibroblasts in tumor progression. For example, conditional inactivation of the TGF-beta type II receptor in mouse fibroblasts results in prostate intraepithelial neoplasia (PIN) and invasive squamous cell carcinoma in the forestomach.34 Thus, inhibition of TGF beta signaling in fibroblasts exerts a paracrine effect on epithelial proliferation. The growth promoting effects of stromal cells on adjacent epithelia have also been observed in other cancer types. Stromal fibroblasts enhance tumorigenicity when co-injected with epithelial cells in mouse models of breast, pancreatic, and prostate carcinomas.35,36,29,4,37 Conversely, blocking growth signaling pathways in stromal fibroblasts can dramatically slow malignant tumor expansion and invasion.35,37,38 These studies indicate that stromal cells can interact with and significantly impact the growth and oncogenic potential of adjacent epithelia.
However, despite the biological insights provided by these genetic mouse model systems, clear evidence that genetic events in fibroblasts contribute to human cancer progression is limited. One example of a genetic loss observed in human neoplastic stroma is in the setting of juvenile hamartomatous polyps. Patients with inherited juvenile polyposis carry a germline mutation of SMAD4 in all of their cells and develop juvenile polyps which display allelic loss at the SMAD4 locus both in the epithelium and in the stromal fibroblasts.39 Evidence that similar biallelic inactivation of tumor suppressor genes are occurring in cancer associated fibroblasts is hard to find and available evidence supports the hypothesis that cancer associated fibroblasts generally do not have tumor suppressor gene inactivation. One would expect that if fibroblasts had biallelic inactivation of a tumor suppressor gene they would become a dominant fibroblast clone within the tumor microenvironment and readily identifiable with appropriate markers. Yet, we don’t observe p53 overexpression in pancreatic cancer fibroblasts and we have not seen evidence for inactivation of other tumor suppressor genes in primary pancreatic cancer stroma. For example, inactivation of the TGF beta pathway through SMAD4 mutation occurs in ~55% of pancreatic cancers and can be reliably identified by demonstrating loss of SMAD4 protein, yet pancreatic cancer associated fibroblasts consistently demonstrate normal expression of SMAD4.40 Similarly, in situ studies of chromosomal alterations in human cancers focus on alterations in cancer epithelial cells and not reported than in tumor stroma. Future studies that utilize in situ or immunohistochemical techniques to investigate cancer alterations can observe patterns in cancer associated fibroblasts to determine if these cells ever display genetic losses or tumor suppressor gene inactivation.
Our results are consistent with previous observations that genomic alterations are not typical of cancer associated stromal cells.9,41 Our findings do not exclude the possibility that the cancer associated fibroblasts of certain patients with inherited cancer syndromes could harbor genetic alterations in their tumor fibroblasts. However, our results indicate that genetic alterations such as widespread LOH are not required for the development and progression of pancreatic ductal adenocarcinomas.
CAFs respond to the tumor environment with widespread transcriptional changes and, may acquire inheritable changes in gene expression arising from epigenetic alterations. Evidence supporting this hypothesis includes a comparison of epithelial and stromal cells from breast tumors using serial analysis of gene expression (SAGE), which found that although gene expression changes occur in all cell types composing invasive breast carcinomas, genetic changes were found only in cancer cells.41 Furthermore, methylation-specific digital karyotyping showed that epigenetic alterations occur in normal epithelial cells, myoepithelial cells, and stromal fibroblasts during breast tumorigenesis.9 Our data are inconsistent with previous work reporting evidence that widespread LOH is a common property of cancer associated stroma. Accurate molecular characterization of the tumor microenvironment is necessary for the understanding of mechanisms of tumor stromal interactions needed to develop rational therapeutics.
Materials and Methods
Culture of cell lines and establishment of fibroblast cultures
Primary cultures of stromal fibroblasts were established from surgically resected pancreatic cancer tissue from seven patients (four males, three females with a mean / standard deviation age of 60/±8 years) with clinically sporadic pancreatic ductal adenocarcinoma. The cancers were all moderate to poorly differentiated with a mean tumor size of 3.6 cm. Fresh pancreas tumor tissue was minced into 1–3mm3 fragments and digested with 0.25% trypsin at 37°C for 30 min. The resulting fragments were centrifuged at 600xg for 5 min. and washed once with DMEM containing 10% fetal bovine serum. The tissue fragments were then plated and allowed to adhere. After incubation at 37°C for several days, fibroblast outgrowth from the tissue fragments occurred. Fibroblasts were sub-cultured by trypsinization for 2–3 passages until free of epithelial cell contamination and maintained in DMEM supplemented with 10% fetal bovine serum, 2% penicillin and streptomycin (Invitrogen). Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. The pancreatic cancer cell lines AsPC-1 and Panc-1were maintained in RPMI medium containing 10% fetal bovine serum, 2% penicillin and streptomycin (Invitrogen). All samples were collected with approval from the Johns Hopkins Committee for Clinical Investigation.
Characterization of fibroblast cultures
The purity of fibroblast cultures was assessed by immunostaining for cytokeratin 19 (mouse monoclonal cytokeratin 19 antibody, Santa Cruz Biotechnology, dilution 1:100), and α-smooth muscle actin (mouse monoclonal α-smooth muscle actin antibody, DAKO, clone IA4, dilution 1:100). Cells were cultured on chamber slides (BD Falcon) and grown until sub-confluent. Cells were then fixed in 70% methanol for 10 min. at RT. Slides were removed from chambers, washed once with PBS, and subjected to immunostaining on a DAKO Autostainer, using a primary antibody incubation time of 60 minutes.
Total genome Copy Number analysis
Genomic DNA was extracted from cultured fibroblasts and matching frozen normal tissue (peripheral blood lymphocytes or normal duodenum) using a DNeasy Blood & Tissue Kit (Qiagen). Matched normal DNA was available for five of the CAF samples. DNA was digested and hybridized to the Affymetrix GeneChip Human Mapping 250K Sty I Array to analyze copy number and to detect regions of loss of heterozygosity (LOH). The 7 CAFs were also compared to the genotypes of 270 individuals from the HapMap collection. Genotype calls were determined using the Affymetrix Chromosome Copy Number Analysis Tool 4.0 (CNAT 4.0). Genotypes and copy number changes were analyzed using Partek® Genomics SuiteTM ver. 6.03 to generate LOH predictions. Copy number profiles were generated using Partek® Genomics SuiteTM ver. 6.03 and significant copy number changes were detected using a Hidden Markov’s model. The copy number patterns of two pancreatic adenocarcinoma cell lines known to have widespread copy number alterations were analyzed for comparison.
Pancreatic adenocarcinoma tissue microarrays and p53 immunohistochemistry
The expression of p53 protein was examined utilizing immunohistochemical labeling of formalin-fixed, paraffin-embedded tissue microarrays (TMAs) using the DAKO Autostainer. Seven tissue microarrays containing a total of 117 different surgically resected pancreatic ductal adenocarcinomas were constructed as previously described.26 Sections were deparaffinized in xylene, hydrated in graded ethanol concentrations, and boiled for 20 minutes in epitope retrieval buffer (DAKO). Immunostaining was then performed on the DAKO Autostainer using a monoclonal mouse anti-human p53 primary antibody (clone D0-7; DAKO) with an overnight incubation, an anti-cytokeratin-19 antibody (Santa Cruz Biotechnology) for 60 minute incubation, or an anti-α-smooth muscle actin antibody (DAKO) also for 60 minute incubation. Labeling was performed according to the manufacturer’s protocol using the Envision Plus Detection Kit (DAKO). Nuclei were counterstained with hematoxylin. Hematoxylin and eosin staining was performed by heating one tissue microarray for 20 minutes at 95°C followed by a 10 minute incubation in hematoxylin and a five minute incubation in eosin.
Acknowledgments
Grant support: Supported by the NCI grants Specialized Programs of Research Excellence in Gastrointestinal Malignancies (CA62924), the Michael Rolfe Foundation.
References
- 1.Infante JR, Matsubayashi H, Sato N, Tonascia J, Klein AP, Riall TA, Yeo C, Iacobuzio-Donahue C, Goggins M. Peritumoral fibroblast SPARC expression and patient outcome with resectable pancreatic adenocarcinoma. J Clin Oncol. 2007;25:319–325. doi: 10.1200/JCO.2006.07.8824. [DOI] [PubMed] [Google Scholar]
- 2.Sato N, Fukushima N, Maehara N, Matsubayashi H, Koopmann J, Su GH, Hruban RH, Goggins M. SPARC/osteonectin is a frequent _target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor-stromal interactions. Oncogene. 2003;22:5021–5030. doi: 10.1038/sj.onc.1206807. [DOI] [PubMed] [Google Scholar]
- 3.Sato N, Maehara N, Goggins M. Gene expression profiling of tumor-stromal interactions between pancreatic cancer cells and stromal fibroblasts. Cancer Res. 2004;64:6950–6956. doi: 10.1158/0008-5472.CAN-04-0677. [DOI] [PubMed] [Google Scholar]
- 4.Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A, Ji B, Evans DB, Logsdon CD. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68:918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricci F, Kern SE, Hruban RH, Iacobuzio-Donahue CA. Stromal responses to carcinomas of the pancreas: juxtatumoral gene expression conforms to the infiltrating pattern and not the biologic subtype. Cancer Biol Ther. 2005;4:302–307. doi: 10.4161/cbt.4.3.1501. [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 7.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, Zhu Z, Hicklin D, Wu Y, Port JL, Altorki N, Port ER, Ruggero D, Shmelkov SV, Jensen KK, Rafii S, Lyden D. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 10.Fukino K, Shen L, Matsumoto S, Morrison CD, Mutter GL, Eng C. Combined total genome loss of heterozygosity scan of breast cancer stroma and epithelium reveals multiplicity of stromal _targets. Cancer Res. 2004;64:7231–7236. doi: 10.1158/0008-5472.CAN-04-2866. [DOI] [PubMed] [Google Scholar]
- 11.Fukino K, Shen L, Patocs A, Mutter GL, Eng C. Genomic instability within tumor stroma and clinicopathological characteristics of sporadic primary invasive breast carcinoma. Jama. 2007;297:2103–2111. doi: 10.1001/jama.297.19.2103. [DOI] [PubMed] [Google Scholar]
- 12.Trimboli AJ, Fukino K, de Bruin A, Wei G, Shen L, Tanner SM, Creasap N, Rosol TJ, Robinson ML, Eng C, Ostrowski MC, Leone G. Direct evidence for epithelial-mesenchy-mal transitions in breast cancer. Cancer Res. 2008;68:937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 13.Patocs A, Zhang L, Xu Y, Weber F, Caldes T, Mutter GL, Platzer P, Eng C. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–2551. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- 14.Weber F, Xu Y, Zhang L, Patocs A, Shen L, Platzer P, Eng C. Microenvironmental genomic alterations and clinicopathological behavior in head and neck squamous cell carcinoma. Jama. 2007;297:187–195. doi: 10.1001/jama.297.2.187. [DOI] [PubMed] [Google Scholar]
- 15.Kurose K, Hoshaw-Woodard S, Adeyinka A, Lemeshow S, Watson PH, Eng C. Genetic model of multi-step breast carcinogenesis involving the epithelium and stroma: clues to tumour-microenvironment interactions. Hum Mol Genet. 2001;10:1907–1913. doi: 10.1093/hmg/10.18.1907. [DOI] [PubMed] [Google Scholar]
- 16.Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32:355–357. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 17.Moinfar F, Man YG, Arnould L, Bratthauer GL, Ratschek M, Tavassoli FA. Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res. 2000;60:2562–2566. [PubMed] [Google Scholar]
- 18.Coudry RA, Meireles SI, Stoyanova R, Cooper HS, Carpino A, Wang X, Engstrom PF, Clapper ML. Successful application of microarray technology to microdissected formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2007;9:70–79. doi: 10.2353/jmoldx.2007.060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods. 2001;25:409–418. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- 20.Xu C, Houck JR, Fan W, Wang P, Chen Y, Upton M, Futran ND, Schwartz SM, Zhao LP, Chen C, Mendez E. Simultaneous isolation of DNA and RNA from the same cell population obtained by laser capture microdissection for genome and transcriptome profiling. J Mol Diagn. 2008;10:129–134. doi: 10.2353/jmoldx.2008.070131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Craven RA, Totty N, Harnden P, Selby PJ, Banks RE. Laser capture microdissection and two-dimensional polyacrylamide gel electrophoresis: evaluation of tissue preparation and sample limitations. Am J Pathol. 2002;160:815–822. doi: 10.1016/S0002-9440(10)64904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burton MP, Schneider BG, Brown R, Escamilla-Ponce N, Gulley ML. Comparison of histologic stains for use in PCR analysis of microdissected, paraffin-embedded tissues. Biotechniques. 1998;24:86–92. doi: 10.2144/98241st01. [DOI] [PubMed] [Google Scholar]
- 23.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Bischoff FZ, Yim SO, Pathak S, Grant G, Siciliano MJ, Giovanella BC, Strong LC, Tainsky MA. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: aneuploidy and immortalization. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 25.Liu PK, Kraus E, Wu TA, Strong LC, Tainsky MA. Analysis of genomic instability in Li-Fraumeni fibroblasts with germline p53 mutations. Oncogene. 1996;12:2267–2278. [PMC free article] [PubMed] [Google Scholar]
- 26.Koopmann J, Fedarko NS, Jain A, Maitra A, Iacobuzio-Donahue C, Rahman A, Hruban RH, Yeo CJ, Goggins M. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13:487–491. [PubMed] [Google Scholar]
- 27.Calhoun ES, Hucl T, Gallmeier E, West KM, Arking DE, Maitra A, Iacobuzio-Donahue CA, Chakravarti A, Hruban RH, Kern SE. Identifying Allelic Loss and Homozygous Deletions in Pancreatic Cancer without Matched Normals Using High-Density Single-Nucleotide Polymorphism Arrays. Cancer Res. 2006;66:7920–7928. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 28.Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA. Laser-capture micro-dissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14:272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 29.Cunha GR, Hayward SW, Wang YZ. Role of stroma in carcinogenesis of the prostate. Differentiation. 2002;70:473–485. doi: 10.1046/j.1432-0436.2002.700902.x. [DOI] [PubMed] [Google Scholar]
- 30.Miller CR, Joyce P, Waits LP. Assessing allelic dropout and genotype reliability using maximum likelihood. Genetics. 2002;160:357–366. doi: 10.1093/genetics/160.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y. Induction of genetic instability by gain-of-function p53 cancer mutants. Oncogene. 2008;25:3501–3507. doi: 10.1038/sj.onc.1211023. [DOI] [PubMed] [Google Scholar]
- 33.Shiao YH, Buzard GS, Weghorst CM, Rice JM. DNA template as a source of artifact in the detection of p53 gene mutations using archived tissue. Biotechniques. 1997;22:608–610. doi: 10.2144/97224bm07. 12. [DOI] [PubMed] [Google Scholar]
- 34.Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, Washington MK, Neilson EG, Moses HL. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 35.Verona EV, Elkahloun AG, Yang J, Bandyopadhyay A, Yeh IT, Sun LZ. Transforming growth factor-beta signaling in prostate stromal cells supports prostate carcinoma growth by up-regulating stromal genes related to tissue remodeling. Cancer Res. 2007;67:5737–5746. doi: 10.1158/0008-5472.CAN-07-0444. [DOI] [PubMed] [Google Scholar]
- 36.Barcellos-Hoff MH, Ravani SA. Irradiated mammary gland stroma promotes the expression of tumorigenic potential by unirradiated epithelial cells. Cancer Res. 2000;60:1254–1260. [PubMed] [Google Scholar]
- 37.Ohuchida K, Mizumoto K, Murakami M, Qian LW, Sato N, Nagai E, Matsumoto K, Nakamura T, Tanaka M. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumor-stromal interactions. Cancer Res. 2004;64:3215–3222. doi: 10.1158/0008-5472.can-03-2464. [DOI] [PubMed] [Google Scholar]
- 38.Pietras K, Pahler J, Bergers G, Hanahan D. Functions of paracrine PDGF signaling in the proangiogenic tumor stroma revealed by pharmacological _targeting. PLoS Med. 2008;5:e19. doi: 10.1371/journal.pmed.0050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodford-Richens K, Williamson J, Bevan S, Young J, Leggett B, Frayling I, Thway Y, Hodgson S, Kim JC, Iwama T, Novelli M, Sheer D, Poulsom R, Wright N, Houlston R, Tomlinson I. Allelic loss at SMAD4 in polyps from juvenile polyposis patients and use of fluorescence in situ hybridization to demonstrate clonal origin of the epithelium. Cancer Res. 2000;60:2477–2482. [PubMed] [Google Scholar]
- 40.Tascilar M, Skinner HG, Rosty C, Sohn T, Wilentz RE, Offerhaus GJ, Adsay V, Abrams RA, Cameron JL, Kern SE, Yeo CJ, Hruban RH, Goggins M. The SMAD4 protein and prognosis of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2001;7:4115–4121. [PubMed] [Google Scholar]
- 41.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]