Abstract
Deficient repair activity for 8-hydroxy-2′-deoxyguanine (8-oxoguanine), a premutagenic oxidative DNA damage, has been observed in affected tissues in neurodegenerative diseases of aging, such as Alzheimer’s disease, and in ischemia/reperfusion injury, type 2 diabetes mellitus, and cancer. These conditions have in common the accumulation of oxidative DNA damage, which is believed to play a role in disease progression, and loss of intracellular calcium regulation. These observations suggest that oxidative DNA damage repair capacity may be influenced by fluctuations in cellular calcium. We have identified human 8-oxoguanine-DNA glycosylase 1 (OGG1), the major 8-oxoguanine repair activity, as a specific _target of the Ca2+-dependent protease Calpain I. Protein sequencing of a truncated partially calpain-digested OGG1 revealed that calpain recognizes OGG1 for degradation at a putative PEST (Proline, Glutamic acid, Serine, Threonine) sequence in the C-terminus of the enzyme. Co-immunoprecipitation experiments showed that OGG1 and Calpain I are associated in human cells. Exposure of HeLa cells to hydrogen peroxide or cisplatin resulted in the degradation of OGG1. Pretreatment of cells with the calpain inhibitor calpeptin resulted in inhibition of OGG1 proteolysis and suggests that OGG1 is a _target for calpain-mediated degradation in vivo during oxidative stress- and cisplatin-induced apoptosis. Polymorphic OGG1 S326C was comparatively resistant to calpain digestion in vitro, yet was also degraded by a calpain-dependent pathway in vivo following DNA damaging agent exposure. The degradation of OGG1 by calpain may contribute to decreased 8-oxoguanine repair activity and elevated levels of 8-oxoguanine reported in tissues undergoing chronic oxidative stress, ischemia/reperfusion and other cellular stressors known to produce perturbations of intracellular calcium homeostasis which activate calpain.
Keywords: OGG1, calpain, 8-oxoguanine, calcium, PEST
1. Introduction
Exposure to DNA damaging agents and reactive oxygen species (ROS) frequently results in the activation of Ca2+-dependent proteases and the subsequent proteolytic modification or destruction of numerous cellular proteins leading to irreversible activation of the apoptotic pathway. The caspase and calpain families are major Ca2+-dependent proteases involved in apoptotic and necrotic cell death [reviewed in (1)]. Much attention has been focused on caspases due to their established role in the progression of classical apoptosis (2). Initially implicated in the necrotic process, activation of calpains has since been shown to play a prominent role in apoptosis. Significant cross-talk occurs between caspases and calpains in cell death pathways (3,4) and calpain activation may occur upstream, downstream, or be independent of caspases in the apoptotic cascade, depending upon the cell type and nature of cell damage induced (5–8). More recently, dysregulation of calpain has been identified as a central factor in the progression of numerous pathological conditions, including Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, cataracts, ischemic neurodegeneration (stroke), type 2 diabetes mellitus, and cancer (9).
Alzheimer’s disease, type 2 diabetes, ischemia/reperfusion injury, and cancer are all associated with increased oxidative stress, elevated intracellular calcium and calpain hyperactivation (10–15). In affected tissues in these conditions, elevated 8-oxoguanine in DNA, decreased levels of OGG1 enzyme, and decreased 8-oxoguanine excision activity have been observed (16–21). Evidence suggests that accumulation of oxidative DNA damage in these conditions may be involved in cell death or transformation. A potential link between ROS exposure and calcium overload involves the ROS-mediated formation of reactive aldehydes that inhibit plasma membrane Ca2+ ATPase activity (22). An oxidative stress-mediated decrease in Ca2+ ATPase activity would inhibit the removal of calcium from the cell resulting in intracellular calcium accumulation that promotes activation of calcium-dependent proteases. In cortical neurons, β-amyloid protein increases the amplitude of voltage-dependent Ca2+ channel currents resulting in elevated free intracellular calcium (23). Calcium released from endogenous endoplasmic reticulum and mitochondrial calcium stores may also contribute to increased free intracellular calcium following cellular insults such as ROS exposure (1). Since some conditions associated with intracellular calcium dysregulation are associated with an oxidative DNA damage repair deficit, DNA glycosylases which recognize and excise oxidative DNA damage, such as OGG1, may represent potential _targets for calcium-dependent proteolytic modification or destruction. We examined this possibility by inducing calpain activity with ROS or cisplatin exposure in human cells and observed proteolysis of OGG1. The degradation of OGG1 in cells with activated calpain suggests that proteolysis of the major 8-oxoguanine repair activity in cells with loss of intracellular calcium regulation may contribute to the associated impaired oxidative DNA damage repair capacity and accumulation of oxidative DNA damage.
2. Materials and methods
2.1 In vitro cleavage of OGG1 by calpain
Purified human OGG1–1a was prepared as described previously (24). Purified Calpain I was obtained from Calbiochem. OGG1 was reacted with calpain in 20 mM Tris-HCl, pH 7.4, 2 mM CaCl2 at 30°C. Calpastatin, a specific calpain inhibitor, was obtained from Calbiochem. Reactions were terminated by adding SDS sample buffer and heating to 95°C for 5 min. Samples were analyzed by electrophoresis on 4–20% acrylamide gels (Invitrogen) and Coomassie staining (Bio-Rad).
2.2 Induction of calpain activity in human cells by ROS and cisplatin exposure
HeLa cells were plated at 0.5×106 cells per dish in 100 mm dishes and transfected with 0.25 μg of previously described pCMVF-WT OGG1 mammalian OGG1 expression vector (24) or pCMVF vector using Fugene 6 transfection reagent (Roche) according to the manufacturer’s instructions. Twenty-four hours later, medium was replaced and cells were pretreated with 0, 5, or 10 μM calpeptin (Calbiochem), a cell-permeable calpain inhibitor, for 2 hrs prior to being exposed to hydrogen peroxide (Sigma) or cisplatin (Calbiochem) for 24 hrs. Cells were trypsinized and whole cell extracts prepared with M-PER reagent (Pierce) containing HALT protease inhibitor cocktail (Pierce) were analyzed for intracellular OGG1 levels by anti-FLAG (M2, Sigma) western blotting of whole cell extracts. Untransfected cells were similarly treated and cell extracts were analyzed for degradation of endogenous OGG1–1a and –2a by western blot using a polyclonal OGG1/2 antibody (H-300, Santa Cruz). Apoptosis was measured with an Annexin V-FITC apoptosis detection kit I (BD Pharmingen) and flow cytometry analysis on a BD FACSCalibur flow cytometer (BD Biosciences).
2.3 Co-immunoprecipitation of OGG1 and calpain
HeLa cells were plated at 6×106 cells per dish on 150 mm dishes. Twenty-four hours later, cells were transfected with 10 μg of pCMVF-WT OGG1 vector. Twenty-four hours after transfection, cells were harvested and whole cell extracts were prepared using M-PER reagent containing HALT protease inhibitor cocktail. M-PER was made 20 mM Tris-HCl 7.4, 150 mM NaCl prior to adding to cell pellets. Extracts were precleared with protein G agarose (Santa Cruz) for 2 hrs. Anti-FLAG, anti-Calpain I (N-19, Santa Cruz) antibody or normal mouse IgG (Santa Cruz) were incubated with cell extract at 4°C for 24 hrs with rotation. Protein G agarose was then added to each incubation for an additional 24 hrs with rotation. Beads were washed in PBS and suspended in 40 μl of 1x SDS buffer. Five μg of whole cell extract and 10 μl of each incubation reaction were analyzed by western blot using either anti-FLAG or anti-Calpain I antibodies.
3. Results
3.1 OGG1 is cleaved by calpain in vitro
An examination of the OGG1 amino acid sequence using the PESTfind program (25,26) revealed putative low-score PEST sequences in the enzyme that could represent consensus cleavage sites for the calpain family of calcium-dependent proteases (Fig. 1). We tested the possibility that OGG1 might be a calpain substrate by reacting purified OGG1 with human Calpain I. Calpain cleaved OGG1 (Fig. 2), removing approximately 2 kDa from the full-length protein and degrading the enzyme. An extended exposure of OGG1 to calpain resulted in complete cleavage and partial degradation of the enzyme (Fig. 2, lane 5). To ensure the specificity of OGG1 cleavage by calpain, cleavage reactions were carried out in the presence of EDTA or calpastatin. In the absence of calcium (Fig. 2, lane 6), calpain failed to cleave OGG1. In the presence of calpastatin, (Fig. 2, lane 7), OGG1 was protected from calpain cleavage. N-terminal protein sequencing of partially calpain-degraded OGG1 showed an unmodified N-terminus (data not shown). C-terminal amino acid sequencing (Mayo Proteomics Research Center, Rochester, NY) of partially calpain-digested OGG1 produced the sequence –QSRHA, indicating that calpain cleaves OGG1 between amino acids A329 and Q330 during OGG1 proteolysis (Figs. 1 and 2).
Figure 1.
PEST sequences in the OGG1 enzyme. The OGG1 amino acid sequence was analyzed for consensus calpain cleavage sites by the PESTfind program (https://emb1.bcc.univie.ac.at/toolbox/pestfind/pestfind-analysis-webtool.htm) (25). Poor PEST sequences identified by PESTfind are dash underlined. An experimentally determined PEST sequence where OGG1 is cleaved by Calpain I is shown in bold. Arrows indicate a site of calpain cleavage between A329 and Q330 in the OGG1 C-terminus. Serine 326 is underlined in bold.
Figure 2.
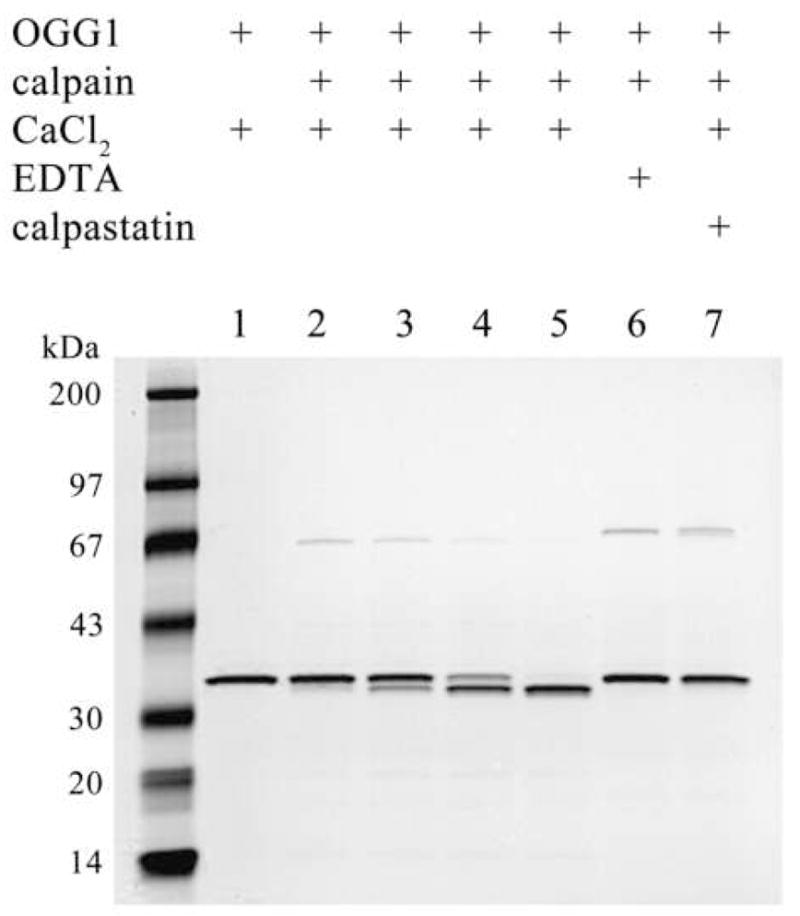
In vitro proteolysis of OGG1 by Calpain I. OGG1 (1 μg) was reacted with 200 ng of Calpain I for 15, 30 or 60 min (lanes 2–4) or 120 min (lanes 5–7) at 30°C. Reactions were supplemented with 2 mM CaCl2, 2 mM EDTA, or 1μg calpastatin as indicated. Reactions were terminated with SDS sample buffer and analyzed by SDS-PAGE and Coomassie staining.
3.2 In vivo degradation of OGG1 by calpain following ROS and cisplatin exposure
With the identification of OGG1 as a calpain substrate in vitro, we tested whether OGG1 is degraded by calpain in cells. Human cells transfected with FLAG-tagged OGG1 were exposed to various concentrations of hydrogen peroxide (Fig. 3A) and cisplatin (Fig. 3B). After a 24 hr exposure, the percentage of apoptotic cells was determined for each treatment condition (Fig. 3). Levels of OGG1 in whole cell extracts of OGG1-transfected normal and treated cells were analyzed by western blot. In apoptotic cells with activated calpain, OGG1 was degraded and no significant amount of truncated product was detected (Fig. 3A and B). Roughly half of endogenous OGG1–1a was degraded following treatment of untransfected cells with hydrogen peroxide (Fig. 5, lane 2), while no significant change was observed in OGG1–2a level (Fig. 5, lane 2). Cisplatin exposure resulted in significant degradation of both OGG1–1a and OGG1–2a in untransfected cells (Fig. 5, lane 5). The complete (Fig. 3A and Fig. 5, lanes 3 and 4) or partial (Fig. 3B and Fig. 5, lanes 6 and 7) inhibition of OGG1 proteolysis in cells by a specific calpain inhibitor and the cleavage and degradation of OGG1 by calpain in vitro (Fig. 2) suggest a major role for calpain in OGG1 degradation in vivo.
Figure 3.
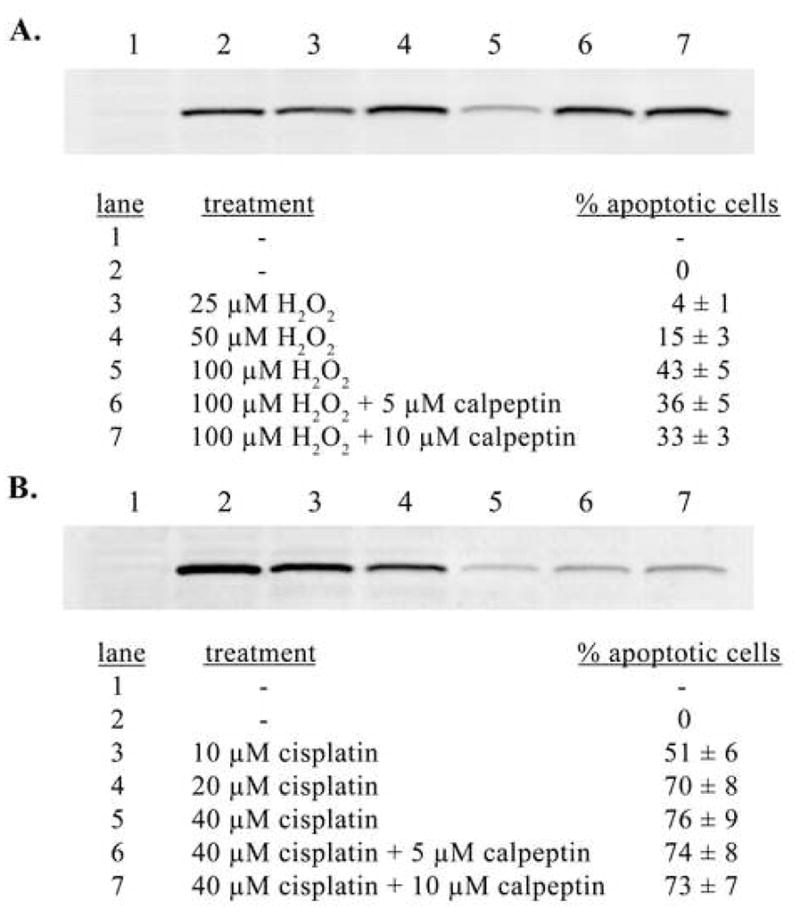
In vitro degradation of OGG1 by calpain during ROS- and cisplatin-induced apoptosis. A. Anti-FLAG western blot of 20 μg of whole cell extract from cells transfected with pCMVF vector (lane 1) or pCMVF-WT OGG1 (lanes 2–7) and exposed to hydrogen peroxide (H2O2) for 24 hrs. Cell treatments and percent apoptotic cells (normalized) for cells used to prepare extracts analyzed for OGG1 levels by western are shown for each lane in panel A. An identical experiment using cisplatin exposure is shown in panel B.
Figure 5.
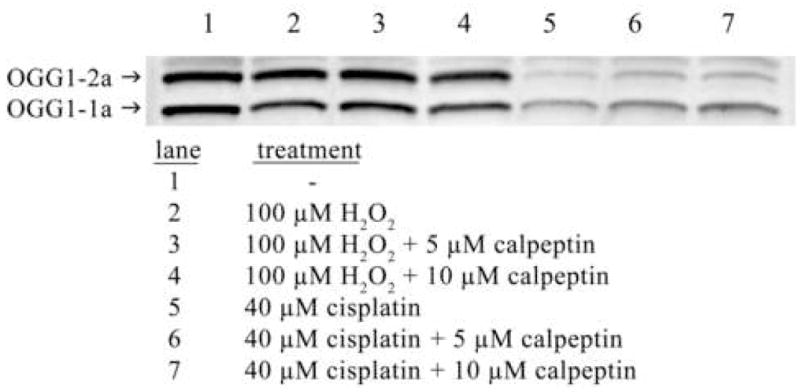
Degradation of endogenous OGG1s by calpain following hydrogen peroxide and cisplatin exposure. Anti-OGG1/2 polyclonal antibody western blot of 20 μg of whole cell extract from untransfected HeLa cells. Cells were untreated or exposed to 100 μM hydrogen peroxide or 40 μM cisplatin for 24 hrs with or without 2 hr calpeptin pretreatment as indicated.
3.3 Interaction of OGG1 and Calpain I in human cells
Since OGG1 is a calpain substrate, interacts with calpain at least transiently in vitro and is degraded by a calpain-dependent mechanism in vivo, we carried out co-immunoprecipitation experiments to determine if the proteins are associated in cells. Using whole cell extracts from human cells overexpressing N-terminally FLAG-tagged OGG1, we specifically pulled down OGG1 and Calpain I using anti-Calpain I and anti-FLAG antibodies, respectively, in reciprocal experiments (Fig. 4). These results suggest that OGG1 and calpain form a complex in vivo and further support the direct degradation of OGG1 by calpain in cells.
Figure 4.
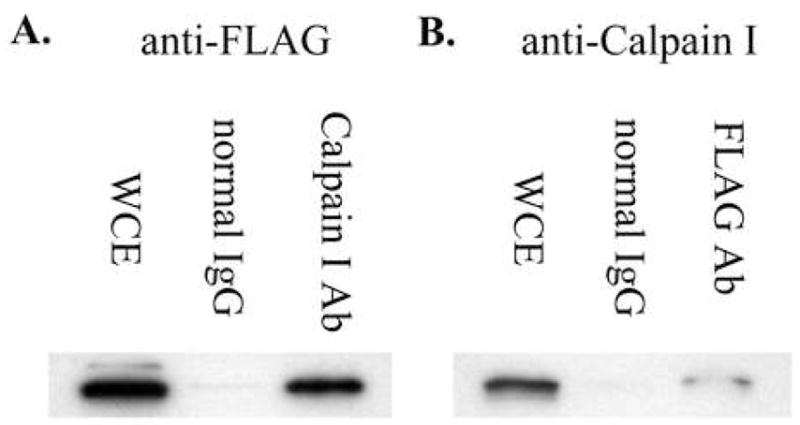
Co-immunoprecipitation of OGG1 and Calpain I. A. Anti-FLAG (OGG1) western blot of HeLa whole cell extract (WCE) (lane 1), proteins pulled down from WCE with normal mouse IgG (lane 2), or Calpain I antibody (lane 3). B. Anti-Calpain I western blot of HeLa WCE (lane 1), proteins pulled down from WCE with normal mouse IgG (lane 2), or FLAG antibody (lane 3).
3.4 Proteolysis of polymorphic OGG1 S326C by calpain
The calpain cleavage (PEST) sequence in the OGG1 C-terminus identified by C-terminal sequencing of calpain-digested OGG1 encompasses amino acid serine 326, the site of a prevalent cancer-associated serine to cysteine OGG1 S326C (rs1052133) polymorphism (24). Amino acid substitutions in PEST sequences frequently alter or abolish calpain cleavage at such sites. Using conditions identical to those used for the wild-type enzyme, OGG1 S326C was reacted with purified Calpain I (Fig. 6A). The substitution of serine for cysteine at position 326 in OGG1 greatly reduced in vitro cleavage of the enzyme by calpain, with roughly 10% cleavage occurring after 2 hrs (Fig. 6A, lane 5), compared to complete cleavage of the wild-type enzyme in an identical reaction (Fig. 2, lane 5). Similar to the wild-type enzyme, OGG1 S326C expressed in human cells was degraded following exposure to hydrogen peroxide (Fig. 6B) or cisplatin (Fig. 6C). Inhibition of OGG1 S326C degradation by calpeptin pretreatment suggests that the isoform is also degraded by a calpain-dependent mechanism in vivo. The percentage of apoptotic cells following treatment with either hydrogen peroxide or cisplatin was similar to that observed in wild-type OGG1-transfected cells (data not shown).
Figure 6.
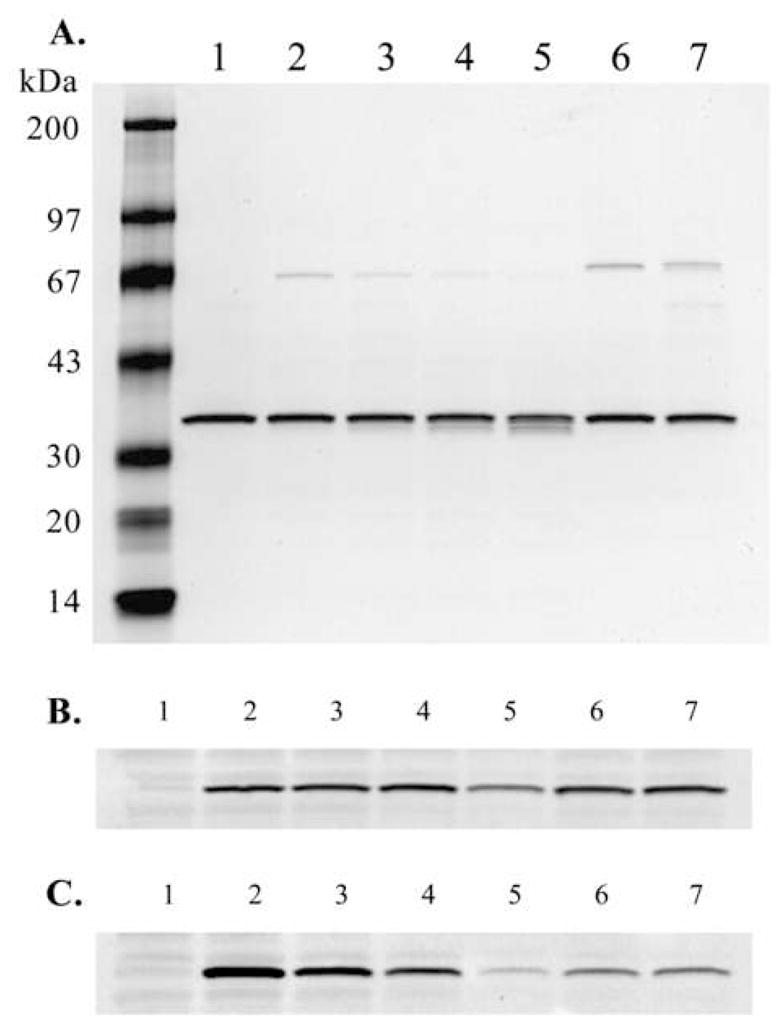
In vitroand in vivo proteolysis of polymorphic OGG1 S326C by calpain. A. In an experiment identical to that shown in Fig. 2, OGG1 S326C was reacted with purified Calpain I. B. HeLa cells transfected with OGG1 S326C were exposed to hydrogen peroxide for 24 hrs in an experiment identical to that shown in Fig. 3A. C. HeLa cells transfected with OGG1 S326C were exposed to cisplatin for 24 hrs in an experiment identical to that shown in Fig. 3B.
Discussion
We show that OGG1 is cleaved and degraded by calpain in vitro, interacts with calpain in cells, and is degraded by calpain-dependent proteolysis in vivo after exposure to hydrogen peroxide and cisplatin. The upregulation of calpain activity, influx of Ca2+, and induction of apoptosis are well characterized effects of oxidative stress (27). Hydrogen peroxide exposure results in oxidative stress by direct production of damaging oxygen radicals in cells via the Fenton reaction (28). The toxicity of cisplatin, a common chemotherapeutic agent known to induce apoptosis and activate intracellular calpain (4), is also mediated partly through ROS production. Cisplatin inactivates glutathione and antioxidant enzymes superoxide dismutase, catalase, glutathione peroxidase and glutathione reductase leading to intracellular accumulation of endogenously produced ROS and thereby elevated genomic 8-oxoguanine, calcium influx, and calpain activation (29–31). The significant inhibition of ROS-induced apoptosis by calpeptin (Fig. 3A) highlights the major role of calpains in cell death pathways following ROS exposure.
The failure of the PESTfind program to identify OGG1 as a high-score consensus calpain substrate underscores the variability in calpain cleavage sequences. Many known high affinity calpain substrates do not possess PEST sequences and many proteins containing consensus PEST sequences are not calpain substrates (32). Calpains may then recognize a localized conformation that may occur preferentially, though not always, at PEST sequences. Although OGG1 was slightly degraded by calpain in vitro, our C-terminal sequencing of a partially calpain-digested OGG1 showed a preferential cleavage that removes the C-terminal 16 amino acids (Figs. 1 and 2). The 4 poor PEST sequences identified within the OGG1 enzyme by the PESTfind program (Fig. 1) may be other points at which OGG1 is degraded by calpain. That OGG1 was degraded rather than truncated in cells with activated calpain may indicate differences between the in vitro and in vivo specificities of calpain or suggest the action of additional proteases in calpain-dependent OGG1 degradation. The latter possibility is supported by the observation that mouse OGG1, which shares significant homology with human OGG1, was cleaved in vivo by a caspase-dependent mechanism during apoptosis induced by etoposide or mitomycin C (33).
We previously demonstrated that removal of the C-terminal 20 amino acids of OGG1 results in a fully functional enzyme with reduced DNA binding affinity (24). However, the C-terminal 11 amino acids comprise the OGG1 nuclear localization signal (NLS) (34). Since OGG1 contains both an N-terminal mitochondrial localization sequence (34) and a C-terminal NLS, low-level intracellular calpain activation and cleavage of OGG1 could result in a truncated form of OGG1 that would retain repair activity and localize to mitochondria. The predominant mitochondrial OGG1 isoform, OGG1–2a, was shown to be devoid of enzymatic activity (35). Since roughly 10% of OGG1–1a resides in mitochondria, it was proposed that nuclear OGG1–1a may also be the major mitochondrial OGG1 activity (35). Proteolytically modified OGG1–1a species lacking an NLS may serve such a role. This possibility is supported by the detection of multiple OGG1 species, smaller in size than the full-length OGG1–1a, in mitochondria (35).
Mitochondrial OGG1–2a is identical to OGG1–1a in amino acids 1–316 but lacks the experimentally identified OGG1 PEST sequence in the OGG1–1a C-terminus (residues 324–333, Fig. 1). Accordingly, it is anticipated that the mitochondrial isoform would be less sensitive to degradation during ROS-induced calpain activation. We determined the effect of hydrogen peroxide and cisplatin exposure on levels of endogenously expressed OGG1–1a and –2a (Fig. 5). After hydrogen peroxide exposure, OGG1–1a was decreased by roughly half while no significant change was observed in the mitochondrial enzyme (Fig 5, lane 2). In contrast, cisplatin was more than twice as cytotoxic as hydrogen peroxide at the highest dose used (Fig. 3) and resulted in significant degradation of both OGG1–1a and –2a (Fig. 5 lane 5). In cells exposed to hydrogen peroxide, pretreatment with calpeptin resulted in complete protection of OGG1–1a (Fig. 3A, lane 6 and 7, Fig. 5, lanes 3 and 4). However, calpeptin pretreatment prior to cisplatin exposure only partially protected either OGG1 isoform (Fig. 3B, lanes 6 and 7, Fig. 5, lanes 6 and 7) and suggests that a significant fraction of OGG1 degradation following cisplatin exposure is independent of calpain activity.
The amino acid substitution of the OGG1 S326C variant greatly reduced cleavage of the enzyme in vitro (Fig. 6A) and confirms the involvement of the 324–333 region in the cleavage of OGG1 by calpain in vitro. Following hydrogen peroxide exposure, OGG1 S326C in cells was degraded less than the wild-type enzyme in an equivalent experiment (compare Fig. 3A, lane 5 and Fig. 6B, lane 5). OGG1 S326C was degraded similarly to wild-type OGG1 after cisplatin exposure (Fig. 6C). These results suggest that S326C may be moderately less sensitive to calpain-mediated degradation in vivo under conditions of oxidative stress.
Our finding of calpain degradation of OGG1 is the first report of proteolysis of the human enzyme. Interestingly, other components of the base excision repair pathway, including DNA ligase III and Poly (ADP-ribose) polymerase (PARP), have been identified as _targets for calpain degradation (36,37). While some calpain substrates, such as PARP and DNA polymerase ε, are discretely cleaved in vivo (37,38), others such as DNA ligase III (36) and OGG1 may be completely degraded upon activation of calpain. Further studies are warranted to elucidate the physiological significance of OGG1 proteolysis. Increases in cellular ROS, such as those produced by hydrogen peroxide or cisplatin exposure, result in elevated 8-oxoguanine in DNA, the repair of which may be negatively regulated by ROS-induced OGG1 proteolysis. Cleavage of OGG1 by calpain may influence intracellular localization of the enzyme in response to oxidative stress. Calpain-mediated degradation of OGG1 may underlie or contribute to elevated levels of 8-oxoguanine, low levels of OGG1 enzyme and decreased 8-oxoguanine repair activity observed in tissues with disruption of intracellular calcium regulation and calpain overactivation due to chronic oxidative stress.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging and National Cancer Institute grants CA73629 and CA090898.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harwood SM, Yaqoob MM, Allen DA. Caspase and calpain function in cell death: bridging the gap between apoptosis and necrosis. Ann Clin Biochem. 2005;42:415–31. doi: 10.1258/000456305774538238. [DOI] [PubMed] [Google Scholar]
- 2.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–6. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 3.Bizat N, Hermel JM, Humbert S, Jacquard C, Creminon C, Escartin C, Saudou F, Krajewski S, Hantraye P, Brouillet E. In vivo calpain/caspase cross-talk during 3-nitropropionic acid-induced striatal degeneration: implication of a calpain-mediated cleavage of active caspase-3. J Biol Chem. 2003;278:43245–53. doi: 10.1074/jbc.M305057200. [DOI] [PubMed] [Google Scholar]
- 4.Del Bello B, Moretti D, Gamberucci A, Maellaro E. Cross-talk between calpain and caspase-3/-7 in cisplatin-induced apoptosis of melanoma cells: a major role of calpain inhibition in cell death protection and p53 status. Oncogene. 2007;26:2717–26. doi: 10.1038/sj.onc.1210079. [DOI] [PubMed] [Google Scholar]
- 5.Waterhouse NJ, Finucane DM, Green DR, Elce JS, Kumar S, Alnemri ES, Litwack G, Khanna K, Lavin MF, Watters DJ. Calpain activation is upstream of caspases in radiation-induced apoptosis. Cell Death Differ. 1998;5:1051–61. doi: 10.1038/sj.cdd.4400425. [DOI] [PubMed] [Google Scholar]
- 6.Varghese J, Radhika G, Sarin A. The role of calpain in caspase activation during etoposide induced apoptosis in T cells. Eur J Immunol. 2001;31:2035–41. doi: 10.1002/1521-4141(200107)31:7<2035::aid-immu2035>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 7.McCollum AT, Nasr P, Estus S. Calpain activates caspase-3 during UV-induced neuronal death but only calpain is necessary for death. J Neurochem. 2002;82:1208–20. doi: 10.1046/j.1471-4159.2002.01057.x. [DOI] [PubMed] [Google Scholar]
- 8.Kim KA, Lee YA, Shin MH. Calpain-dependent calpastatin cleavage regulates caspase-3 activation during apoptosis of Jurkat T cells induced by Entamoeba histolytica. Int J Parasitol. 2007;37:1209–19. doi: 10.1016/j.ijpara.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Zatz M, Starling A. Calpains and disease. N Engl J Med. 2005;352:2413–23. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- 10.Camins A, Verdaguer E, Folch J, Pallas M. Involvement of calpain activation in neurodegenerative processes. CNS Drug Rev. 2006;12:135–48. doi: 10.1111/j.1527-3458.2006.00135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris F, Biswas S, Singh J, Dennison S, Phoenix DA. Calpains and their multiple roles in diabetes mellitus. Ann NY Acad Sci. 2006;1084:452–80. doi: 10.1196/annals.1372.011. [DOI] [PubMed] [Google Scholar]
- 12.Raynaud F, Marcilhac A. Implication of calpain in neuronal apoptosis. A possible regulation of Alzheimer’s disease. FEBS J. 2006;273:3437–43. doi: 10.1111/j.1742-4658.2006.05352.x. [DOI] [PubMed] [Google Scholar]
- 13.Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121:2381–6. doi: 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- 14.Monteith GR, McAndrew D, Faddy HM, Roberts-Thomson SJ. Calcium and cancer: _targeting Ca2+ transport. Nat Rev Cancer. 2007;7:519–30. doi: 10.1038/nrc2171. [DOI] [PubMed] [Google Scholar]
- 15.Carragher NO, Frame MC. Calpain: a role in cell transformation and migration. Int J Biochem Cell Biol. 2002;34:1539–43. doi: 10.1016/s1357-2725(02)00069-9. [DOI] [PubMed] [Google Scholar]
- 16.Lovell MA, Xie C, Markesbery WR. Decreased base excision repair and increased helicase activity in Alzheimer’s disease brain. Brain Res. 2000;855:116–23. doi: 10.1016/s0006-8993(99)02335-5. [DOI] [PubMed] [Google Scholar]
- 17.Markesbery WR, Lovell MA. DNA oxidation in Alzheimer’s disease. Antioxid Redox Signal. 2006;8:2039–45. doi: 10.1089/ars.2006.8.2039. [DOI] [PubMed] [Google Scholar]
- 18.Tsuruya K, Furuichi M, Tominaga Y, Shinozaki M, Tokumoto M, Yoshimitsu T, Fukuda K, Kanai H, Hirakata H, Iida M, Nakabeppu Y. Accumulation of 8-oxoguanine in the cellular DNA and the alteration of the OGG1 expression during ischemia-reperfusion injury in the rat kidney. DNA Repair (Amst) 2003;2:211–29. doi: 10.1016/s1568-7864(02)00214-8. [DOI] [PubMed] [Google Scholar]
- 19.Sakuraba H, Mizukami H, Yagihashi N, Wada R, Hanyu C, Yagihashi S. Reduced beta-cell mass and expression of oxidative stress-related DNA damage in the islet of Japanese Type II diabetic patients. Diabetologia. 2002;45:85–96. doi: 10.1007/s125-002-8248-z. [DOI] [PubMed] [Google Scholar]
- 20.Inoue M, Osaki T, Noguchi M, Hirohashi S, Yasumoto K, Kasai H. Lung cancer patients have increased 8-hydroxydeoxyguanosine levels in peripheral lung tissue DNA. Jpn J Cancer Res. 1998;89:691–695. doi: 10.1111/j.1349-7006.1998.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mambo E, Chatterjee A, de Souza-Pinto NC, Mayard S, Hogue BA, Hoque MO, Dizdaroglu M, Bohr VA, Sidransky D. Oxidized guanine lesions and hOgg1 activity in lung cancer. Oncogene. 2005;24:4496–4508. doi: 10.1038/sj.onc.1208669. [DOI] [PubMed] [Google Scholar]
- 22.Siems W, Capuozzo E, Lucano A, Salerno C, Crifo C. High sensitivity of plasma membrane ion transport ATPases from human neutrophils towards 4-hydroxy-2,3-trans-nonenal. Life Sci. 2003;73:2583–2590. doi: 10.1016/s0024-3205(03)00661-1. [DOI] [PubMed] [Google Scholar]
- 23.MacManus A, Ramsden M, Murray M, Henderson Z, Pearson HA, Campbell VA. Enhancement of 45Ca2+ influx and voltage-dependent Ca2+ channel activity by β-amyloid-(1–40) in rat cortical synaptosomes and cultured cortical neurons. Modulation by the proinflammatory cytokine interleukin-1beta. J Biol Chem. 2000;275:4713–8. doi: 10.1074/jbc.275.7.4713. [DOI] [PubMed] [Google Scholar]
- 24.Hill JW, Evans MK. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res. 2006;34:1620–32. doi: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 26.Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. TIBS. 1996;21:267–271. [PubMed] [Google Scholar]
- 27.Ray SK, Fidan M, Nowak MW, Wilford GG, Hogan EL, Banik NL. Oxidative stress and Ca2+ influx upregulate calpain and induce apoptosis in PC12 cells. Brain Res. 2000;852:326–34. doi: 10.1016/s0006-8993(99)02148-4. [DOI] [PubMed] [Google Scholar]
- 28.Marlatt M, Lee HG, Perry G, Smith MA, Zhu X. Sources and mechanisms of cytoplasmic oxidative damage in Alzheimer’s disease. Acta Neurobiol Exp (Wars) 2004;64:81–7. doi: 10.55782/ane-2004-1493. [DOI] [PubMed] [Google Scholar]
- 29.Salganik RI. The benefits and hazards of antioxidants: controlling apoptosis and other protective mechanisms in cancer patients and the human population. J Am Coll Nutr. 2001;20:464S–472S. doi: 10.1080/07315724.2001.10719185. [DOI] [PubMed] [Google Scholar]
- 30.Rybak LP, Husain K, Morris C, Whitworth C, Somani S. Effect of protective agents against cisplatin ototoxicity. Am J Otol. 2000;21:513–20. [PubMed] [Google Scholar]
- 31.Conklin KA. Cancer chemotherapy and antioxidants. J Nutr. 2004;134:3201S–3204S. doi: 10.1093/jn/134.11.3201S. [DOI] [PubMed] [Google Scholar]
- 32.Molinari M, Anagli J, Carafoli E. PEST sequences do not influence substrate susceptibility to calpain proteolysis. J Biol Chem. 1995;270:2032–5. doi: 10.1074/jbc.270.5.2032. [DOI] [PubMed] [Google Scholar]
- 33.Hirano T, Kawai K, Ootsuyama Y, Orimo H, Kasai H. Detection of a mouse OGG1 fragment during caspase-dependent apoptosis: oxidative DNA damage and apoptosis. Cancer Sci. 2004;95:634–8. doi: 10.1111/j.1349-7006.2004.tb03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell. 1999;10:1637–52. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashiguchi K, Stuart JA, de Souza-Pinto NC, Bohr VA. The C-terminal alphaO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial beta-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bordone L, Campbell C. DNA ligase III is degraded by calpain during cell death induced by DNA-damaging agents. J Biol Chem. 2002;277:26673–80. doi: 10.1074/jbc.M112037200. [DOI] [PubMed] [Google Scholar]
- 37.McGinnis KM, Gnegy ME, Park YH, Mukerjee N, Wang KK. Procaspase-3 and poly(ADP)ribose polymerase (PARP) are calpain substrates. Biochem Biophys Res Commun. 1999;263:94–9. doi: 10.1006/bbrc.1999.1315. [DOI] [PubMed] [Google Scholar]
- 38.Liu W, Linn S. Proteolysis of the human DNA polymerase epsilon catalytic subunit by caspase-3 and calpain specifically during apoptosis. Nucleic Acids Res. 2000;28:4180–8. doi: 10.1093/nar/28.21.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]



