Abstract
Throughout the last decade, great advances have been made in our understanding of how DNA-templated cellular processes occur in the native chromatin environment. Proteins that regulate transcription, replication, DNA repair, mitosis and other processes must be _targeted to specific regions of the genome and granted access to DNA, which is normally tightly packaged in the higher-order chromatin structure of eukaryotic nuclei. Massive multiprotein complexes have been discovered, which facilitate access to DNA and recruitment of downstream effectors through three distinct mechanisms: chemical modification of histone amino-acid residues, ATP-dependent chromatin remodeling and histone exchange. The yeast Spt-Ada-Gcn5-Acetyl transferase (SAGA) transcriptional co-activator complex regulates numerous cellular processes through coordination of multiple histone post-translational modifications. SAGA is known to generate and interact with a number of histone modifications, including acetylation, methylation, ubiquitylation and phosphorylation. Although best characterized for its role in regulating transcriptional activation, SAGA is also required for optimal transcription elongation, mRNA export and perhaps nucleotide excision repair. Here, we discuss findings from recent years that have elucidated the function of this 1.8-MDa complex in multiple cellular processes, and how misregulation of the homologous complexes in humans may ultimately play a role in development of disease.
Keywords: SAGA, histone, modification, acetyl transferase, chromatin, transcription
Nucleosomal acetylation: Gcn5 needs a little help from its friends
Over a decade ago, Brownell and Allis (1995) discovered the first nuclear histone acetyltransferase (HAT) in the ciliate Tetrahymena thermophila. Originally named p55, this HAT was shown to be highly homologous to the Saccharomyces cerevisiae transcriptional co-activator Gcn5, which was, soon thereafter, also shown to possess HAT activity (Brownell et al., 1996). Thus, histone acetylation was directly linked to activation of gene expression. A puzzling observation from early in vitro studies was the apparent inability of recombinant yeast Gcn5 to efficiently acetylate nucleosomal histones, in stark contrast to its potent HAT activity on recombinant histones. Workman and colleagues subsequently showed that native yeast Gcn5 exists in a number of high-molecular-weight complexes, multiple subunits of which enable Gcn5 to acetylate nucleosomes (Grant et al., 1997). The more notable of these complexes was named SAGA, for Spt-Ada-Gcn5-Acetylatransferase, and preferentially acetylates multiple lysine residues on the N-terminal tails of histones H3 and H2B. Further analysis of the composition and function of the SAGA complex and its orthologs in higher eukaryotes has served as a paradigm for our understanding of the coupling of histone modifications with transcriptional co-activation from yeast to humans. Here, we review the organization and function of the S. cerevisiae SAGA complex, and highlight recent insights into the rapidly expanding cellular role of this transcriptional co-activator. The highly conserved mammalian GCN5/PCAF complexes are discussed more extensively in a separate article in this issue (Nagy and Tora, 2007).
The discovery of SAGA clarified years of genetic and biochemical studies on transcriptional regulation by showing that Gcn5 exists in complex with at least three protein families known to function in gene expression: the Ada and Spt protein families, and a subset of TATA-binding protein (TBP)-associated factors (TAFs; Grant et al., 1997, 1998a). The Ada proteins (alteration/deficiency in activation) are encoded by genes that when mutated alleviate the toxicity of the chimeric activator Gal4-VP16 (Berger et al., 1992). The Ada1–5 proteins are components of SAGA (Timmers and Tora, 2005). Ada4 is identical to Gcn5, and has been shown to interact with Ada2 in vivo and in vitro, establishing a physical and genetic link between these transcriptional adaptors (Marcus et al., 1994). Ada5 is identical to Spt20, which along with Spt3, -7 and -8 make up the second protein family in SAGA (Marcus et al., 1996; Timmers and Tora, 2005). The SPT genes (suppressor of Ty) were identified as suppressors of transcriptional defects caused by insertion of Ty transposable elements at the 5′ regions of genes (reviewed in Winston and Carlson, 1992). Although the roles of the Spt proteins in transcription remain somewhat elusive, they are believed to facilitate interactions between SAGA and TBP at certain gene promoters; interestingly, TBP itself was also isolated as the product of an SPT gene in this genetic screen (Eisenmann et al., 1989, 1994; Dudley et al., 1999; Sterner et al., 1999; Larschan and Winston, 2001). The finding that proteins from the Spt and Ada families exist in a multiprotein complex and cooperate to allow nucleosomal acetylation by Gcn5 was a large step forward in understanding both the genetics and biochemistry of transcriptional activation.
TAFs 5, 6, 9, 10 and 12 were also subsequently shown to be integral components of SAGA (Grant et al., 1998a). TAFs are highly conserved proteins that, along with TBP, make up the general transcription initiation factor, TFIID (reviewed in Green, 2000). TFIID is required for promoter recognition in RNA polymerase II (Pol II)-catalyzed transcription at most genes (reviewed in Burley and Roeder, 1996). The identification of TAFs in SAGA was the first discovery that these proteins function outside the context of TFIID, and physically links the members of three distinct families of gene products known to be involved in transcriptional activation (Grant et al., 1998a). Genome-wide expression analysis indicates that SAGA and TFIID, despite sharing a number of common subunits, are responsible for expression of different subsets of genes. While SAGA functions mostly at highly regulated genes that respond to environmental stresses, such as metabolic starvation, DNA damage and heat, TFIID plays a more general housekeeping role (Huisinga and Pugh, 2004).
Altogether, SAGA is a 1.8-MDa complex consisting of more than 20 polypeptide subunits (Table 1). Although it remains unclear, how the other subunits of SAGA enable Gcn5 to acetylate nucleosomes, it is believed that Gcn5, Ada2 and Ada3 compose a catalytic core minimally capable of nucleosomal acetylation (Balasubramanian et al., 2002). However, defects to other SAGA components have been shown to compromise native Gcn5 activity, for instance, mutation of TAF12 greatly reduces the ability of SAGA to acetylate nucleosomes and activate transcription in vitro (Grant et al., 1998a). The mechanistic dependence of nucleosomal acetylation on TAF12, as well as other SAGA subunits, remains unclear. Additionally, several complex components are known to be required for the overall stability of SAGA. Mutation of the Ada1, Spt7 or Spt20 subunits results in complete disruption of the SAGA complex (Grant et al., 1997; Sterner et al., 1999). These findings, in combination with mutational analyses of the structurally nonessential components, indicate that SAGA is divided into several discrete modules responsible for nucleosome acetylation, TBP interaction and complex integrity (Sterner et al., 1999).
Table 1.
Molecular composition of the SAGA/SLIK HAT complexes
| SAGA/SLIK complex Biological function component | ||
|---|---|---|
| Gcn5(Ada4) | Acetylation of lysines on H3 and H2B; transcriptional activation; NER recognition of acetylated lysine via bromodomain |
|
| Ada1 | Complex stability | |
| Ada2 | Required for nucleosomal acetylation by Gcn5 | |
| Ada3 | ||
| Spt3 | TBP interaction, transcriptional repression at | |
| Spt8 | HIS3 and ARG1 loci | |
| Spt7 | Complex stability | |
| Spt20(Ada5) | ||
| TAF5 | ||
| TAF6 | Structural integrity of complex | |
| TAF9 | Interaction with basal transcription machinery | |
| TAF10 | ||
| TAF12 | Required for nucleosomal acetylation by Gcn5 Interaction with transcriptional activators |
|
| Tra1 | Interaction with transcriptional activators | |
| Ubp8 | Deubiquitylation of H2B Lys 123; transcriptional activation | |
| Rtg2 | SLIK stability; links complex to retrograde response pathway | |
| Chd1 | Recognition of H3 Lys 4 methylation via chromodomain; potentiation of histone acetylation by Gcn5 |
|
| Sus1 | mRNA export | |
| Sgf11 | Required for association of Ubp8 and Sus1 with SAGA |
|
| Sgf29 | ? | |
| Sca7 | Poly(Q) expansion inhibits nucleosomal acetylation by Gcn5 |
|
Abbreviations: NER, nucleotide excision repair; SAGA, Spt-Ada-Gcn5-Acetyl transferase; TAF, TBP-associated factors; TBP, TATA-binding protein. Protein subunits of the SAGA/SLIK histone acetyltransferase complexes in Saccharomyces cerevisiae are listed and grouped by gene family where appropriate. Biological functions of proteins are listed for subunits that have been characterized.
Electron microscopy and immunolabeling have provided a low-resolution three-dimensional model of SAGA, further proving that the complex components are indeed grouped into distinct modules, which may differentially contribute to SAGA function (Wu et al., 2004). Image reconstruction from this study indicates that SAGA is composed of five modular domains. The first of these contains the essential Tra1 protein, which contributes to SAGA interaction with acidic activators (see below). Domains II, III and IV contain several TAFs; the central domain III additionally contains Gcn5, and thus harbors the HAT activity of SAGA. Finally the more flexible domain V contains Spt20, Spt3 and possibly Spt8, potentially representing a surface for TBP interaction. These findings are reviewed further by Timmers and Tora (2005).
_targeting SAGA to gene promoters
For SAGA to function effectively as a transcriptional co-activator at inducible genes, it must be selectively _targeted to the promoter regions of these genes. The _targeting of SAGA is mediated primarily through direct interaction with acidic activator domains of transcriptional activators, such as Gal4 and Gcn4 (Utley et al., 1998; Ikeda et al., 1999). The interaction of SAGA with promoter-bound activators is facilitated largely by Tra1, a >400KDa subunit of the complex (Grant et al., 1998b; Brown et al., 2001). Given its immense size, Tra1 may also serve as a structural scaffold of SAGA essential to the stability of the complex. Tra1 is the yeast homolog of the human transformation/transcription domain-associated protein (TRRAP), which is required for c-Myc- and E1A-mediated oncogenic transformation (McMahon et al., 1998; Grant et al., 1998b). In yeast, Tra1 is similarly necessary for the growth phenotype observed on expression of the N-terminal 81 amino acids of E1A (Kulesza et al., 2002).
S. cerevisiae Tra1 is stably associated not only with SAGA but also with the NuA4 HAT complex, indicating that it may play a general role in recruiting transcriptional co-activators through interaction with acidic activators (Grant et al., 1998b). Indeed, Tra1 has been shown through various methods to interact with a number of acidic activators, including Gcn4, VP16, Gal4 and Hap4, in the context of both SAGA and NuA4 (Brown et al., 2001). More recently, fluorescence resonance energy transfer (FRET) has provided direct in vivo evidence that Tra1 interacts with the Gal4 activator protein. Interestingly, deletion of SPT20 results in a loss of FRET signal, indicating that Tra1 associated with SAGA alone, and not NuA4, is responsible for the interaction with Gal4 (Bhaumik et al., 2004). This finding also suggests that the interaction between Tra1 and Gal4 is facilitated in part by other SAGA subunits. In agreement, the SAGA subunit TAF12 was recently shown to interact with both Gal4 and Gcn4 (Reeves and Hahn, 2005). Interestingly, human TRRAP similarly functions to recruit the SAGA homolog STAGA (Spt3-Taf9-Ada-Gcn5 acetyltransferase) to gene promoters through interaction with the c-Myc transactivation domain (McMahon et al., 2000; Liu et al., 2003). Thus TRRAP’s role in oncogenic transformation may be achieved primarily through aberrant gene expression due to avhieved improper STAGA recruitment. The role of TRRAP in chromatin-based processes is further discussed in a separate article in this issue (Murr et al., 2007).
Ubp8: modulating the ‘trans-histone’ modification pathway
SAGA also contains a number of subunits that contribute to its role in transcriptional activation without directly affecting complex stability or HAT activity. One such protein, Ubp8, is a ubiquitin protease that specifically removes monoubiquitin from lysine 123 of the H2B C-terminal tail (Henry et al., 2003; Daniel et al., 2004). Monoubiquitylation of H2B Lys 123 is catalyzed by the Rad6/Bre1 ubiquitin-conjugating/ligating enzymes, and is required for Set1-mediated di- and trimethylation of H3 lysine 4, a modification that has been well characterized as a mark of actively transcribed chromatin (Dover et al., 2002; Santos-Rosa et al., 2002; Sun and Allis, 2002; Hwang et al., 2003; Wood et al., 2003). This coordination of multiple post-translational modifications on different histone tails is referred to as the ‘trans-histone’ regulatory pathway (Briggs et al., 2002).
It is anticipated that loss of Ubp8 activity would lead to an increase of cellular-ubiquitylated H2B and, consequently, affect the dynamics of H3 Lys 4 methylation. Indeed, H3 Lys 4 trimethylation increases at the GAL1 promoter when UBP8 is deleted; this finding is expected given the dependence of H3 trimethylation on H2B ubiquitylation (Henry et al., 2003). However, contradictory evidence has shown that deletion of UBP8 results in a loss of H3 Lys 4 trimethylation at the GAL1–10 upstream activating sequence (UAS), indicating that the removal of ubiquitin rather than just its conjugation to H2B is important for H3 methylation (Daniel et al., 2004). This result points to a more causal role of Ubp8 in the transition to highly methylated H3 Lys 4, and hence, to transcriptionally active chromatin.
Further complicating the rapidly unfolding scenario of histone modifications at gene promoters, recent experiments using a highly reconstituted human in vitro transcription system indicate that H2B monoubiquitylation is required for transcriptional elongation independent of H3 Lys 4 methylation status (Pavri et al., 2006). This finding suggests regional differences in the requirements of H2B ubiquitylation and H3 Lys 4 methylation within an actively transcribing gene. It is possible, however, that the absence of downstream effectors for the trimethyl moiety on H3 Lys 4 in this in vitro system may account for the lack of a heightened elongation efficiency when the modification is present (Pavri et al., 2006). Further experiments are required to resolve the details of these distinct results; however, it is clear that Ubp8 function is directly linked to the methylation state of H3 Lys 4, and plays an important role in transcriptional activation of SAGA-regulated genes.
Recently, Ubp8 was shown to form a distinct module within the SAGA complex with another structurally nonessential component, Sgf11. Sgf11 (11-kDa SAGA-associated factor) was identified by mass spectrometry as a component of SAGA (Powell et al., 2004; Ingvarsdottir et al., 2005; Lee et al., 2005b). Although deletion of SGF11 has essentially no effect on complex stability or HAT activity, microarray analyses indicate that its loss alters transcription of a number of SAGA-regulated genes (Powell et al., 2004). Sgf11 is required for the association of Ubp8 with SAGA and, conversely, dependent on Ubp8 for its association with the complex; thus, as expected, sgf11Δ negatively affects Ubp8 function and results in an increase in cellular ubiquitylated H2B (Ingvarsdottir et al., 2005; Lee et al., 2005b; Shukla et al., 2006a). Interestingly, Ubp8 is able to deubiquitylate H2B only in the context of SAGA, and is thus similar to Gcn5 in that both enzymes require association with the entire complex for optimal activity (Grant et al., 1997; Lee et al., 2005b).
The proteasome 19S regulatory particle facilitates loading of SAGA onto chromatin
The relationship of SAGA with H2B ubiquitylation and H3 methylation may be further mediated by the 19S regulatory particle (19S RP) of the proteasome. The proteasome is well known for its function in ubiquitin-mediated protein degradation. The 19S RP along with the 20S catalytic core assemble in a barrel-like structure consisting of one 20S core sandwiched between two 19S RPs. This structure in whole is referred to as the 26S holoenzyme, and facilitates degradation of proteins. Polyubiquitin on _target proteins is recognized by the 19S RP, which directs these _target proteins to the 20S core for processing (reviewed in Kinyamu et al., 2005).
In recent years, our understanding of proteasome function has expanded beyond its role in protein degradation. Particularly, the 19S RP functions in multiple cellular processes including transcription and nucleotide excision repair (NER; reviewed in Muratani and Tansey, 2003). Coimmunoprecipitation has linked the proteasome to RNA Pol II, and genetic studies implicate the 19S RP subunits Sug1 and -2 in transcriptional elongation (Ferdous et al., 2001; Gillette et al., 2004). Additionally, the 19S RP interacts with the ubiquitin-like domain of Rad23, which recognizes damaged DNA in the NER pathway (Schauber et al., 1998). Optimal NER in vitro requires the activity of the proteasomal ATPase Sug1, while inhibition of proteolysis has no effect on NER, indicating that this activity is completely independent of the protein degradation pathway (Russell et al., 1999).
Interestingly, the 19S RP interacts directly with the SAGA complex and mediates histone modifications that are known to be affected by SAGA (Ezhkova and Tansey, 2004; Lee et al., 2005a). As mentioned previously, ubiquitylation of H2B Lys 123 is required for H3 Lys 4 methylation (Dover et al., 2002; Sun and Allis, 2002). This dependence is mediated by the 19S RP, as inhibition of proteasomal ATPases results in a loss of H3 Lys 4 methylation, but not H2B ubiquitylation (Ezhkova and Tansey, 2004). However, it is not known how the 19S RP regulates this trans-histone modification pathway. Recently, it was shown that the 19S RP facilitates loading of SAGA onto chromatin, and is required for optimal induction of the GAL1–10 UAS (Lee et al., 2005a). This finding not only elucidates another degradation-independent role for the proteasome, but may also explain its role in regulation of multiple modifications. The SAGA subunit Ubp8 is important for increased H3 Lys 4 trimethylation upon galactose induction (Daniel et al., 2004). Perhaps, loss of trimethylation at H3 Lys 4 upon inhibition of the 19S RP is due in part to the inability of SAGA to properly load onto chromatin. Furthermore, given that the 19S RP is associated with RNA Pol II and required for efficient transcription elongation, perhaps removal of the ubiquitin moiety on H2B Lys 123 by SAGA facilitates progression of the 19S RP from promoter to coding region within a transcribing gene (Ferdous et al., 2001; Gillette et al., 2004; Baker and Grant, 2005). These predictions would not explain, however, the loss of Lys 79 methylation or lower methylation states of Lys 4 observed upon proteasomal inhibition; thus there are undoubtedly multiple mechanisms by which the proteasome regulates modification of histones.
Sus1 couples transcription to mRNA export
In addition to its roles in transcriptional activation, SAGA has been linked to nuclear export of transcribed mRNA through the Sus1 protein (Rodriguez-Navarro et al., 2004). SUS1 (sl gene upstream of Ysa1) was originally identified through a synthetic lethality screen with YRA1, which encodes a component of the highly conserved mRNA export machinery (Sträβer and Hurt, 2000). Sus1 was subsequently shown to interact with several subunits of SAGA, as well as the Sac3 and Thp1 components of the nuclear export machinery (Rodriguez-Navarro et al., 2004). The interaction of Sac3–Thp1 with the nuclear pore complex (NPC) is mediated by docking with specific nucleoporins at the NPC entrance and is required for mRNA export (Fischer et al., 2002). Additionally, dynamic motility studies of the SAGA-regulated genes GAL1, GAL7 and GAL10 have shown that they are confined to the nuclear periphery when actively transcribed and that this localization is dependent on Sus1 (Cabal et al., 2006). Thus, Sus1 provides a physical link between SAGA and the nuclear export machinery.
Remarkably, in addition to defects in mRNA export, deletion of SUS1 also results in altered transcription of approximately 9% of yeast genes. The overlap between those genes affected by sus1Δ with mutation of other SAGA components is similar to that seen between SAGA mutants, indicating that Sus1 may function directly in SAGA-mediated transcriptional activation independently of mRNA export. This prospect is supported by the finding that Sus1 is recruited to the GAL1 promoter upon transcriptional induction in a manner similar to that of the SAGA subunit Ada2 (Rodriguez-Navarro et al., 2004).
Recently, a more thorough biochemical analysis has shown that Sus1 associates with both Ubp8 and Sgf11 within SAGA, and that a subcomplex containing these three proteins can be separated from the rest of the complex by treatment with high salt (Kohler et al., 2006). The association of Sus1 with SAGA is dependent on the presence of Ubp8, and the two subunits are co-dependent in recruitment to the GAL1 promoter, similarly to the relationship observed between Sgf11 and Ubp8 (Ingvarsdottir et al., 2005; Kohler et al., 2006). Additionally, deletion of SUS1 results in an increase of global ubiquitylated H2B and H3 lysine methylation. This is presumably due to loss of Ubp8 association with SAGA; however, co-precipitation experiments show that sus1Δ SAGA is not as tightly associated with histones as wild-type SAGA, indicating Sus1 may play an undescribed role in recognition of histone substrates (Kohler et al., 2006). Importantly, deletion of SGF11, but not UBP8, displays a synthetic mRNA export defect with sus1Δ, indicating that Sgf11 assists in the coupling of transcription and mRNA export (Kohler et al., 2006).
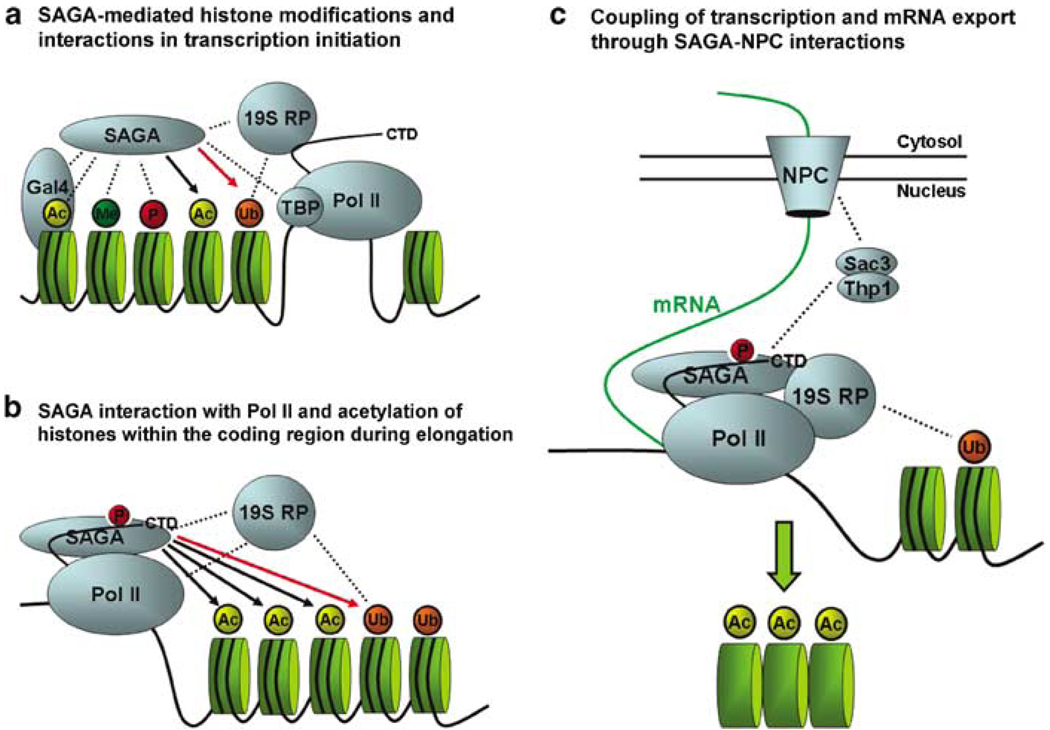
Furthermore, a recent report suggests that SAGA localizes to the coding sequences of genes and that acetylation by Gcn5 promotes nucleosome eviction during transcription elongation. This association with the coding sequence is dependent on phosphorylation of the Pol II C-terminal domain (CTD), indicating that SAGA may interact with the actively transcribing Pol II during elongation (Govind et al., 2007). On the basis of these data, along with the recently described functions of Sus1, Ubp8 and the 19S RP, we can begin to make predictions about the overall role of SAGA in regulation of the entire transcriptional process (Figure 1). As the 19S RP is known to interact with the ubiquitin moiety, it may be _targeted to the promoter region partially through the ubiquitylation of histone H2B. This in turn facilitates trimethylation of H3 Lys 4, which coordinates with transcriptional activation and potentiates the histone acetylation capability of SAGA (Pray-Grant et al., 2005). Removal of ubiquitin by Ubp8 could subsequently allow progression of both SAGA and the 19S RP into the coding region of the gene (Figure 1a). During elongation, SAGA interacts with the phosphorylated CTD of Pol II, and acetylates nucleosomes in the coding region of the transcribed gene. The recent finding that H2B is ubiquitylated during elongation may provide a mechanism for localization of the 19S RP to the coding region, and removal of this modification by SAGA could allow progression along the open reading frame (ORF), although H2B deubiquitylation in the coding region has not been described to date (Figure 1b). The eviction of nucleosomes acetylated by SAGA within the coding region ensures optimal processivity of Pol II, and the interaction of SAGA with the Sac3–Thp1 complex may facilitate a smooth continuum between transcriptional activation, elongation, and the export of newly synthesized mRNA (Figure 1c).
Figure 1. Proposed model of SAGA activities that ensure smooth transition between transcription initiation, elongation, and mRNA export.
(a) Upon transcriptional induction, SAGA is recruited to the gene promoter primarily through interaction with an acidic activator (Gal4 is shown) and loaded onto chromatin by the proteasome 19S RP. 19S RP interaction with monoubiquitin on H2B may be important for transcriptional regulation, as it is required for H3 Lys 4 trimethylation and may aid in _targeting the 19S RP to the promoter region. SAGA interacts with trimethylation, phosphorylation and acetylation on histone H3. These interactions mediate further H3 acetylation by SAGA, which facilitates initiation of transcription. Removal of H2B monoubiquitin by SAGA is also important for transcriptional activation, and could allow progression of the 19S RP into the coding region. (b) Both SAGA and the 19S RP are associated with the coding regions of genes during transcriptional elongation, and SAGA may functionally interact with phosphorylation on the Pol II CTD. Histone H3 acetylation by SAGA in the coding region is important for nucleosome eviction. H2B ubiquitylation is also important for transcriptional elongation, and perhaps its removal within the coding region allows continued progression. (c) Eviction of acetylated nucleosomes allows increased processivity of Pol II. As nascent mRNA is formed, interactions between SAGA and components of the nuclear pore complex (NPC) maintain localization of actively transcribed genes to the nuclear periphery and allow for efficient coupling between synthesis and export. Dotted lines indicate physical interactions, black and red arrows designate addition and removal of post-translational modifications, respectively. 19S RP, 19S regulatory particle; Pol II CTD, polymerase II C-terminal domain; SAGA, Spt-Ada-Gcn5-Acetyl transferase.
SLIK: linking histone acetylation and the retrograde response pathway
Subsequent to the discovery of SAGA, Gcn5 was shown to exist in another multiprotein complex similar in size to SAGA, yet chromatographically distinct. This complex was named SLIK, for SAGA-like (also called SALSA; SAGA altered, Spt8 absent; Pray-Grant et al., 2002; Sterner et al., 2002). Despite containing the vast majority of polypeptides found in SAGA, SLIK composition is divergent in a number of ways. Most notably, the protein Rtg2, which is a core component of SLIK but not SAGA, is essential to the stability of SLIK and links histone acetylation to the retrograde response pathway (Pray-Grant et al., 2002). Retrograde response signaling is responsible for communicating to the nucleus the need to make metabolic adjustments in times of mitochondrial dysfunction. More specifically, retrograde response induces expression of genes whose products compensate for defects in the tricarboxylic acid cycle and allows use of acetate as a carbon source (reviewed in Butow and Avadhani, 2004). Thus, the biological function of SLIK is consistent with SAGA’s role in regulating stress–response genes.
There are three known positive regulators of the retrograde response pathway, RTG1–3. The products of these genes are required for transcriptional induction of CIT2, expression of which is a hallmark of mitochondrial dysfunctions (Butow and Avadhani, 2004). Rtg1 and -3 form a heterodimeric complex that binds the CIT2 UASr element and is required for CIT2 expression (Jia et al., 1997). The role of Rtg2 in CIT2 expression, however, is not completely understood. Rtg2 exists predominantly in the cytoplasm, where it is thought to function as a sensor of mitochondrial stress and to facilitate entry of the Rtg1–3 complex into the nucleus (Sekito et al., 2000). The finding that Rtg2 is also an essential component of the SLIK co-activator complex and occupies chromatin at the CIT2 promoter expands its role in retrograde response signaling to a more direct effect in transcriptional induction and illuminates a previously undescribed pool of nuclear Rtg2 (Pray-Grant et al., 2002).
SLIK also differs from SAGA in that it lacks the Spt8 protein component (Pray-Grant et al., 2002; Sterner et al., 2002). Interestingly, deletion of Spt8 causes derepression of some inducible genes, such as HIS3. This effect is strikingly different from deletion of SAGA subunits that completely disrupt the complex, such as spt20Δ, which result in transcriptional defects at these loci. These findings indicate that SAGA plays both a positive and negative role in expression of these genes (Belotserkovskaya et al., 2000). SAGA has similarly been suggested to function in transcriptional repression of the ARG1 gene in rich medium, in a manner directly related to Gcn5 HAT activity (Ricci et al., 2002). The inhibitory role of SAGA at HIS3 occurs specifically through its Spt3 and -8 subunits; indeed, in vitro studies show that SAGA inhibits binding of TBP at the HIS3 promoter, and that this inhibition is lost in deletions of spt3 and -8. Furthermore, induction of these genes results in accumulation of SAGA lacking Spt8, previously termed SAGAalt (Belotserkovskaya et al., 2000). These findings indicate that SAGA plays an active inhibitory role to transcription of a subset of genes through its Spt8 subunit in the absence of inducing conditions, and that SLIK, in lacking Spt8, may potentially be involved in activation of these genes.
SLIK also contains a form of Spt7 which is C-terminally truncated (Pray-Grant et al., 2002; Sterner et al., 2002). Biochemical analyses of Spt7 truncations have shown that removal of different portions of the C terminus results in differential association with SAGA components and various phenotypic severities; thus Spt7 is thought to serve as a structural scaffold of SAGA. Interestingly, partial removal of the Spt7 C terminus results in loss of Spt8 from the complex, consistent with the biochemical studies of SLIK (Wu and Winston, 2002). As expected, truncation of Spt7 results in an increase in basal transcription of HIS3 due to a loss of Spt8 at the promoter, leading to de-repression consistent with the spt8Δ phenotype (Belotserkovskaya et al., 2000; Sterner et al., 2002; Wu and Winston, 2002). However, no change in HIS3 expression is seen under inducing conditions when Spt7 processing is inhibited, leaving unanswered the question of how the absence of Spt8 in SLIK may function in positive regulation of this gene (Wu and Winston, 2002). One possibility is that multiple alterations occur to SLIK in vivo and cooperate to allow expression of HIS3. For example, in addition to Spt7 processing, post-translational modifications to other SAGA/SLIK subunits, as well as association of additional components, may all play a role in HIS3 expression. A discernible his− phenotype may not be apparent unless multiple aspects of this regulation are perturbed.
How is the biological function of SLIK related to that of SAGA? The difficulty in answering this question lies primarily in the fact that mutation of most SAGA subunits undoubtedly compromises SLIK function as well. It is important to note that the two complexes clearly overlap somewhat in their regulation of stress response, as is seen by the synthetic phenotypes of spt8Δ and rtg2Δ, and the finding that both complexes occupy the GAL10 promoter under inducing conditions (Pray-Grant et al., 2002). However, since only SLIK functions in the retrograde response pathway, it seems the two complexes are exclusively required for regulation of certain subsets of genes. Furthermore, silver stains of these two complexes show that they appear to contain subunits which are unique to one complex or the other (Pray-Grant et al., 2002). Identification and characterization of these subunits may further elucidate the distinct roles that each complex plays in regulation of the yeast genome.
Reading the histone code
In addition to regulating numerous epigenetic marks through the enzymatic activities of Gcn5 and Ubp8, SAGA is also capable of ‘reading’ histone modifications through interactions with multiple bromodomain- and chromodomain-containing subunits. The idea that post-translational histone modifications are read by other proteins, which direct downstream biological functions, is a postulate of the ‘histone code’ hypothesis. In particular, various combinations of lysine acetylation, methylation and ubiquitylation, along with arginine methylation and phosphorylation of serine and threonine residues, are thought to interact with specific protein domains, thereby mediating DNA-based cellular processes (Strahl and Allis, 2000).
The original study identifying Gcn5 as the yeast p55 homolog showed that the two proteins contain a highly conserved bromodomain, which is not found in the cytoplasmic HAT Hat1. This finding indicated that the bromodomain is important specifically for nuclear HATs, perhaps by tethering these proteins to chromatin (Brownell et al., 1996). Since this prediction, it has indeed been shown that bromodomains interact with chromatin, specifically binding to acetylated lysine residues, thus regulating localization of transcription-related proteins to acetylated regions of chromatin (reviewed in Yang, 2004). In addition to Gcn5, the Spt7 subunit of SAGA also contains a bromodomain (Gansheroff et al., 1995). Biochemical analyses have shown that the bromodomain of Gcn5, but not Spt7, is important for anchoring SAGA to acetylated nucleosomes. Notably, this effect is only seen in the presence of acetyl-CoA; thus the interaction between SAGA and chromatin is stabilized by its own HAT activity. Additionally, acetylation of chromatin by SAGA stabilizes the SWI/SNF chromatin remodeling complex, further contributing to a transcriptionally active chromatin environment (Hassan et al., 2002; Mitra et al., 2006). The function of the Spt7 bromodomain remains unknown; perhaps, it further stabilizes SAGA at gene promoters by binding acetylated proteins other than histones that are involved in transcription initiation.
In addition to the bromodomains of Gcn5 and Spt7, the SAGA subunit Chd1 contains tandem chromodomains, which further modulate SAGA activity on chromatin through interaction with methylated Lys4 of H3 (Pray-Grant et al., 2005). The chromodomain has been well characterized in its interaction with methylated lysine residues in a number of cellular processes, including transcriptional activation, formation and maintenance of heterochromatin, gene silencing, and DNA damage response (reviewed in Daniel et al., 2005). Specifically, chromodomain 2 of Chd1 is required for optimal SAGA HAT activity in vitro on peptide substrates, and for inducible acetylation of lysines 9 and 14 of H3 at the GAL1-10 UAS in vivo (Pray-Grant et al., 2005). Human Chd1 has also been shown to bind methylated Lys 4 of H3 via its tandem chromodomains, indicating that Chd1 function in recognizing methylated H3 may be conserved in higher eukaryotes (Flanagan et al., 2005; Sims et al., 2005). However, a number of recent reports have shown using alternative in vitro binding methods that yeast Chd1 has no increased affinity for methylated versus unmodified histone H3 (Flanagan et al., 2005; Sims et al., 2005; Okuda et al., 2007). Despite these contrasting results, it is clear that Chd1 is a stable component of SAGA in yeast and facilitates its HAT activity in transcriptional activation, in a manner that is functionally related to H3 Lys4 methylation.
Finally, histone phosphorylation also plays a role in SAGA-mediated gene expression. Phosphorylation of H3 Ser 10 is required for optimal recruitment of SAGA to the INO1 promoter and precedes acetylation of H3 Lys 14 by Gcn5 at multiple gene promoters (Lo et al., 2001, 2005). Ser 10 phosphorylation is catalyzed in yeast by the kinase Snf1, and functions in transcriptional activation of a number of inducible genes (Lo et al., 2001). It is currently not known how SAGA recruitment is mediated by Ser 10 phosphorylation. In vitro HAT assays have shown that recombinant Gcn5 prefers to acetylate Ser 10-phosphorylated peptides versus unmodified peptides. Mutation of a positively charged arginine residue in Gcn5 abolishes this preference, and thus it is thought that the negatively charged phosphate serves to stabilize Gcn5 on chromatin through interaction with this arginine (Cheung et al., 2000; Lo et al., 2000). Structural studies using the Gcn5 homolog from T. thermophila affirm that phosphorylation of S10 promotes additional interactions between Gcn5 and histone residues in comparison with the unmodified peptide (Clements et al., 2003). It remains to be seen, however, whether this interaction is sufficient to recruit SAGA to promoters in vivo, or whether other subunits of the complex may be involved. In mammalian cells, proteins containing the 14-3-3 domain are known to interact with phosphorylated H3 (Macdonald et al., 2005). Perhaps SAGA contains an uncharacterized 14-3-3 protein, which mediates this interaction. There are two 14-3-3 proteins known to exist in yeast, Bmh1 and -2, and disruption of these proteins results in increased sensitivity to environmental stresses, consistent with disruption of SAGA (van Hemert et al., 2001). Bmh2 has been shown to associate with SLIK subunits, including Rtg2, by proteomic analysis of protein complexes (Gavin et al., 2002). It is thus tempting to speculate that Bmh1 or -2 may influence interaction of SLIK/SAGA with phosphorylated H3.
SAGA in disease
What can we learn about development of human disease from a yeast HAT complex? Perhaps, the most direct correlation can be seen in the neurodegenerative disease spinocerebellar ataxia type 7 (SCA7). SCA7 is caused by polyglutamine (polyQ) expansion of the ataxin-7 protein, which leads to neurological dysfunction and blindness (David et al., 1998). Initial attempts to describe the onset of the disease were difficult due to the unknown function of the Sca7 protein. More recently, Sca7 and its homolog in yeast (also called Sgf79; 79-kDa SAGA-associated factor) were identified as components of the SAGA family of transcriptional co-activators in both species (Helmlinger et al., 2004; McMahon et al., 2005; Palhan et al., 2005). In yeast, pathogenic (60Q) Sca7 readily associates with SAGA, but diminishes the presence of other complex components, including Ada2, -3 and TAF12. This correlates with a loss of nucleosomal HAT activity in vitro, and thus is thought to have a transdominant-negative affect, whereby pathogenic SAGA is recruited to gene promoters but is rendered inactive (McMahon et al., 2005). Additionally, deletion of SCA7 results in a loss of both SAGA recruitment and preinitiation complex (PIC) formation at multiple gene promoters, indicating that nonpathogenic Sca7 plays a positive role in regulation of SAGA-mediated genes, perhaps through maintenance of complex integrity (Shukla et al., 2006b). Consistent with these results, human Sca7 is an integral component of the mammalian complexes STAGA and TFTC (TBP-free TAF complex), both SAGA homologs (Helmlinger et al., 2004; Palhan et al., 2005). STAGA acetylates H3 at promoters of cone-rod homeobox (CRX) _target genes, and this acetylation is inhibited by polyQ expanded ataxin-7 in a dominant-negative manner (Palhan et al., 2005). Thus, a contributing factor to retinal degeneration observed in SCA7 patients is probably due to transcriptional dysregulation of CRX _target genes caused by altered STAGA functionality.
It is also important to note that SAGA may function in preservation of genomic integrity outside of its role in gene expression. For example, lysines 9 and 14 of H3 are acetylated by Gcn5 in response to ultraviolet (UV) irradiation, and deletion of GCN5 results in impaired NER. At the yeast MFA2 promoter, UV-induced H3 acetylation does not coincide with transcriptional activity, and hence describes a cellular function of Gcn5 outside its canonical role in gene expression (Yu et al., 2005). Additionally, both Gcn5 and Ada2 have been shown to facilitate NER at the MET16 locus, in a manner directly related to the rate of transcription of this gene (Ferreiro et al., 2006). Thus it seems Gcn5 HAT activity may play a role in regulation of NER in both a transcription-dependent and -independent manner. Given that Ada2 functions in NER as well, this activity probably occurs in the context of SAGA and/or SLIK.
Interestingly, histone acetylation in response to UV irradiation occurs even in the absence of the damage recognition factors Rad4 and -14, indicating that recruitment of other NER proteins is not necessary for UV-induced acetylation by Gcn5 (Yu et al., 2005). However, it is still possible that SAGA interacts with NER proteins in the response to UV damage. The human TFTC complex contains the DNA damage recognition factor Sap130, and acetylates histones more efficiently on damaged DNA templates (Brand et al., 2001). STAGA also contains Sap130 as well as the UV-damaged DNA binding protein DDB1. DDB1 is a component of the NER machinery, but is not required for transcription-coupled repair, supporting a role for this protein in _targeting nucleosomal acetylation at sites of DNA damage (Martinez et al., 2001). Given the high degree of conservation between SAGA and the homologous human HAT complexes, it is possible that yeast SAGA contains subunits that specifically interact with damaged DNA. Importantly, loss of NER function in humans results in the rare autosomal disease Xeroderma pigmentosum (XP). XP individuals experience a heightened sensitivity to UV light, resulting in genomic instability and a higher incidence of cancer (Cleaver, 2000). It is possible that loss of certain NER genes in XP patients leads to failed _targeting of TFTC/STAGA to sites of damaged DNA, causing a loss of acetylation at these regions, and thereby preventing access of the necessary repair proteins to damaged chromatin. Thus, understanding SAGA function in the NER pathway may shed light on the development of genomic instability and cancer in some individuals.
Conclusion
SAGA plays a vital role in expression of stress–response genes, and is subject to regulation on multiple levels. This hierarchy of regulation probably becomes exponentially more critical when considering selective pressures on yeast growing in the wild, where a wide variety of stresses, including metabolic starvation, UV exposure, temperature and others, force a constant flux of gene expression required for cell survival. The interaction of Tra1 with acidic activators is probably most important, as Tra1 is essential in yeast and _targets SAGA to an array of gene promoters (Saleh et al., 1998; Brown et al., 2001; Bhaumik et al., 2004). SAGA is also regulated by the 19S proteasome RP, which facilitates loading of the complex onto promoter DNA (Lee et al., 2005a). Additionally, a number of histone modifications, including H3 lysine acetylation and methylation, and serine phosphorylation, facilitate SAGA function. This regulation most likely occurs in a promoter-specific manner through interaction with SAGA’s numerous highly conserved binding domains (Hassan et al., 2002; Lo et al., 2005; Pray-Grant et al., 2005). Furthermore, the presence of several modifications which regulate SAGA HAT activity are in turn mediated by SAGA itself. For instance, H3 Lys 4 trimethylation, which interacts with the SAGA subunit Chd1, is regulated by the ubiquitin protease activity of Ubp8 (Henry et al., 2003; Daniel et al., 2004; Pray-Grant et al., 2005; Shukla et al., 2006a). Additionally, the Gcn5 bromodomain interacts with lysine acetylation which it generates itself (Hassan et al., 2002). Our picture of how the SAGA family of transcriptional co-activators is regulated is just emerging, and there may be other mechanisms, which fine-tune the activity of this machine in vivo, perhaps through post-translational modification of SAGA subunits by other transcription-related proteins, or even by other components within the complex.
Given that SAGA is such a highly regulated molecular machine, cells are probably sensitive to even slight dysregulation of its function. Thus it is easy to imagine how perturbation of SAGA activity could lead to multiple regulatory problems on a cellular level. In yeast, SAGA is thought to control transcription of approximately 10% of genes, most of which are involved in response to external stresses (Huisinga and Pugh, 2004). In humans, the role of SAGA homologs in transcription genome-wide is not known; however, given the high degree of conservation of these complexes, it is likely that they play a similar role in modulating highly regulated genes. For example, H3 at the promoters of human immediate early genes, such as c-fos, is phosphorylated at Ser 10 in response to epidermal growth factor. This modification occurs in a mitogen-activated protein kinase-dependent manner and, as in yeast, promotes acetylation of H3 by Gcn5 (Cheung et al., 2000; Lo et al., 2001). This represents a highly conserved mechanism of gene regulation, and it is not difficult to imagine that other mechanisms of SAGA regulation are conserved in higher eukaryotes. Abnormal SAGA function in humans could thus have a dramatic effect in gene expression at numerous inducible loci, and may play an important role in multiple cellular processes such as development. Consistent with this idea, Gcn5 HAT activity is important for neural tube closure in mice, and complete loss of Gcn5 leads to embryonic lethality (Xu et al., 2000; Bu et al., 2007).
In addition to gene expression, other activities of SAGA are probably conserved in its human homologs. As STAGA and TFTC both contain DNA damage binding proteins, they are likely integral in the recognition and/or repair of damaged DNA (Brand et al., 2001; Martinez et al., 2001). This effect could perhaps be coupled with SAGA’s role in regulation of transcription. As SAGA is known to regulate primarily stress response genes, it may be indirectly involved in the response to DNA damage through induction of DNA repair genes. This could lead to an additive affect when SAGA activity is compromised, whereby efficient repair is hindered by a lack of access to damaged DNA as well as low levels of DNA repair proteins, both due to loss of Gcn5-mediated histone acetylation. By analogy, a role for histone H4 in the repair of double strand breaks (DSBs) is now well established (reviewed in Altaf et al., 2007). Compiling these effects would most likely lead to an increase in genomic instability and, in mammals, development of cancer. A deeper understanding of SAGA’s role in DNA damage repair, along with further clarification of its many roles in transcriptional regulation, may ultimately lead to elucidation of the development processes of such diseases and possible methods of treatment.
Acknowledgements
We apologize to those authors whose work could not be cited due to space limitations. We thank Christine H Baker for helpful comments and technical reading of this manuscript. Research in the Grant lab is funded from National Institutes of Health R01 Grant no. NS049065. SPB is supported by National Institutes of Health predoctoral cancer training Grant no. 5 T32 CA009109-30.
References
- Altaf M, Saksouk N, Côté J. Histone modifications in response to DNA damage. Mutat Res. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Baker SP, Grant PA. The proteasome: not just degrading anymore. Cell. 2005;4:361–363. doi: 10.1016/j.cell.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Pina B, Silverman N, Marcus GA, Agapite J, Regier JL, et al. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo _target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, et al. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, et al. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY. Loss of gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Cleaver JE. Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum. J Dermatol Sci. 2000;23:1–11. doi: 10.1016/s0923-1811(99)00088-2. [DOI] [PubMed] [Google Scholar]
- Clements A, Poux AN, Lo WS, Pillus L, Berger SL, Marmorstein R. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol Cell. 2003;12:461–473. doi: 10.1016/s1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Pray-Grant MG, Grant PA. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, III, et al. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- David G, Durr A, Stevanin G, Cancel G, Abbas N, Benomar A, et al. Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7) Hum Mol Genet. 1998;7:165–170. doi: 10.1093/hmg/7.2.165. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Dollard C, Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989;58:1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- Ferreiro JA, Powell NG, Karabetsou N, Mellor J, Waters R. Roles for Gcn5p and Ada2p in transcription and nucleotide excision repair at the Saccharomyces cerevisiae MET16 gene. Nucleic Acids Res. 2006;34:976–985. doi: 10.1093/nar/gkj501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Sträβer K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, et al. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Gansheroff LJ, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;425:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. Physical and functional association of RNA polymerase II and the proteasome. Proc Natl Acad Sci USA. 2004;101:5904–5909. doi: 10.1073/pnas.0305411101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltzz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, III, et al. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998a;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Yates JR, III, Workman JL. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998b;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- Green MR. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinki SC, Chandy M, Carrozza MJ, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Steger DJ, Eberharter A, Workman JL. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, et al. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinyamu HK, Chen J, Archer TK. Linking the ubiquitin-proteasome pathway to chromatin remodeling/modification by nuclear receptors. J Mol Endocrinol. 2005;34:281–297. doi: 10.1677/jme.1.01680. [DOI] [PubMed] [Google Scholar]
- Kohler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, et al. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesza CA, Van Buskirk HA, Cole MD, Reese JC, Smith MM, Engel DA. Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1. Oncogene. 2002;21:1411–1422. doi: 10.1038/sj.onc.1205201. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005a;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005b;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278:20405–20412. doi: 10.1074/jbc.M211795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, et al. Snf1 - a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Marcus GA, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, III, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci USA. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol Cell Biol. 2006;26:4095–4110. doi: 10.1128/MCB.01849-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Murr R, Vaissière T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as coactivators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Okuda M, Horikoshi M, Nishimura Y. Structural polymorphism of chromodomains in Chd1. J Mol Biol. 2007;365:1047–1062. doi: 10.1016/j.jmb.2006.10.039. [DOI] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, et al. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci USA. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, et al. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, et al. novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WM, Hahn S. _targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci AR, Genereaux J, Brandl CJ. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol Cell Biol. 2002;22:4033–4042. doi: 10.1128/MCB.22.12.4033-4042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- Saleh A, Schieltz D, Ting N, McMahon SB, Litchfield DW, Yates JR, III, et al. Tra1p is a component of the yeast Ada-Spt transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, et al. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- Sekito T, Thornton J, Butow RA. Mitochondria-tonuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Bajwa P, Bhaumik SR. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 2006b;34:6225–6232. doi: 10.1093/nar/gkl844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol. 2006a;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast Chd1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Belotserkovskaya R, Berger SL. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc Natl Acad Sci USA. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, et al. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sträβer K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Timmers HT, Tora L. SAGA unveiled. Trends Biochem Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Côté J, Steger DJ, Eberharter A, et al. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- van Hemert MJ, van Heusden GP, Steensma HY. Yeast 14-3-3 proteins. Yeast. 2001;18:889–895. doi: 10.1002/yea.739. [DOI] [PubMed] [Google Scholar]
- Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Wu PY, Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol Cell Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci USA. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]