Abstract
Renal excretion of water and major electrolytes exhibits a significant circadian rhythm. This functional periodicity is believed to result, at least in part, from circadian changes in secretion/reabsorption capacities of the distal nephron and collecting ducts. Here, we studied the molecular mechanisms underlying circadian rhythms in the distal nephron segments, i.e., distal convoluted tubule (DCT) and connecting tubule (CNT) and the cortical collecting duct (CCD). Temporal expression analysis performed on microdissected mouse DCT/CNT or CCD revealed a marked circadian rhythmicity in the expression of a large number of genes crucially involved in various homeostatic functions of the kidney. This analysis also revealed that both DCT/CNT and CCD possess an intrinsic circadian timing system characterized by robust oscillations in the expression of circadian core clock genes (clock, bma11, npas2, per, cry, nr1d1) and clock-controlled Par bZip transcriptional factors dbp, hlf, and tef. The clock knockout mice or mice devoid of dbp/hlf/tef (triple knockout) exhibit significant changes in renal expression of several key regulators of water or sodium balance (vasopressin V2 receptor, aquaporin-2, aquaporin-4, αENaC). Functionally, the loss of clock leads to a complex phenotype characterized by partial diabetes insipidus, dysregulation of sodium excretion rhythms, and a significant decrease in blood pressure. Collectively, this study uncovers a major role of molecular clock in renal function.
Keywords: circadian rhythm, homeostasis, renal function
Recent evidence suggests that many if not all specific physiological functions are under the control of the circadian timing system. The mammalian circadian timing system is a hierarchically organized network of molecular oscillators driven by a central pacemaker located in the suprachiasmatic nucleus (SCN) of hypothalamus. This central pacemaker functions in a self-sustained fashion, but is reset each day by exposure to environmental synchronizers, mainly the light/dark cycle. The SCN masterclock drives the rest-activity cycle, which in turn imposes the feeding pattern [reviewed in (1, 2)]. The feeding time seems to be the dominant cue for circadian rhythms in the peripheral tissues (3, 4). Central and peripheral oscillators share a similar molecular core clock based on a set of self-autonomous transcriptional/translational feedback loops. The key molecular components of these loops are the PAS domain transcriptional factors CLOCK, BMAL1, and NPAS2 and the feedback repressors PER1, PER2, CRY1, and CRY2. The orphan nuclear receptors NR1D1 and, probably, NR1D2 form an accessory feedback loop. The core oscillators confer circadian rhythmicity on a set of output genes underlying the tissue-specific functional rhythms. Current estimates indicate that up to 10% of the cellular transcriptome may follow a circadian expression pattern (5–7). Several recent studies have also demonstrated that the transcription of only a minority of these circadian genes is driven by systemic humoral or neuronal circadian signals, whereas the vast majority of them (more than 90%) is dependent on self-autonomous local circadian oscillators (8, 9). This self-autonomous circadian transcription activity is thought to be the main molecular mechanism allowing peripheral tissues to anticipate upcoming circadian environmental challenges (activity, feeding, etc).
The most obvious manifestation of circadian rhythmicity of renal function is a well-marked difference in the volume of urine formation/excretion between the day and the night. The urinary excretion of all major solutes (Na+, K+, Cl−, urea, PO4−, Ca2+, Mg2+) also follows a circadian oscillating pattern. Although, renal excretion rhythms are apparently synchronized with circadian rhythms of activity/feeding, they have been shown to persist over long periods of time under experimental conditions in which external factors such as dietary intake, posture, or sleep were kept constant or were manipulated in a noncircadian manner (10–16). These results have indicated that in addition to the external circadian stimuli (hormones, food, food metabolites) these functional rhythms are also controlled by a self-sustained intrinsic renal clock. Dysfunction of renal excretory rhythms has been proposed as a possible cause for several serious human diseases. For example, the abnormal rhythm of renal sodium reabsorption is considered as one of the major factors leading to the loss of nocturnal dip in the blood pressure which is characteristic for ≈35% of all hypertensive patients (17, 18). This nondipping pattern of blood pressure leads to a significantly increased risk of stroke and end-organ damage. Also, abnormalities in renal calcium or water conservation rhythms have been shown to correlate with development of osteoporosis or nocturnal polyuria, respectively (9, 19–22).
Renal excretory rhythms are driven by circadian changes in both glomerular filtration and tubular reabsorption/secretion (23, 24). Here, we addressed the role of circadian timing system in renal tubular function by studying molecular mechanisms underlying circadian rhythms in mouse distal nephron and collecting ducts. These parts of the renal tubule were chosen because they are responsible for the final adjustment of urine flow and solutes concentration. Our data demonstrate that a large number of genes essential for water and solutes homeostasis follow a well-marked circadian expression pattern. Analysis of renal phenotype in clock(−/−) mice revealed a mild diabetes insipidus, which can be accentuated upon stress condition. The clock(−/−) mice also display a modified pattern of sodium excretion rhythm and a significantly lower blood pressure. Taken together, our study provides evidence for a major role of circadian timing system in renal function.
Results
Temporal Profiling of Distal Convoluted Tubule/Connecting Tubule and Cortical Collecting Duct Transcriptomes.
To identify genes involved in renal circadian rhythms, we examined the temporal profiles of gene expression in mouse distal nephron segments and collecting ducts using Affymetrix oligonucleotide microarrays. The RNA was extracted from microdissected distal convoluted tubule (DCT) and connecting tubule (CNT) samples or cortical collecting duct (CCD) samples. The DCT and CNT were isolated together because of the gradual transition between these two nephron segments in mice (25). The microdissection was performed from the left kidneys of male C57BL/6J mice maintained on a standard laboratory chow diet and adapted to 12-h light/12-h dark cycle for 2 weeks. Animals were killed for microdissection every 4 h, i.e., at ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20 [ZT, Zeitgeber (circadian) time, indicates time of light-on as ZT0 and time of light-off as ZT12]. The microarray hybridization was performed on two pools of RNA composed of equivalent amounts of RNA prepared from five animals at each ZT time point. The quality of microdissection was validated by analysis of abundance of DCT/CNT-specific or CCD-specific transcripts. For example, the microarray hybridization data demonstrate that transcript encoding the DCT-specific sodium-chloride cotransporter (NCC or Slc12a3) is ≈45-fold more abundant in DCT/CNT samples (mean log2 normalized expression value of 10.775) than in CCD samples (mean log2 normalized expression value of 5.281) (see Table S1 and Table S2 for complete circadian transcriptomes of DCT/CNT and CCD, respectively; the in depth analysis of DCT/CNT and CCD transcriptomes is beyond the scope of this manuscript and will be presented elsewhere).
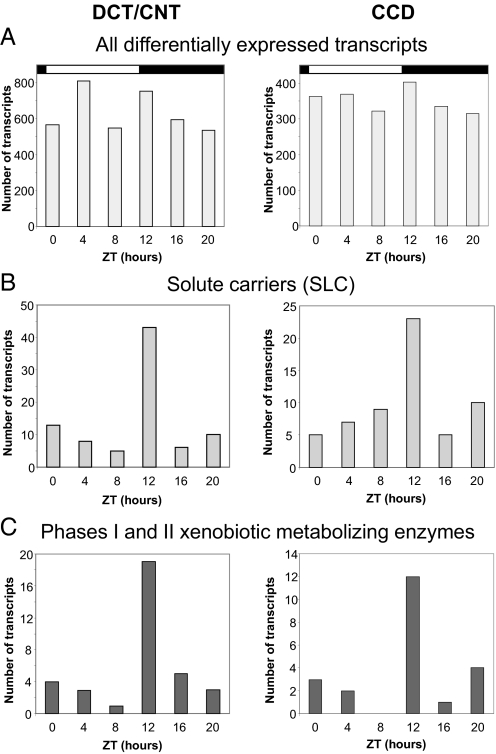
Comparison of gene expression levels between different Zeitgeber time points revealed that a large number of transcripts exhibit marked diurnal expression changes in both DCT/CNT and CCD (5,031 DCT/CNT transcripts and 2,765 CCD transcripts corresponding to 3,814 and 2,112 distinct genes, respectively; fold change >1.8 between the highest and the lowest expression levels; see Table S3 and Table S4, respectively). Temporal expression analysis of these differentially expressed transcripts revealed a random diurnal distribution of acrophases (Fig. 1A). This indicates that the overall transcriptional activity in these parts of the renal tubule is randomly distributed throughout the circadian cycle. The same analysis was also performed on several major gene categories most relevant to the renal tubular function (see Methods). The chosen gene categories were similar to those used by Uawithya et al. for characterization of the inner medullary collecting duct transcriptome (26). Interestingly, this analysis revealed the existence of function-dependent temporal expression patterns for two of the gene categories, namely the genes belonging to the superfamily of solute carriers (Slc) and to the group of enzymes involved in phase I and phase II reactions of metabolism of xenobiotics or other lipophilic compounds. As shown in Fig. 1 B and C, and in the Table S5, the majority of these transcripts exhibit their maximal expression at ZT12.
Fig. 1.
(A) Temporal distribution of acrophases of all differentially expressed transcripts (fold change > 1.8). The fold change was calculated between the expression level of a transcript at each individual Zeitgeber time point and the mean value of expression determined from all six Zeitgeber time points. (B) Temporal distribution of acrophases of differentially expressed solute carriers (Slc, fold change > 1.8), see also Table S5. (C) Temporal distribution of acrophases of differentially expressed enzymes involved in phase I and phase II reactions of metabolism of xenobiotics and other lipophilic compounds (fold change > 1.8), see also Table S5. The analysis was performed on the following phase I/phase II enzymes: CYP450 enzymes, flavin containing monooxygenases, alcohol denydrogenase, epoxide hydrolases, aldehyde dehydrogenases, carboxylesterases, glutathione-S-transferases, N-acetyltransferases, sulfotransfearses, UDP glucoronosyltransferases, NADPH dehydrogenases, glycin-N-acetlytransferase, aldehyde oxidases, aldo-keto reductases, acyl-CoA-synthetases.
Circadian Analysis of DCT/CNT and CCD Transcriptomes.
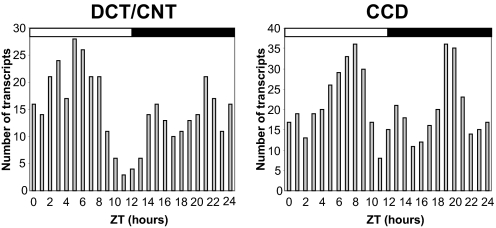
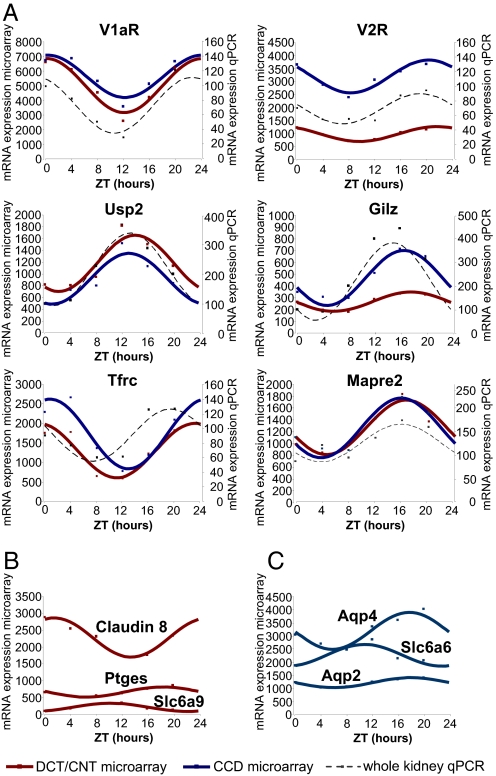
Circadian transcripts were identified by fitting to a cosine curve with a period of 24 h. The circadian variation of transcript expression was considered as significant if it had an adjusted P values < 0.1 and an amplitude higher than 1.5-fold. A total of 356 DCT/CNT transcripts and 504 CCD transcripts met these criteria (Table S6 and Table S7, respectively). Expression of 96 transcripts met the circadian criteria in both DCT/CNT and CCD (Table S8). Temporal analysis of DCT/CNT or CCD circadian transcripts revealed a nonrandom pattern of acrophase distribution characterized by several distinct peaks (Fig. 2). This indicates that transcription of circadian genes in these parts of the renal tubule is controlled by a limited number of circadian transcriptional factors. Importantly, many of the identified circadian transcripts encode proteins that are crucially involved in renal homeostatic function. For example, the V2 vasopressin receptor (V2R) that mediates the antidiuretic effect of vasopressin in the kidney, exhibits circadian expression rhythms in both DCT/CNT and CCD. The aquaporin-2 (aqp-2) and aquaporin-4 (aqp-4) water channels that contribute to water homeostasis by facilitating water movement across cellular membranes also exhibit a circadian expression pattern, albeit only in the CCD [for aqp-2, however, the amplitude of circadian expression (1.36) is below the arbitrary cut-off value of 1.5]. The sets of DCT/CNT and/or CCD circadian transcripts (Table S6 and Table S7 and Table S8) also include genes involved in regulation of transepithelial sodium transport [Usp2 and Gilz (Tsc22d3)], calcium homeostasis (Vdr, Slc8a1, and Calb28k), iron metabolism (Tfrc and Slc40a1), and organic solute transporters involved in maintaining cell volume (Slc6a9 and Slc6a6). Several examples of transcripts exhibiting circadian expression profiles either in both DCT/CNT and CCD or, only in DCT/CNT or CCD are shown in Fig. 3A, B, and C, respectively. For transcripts exhibiting circadian expression in both DCT/CNT and CCD, the circadian expression profile was invariably confirmed by qPCR performed on the whole-kidney RNA samples (Fig. 3A).
Fig. 2.
Temporal distribution of acrophases of transcripts that meet the circadian criteria (fitting to a cosine curve with a period of 24-h, adjusted P value < 0.1, amplitude > 1.8). See also Table S6 and Table S7.
Fig. 3.
Temporal expression profiles of circadian transcripts. (A) Transcripts that meet the circadian criteria in both DCT/CNT and CCD. The red and blue squares show the expression values (arbitrary units of microarray hybridization data) of DCT/CNT and CCD circadian transcripts, respectively. The red and blue solid lines show fitting of the microarray data to the cosine function. The black squares show the qPCR expression values (ZT0 = 100%) of these transcripts in the whole kidney RNA samples. The black dashed line shows fitting of qPCR data to the cosinor function. The qPCR was performed on pools of RNA composed of equivalent amounts of RNA prepared from five animals at each ZT time point. The qPCR data are expressed as arbitrary units normalized for β-actin expression. (B) Transcripts that meet the circadian criteria only in DCT/CNT. (C) Transcripts that meet the circadian criteria only in CCD. Abbreviations used are: V1aR, vasopressin receptor Type 1a; V2R, vasopressin receptor Type 2; Usp2, ubiquitin specific protease 2; Gilz (Tsc22d3), glucocorticoid-induced leucine zipper; Tfrc, transferrin receptor; Mapre2, microtubule-associated protein; Ptges, prostaglandin E synthase; Slc6a9, glycine transporter; Slc6a6, taurine transporter; aqp2, aquaporin 2; aqp4, aquaporin 4.
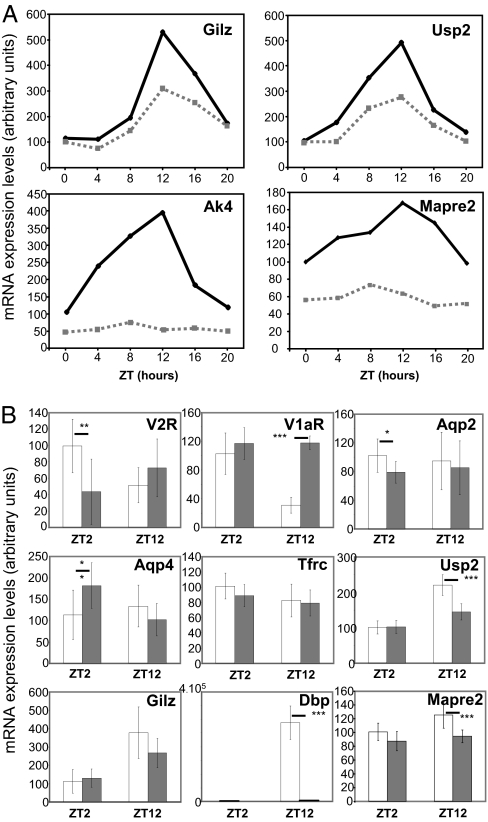
Analysis of circadian transcripts has shown that genes involved in the positive or negative limbs of the circadian clock feedback loop (core clock genes) as well as the clock-controlled Par bZip transcriptional factors (Dbp, Hlf, Tef) exhibit robust circadian expression rhythms in both DCT/CNT and CCD (Fig. S1 A, B, and C, respectively). For example, the amplitude of Dbp RNA oscillation is over 20-fold in DCT/CNT and over 19-fold in CCD. To assess the putative relationship between expression of core clock transcriptional factors or Par bZips on the one hand and circadian output genes on the other hand, we analyzed the expression profiles of several circadian output genes in kidneys of clock knockout mice or mice devoid of dbp/hlf/tef (triple knockout) (27, 28). As estimated by Gachon et al., the DBP/HLF/TEF-dependent genes are expected to show their acrophases in a time-window between ZT12 and ZT16 (28). Accordingly, we tested expression of several genes that meet the criteria in kidneys of dbp/hlf/tef triple knockout mice. As shown in Fig. 4A, all tested genes (Gilz, Usp2, Ak4, and Mapre2) show a significantly reduced expression in kidneys of the triple knockouts. Several other circadian transcripts were tested on RNA extracted at ZT2 or ZT12 from kidneys of wild-type or clock(−/−) mice. As shown in Fig. 4B, genes involved in vasopressin signaling pathway (V2R and V1aR) or in water transport across cellular membranes (aqp-2 and aqp-4) exhibit significant changes in expression at one out of two tested Zeitgeber time points. Interestingly, the expression of V1aR (ZT12) and aqp-4 (ZT2) was increased and not decreased in clock(−/−) mice, indicating an indirect regulation of V1aR and aqp-4 RNA expression by the CLOCK via a clock-controlled transcriptional repressor. The expression levels of Usp2 and Mapre2 RNA were significantly lower in clock(−/−) mice (ZT12), whereas the difference in Gilz and Tfrc RNA levels did not reach statistical significance. The lack of Dbp RNA expression in clock(−/−) mice demonstrates that Par bZips expression in the kidney is controlled by the CLOCK.
Fig. 4.
(A) Temporal expression pattern of Gilz, Usp2, adenylyl kinase 4 (Ak4), and Mapre2 RNA in whole kidneys of wild-type (solid line) or dbp/hlf/tef triple knockouts (dashed line). Data are expressed as the arbitrary units of qPCR amplification normalized for β-actin expression. The qPCR was performed on pools of RNA composed of equivalent amounts of RNA prepared from six animals at each ZT time point. (B) Expression V2R, V1aR, Aqp-2, Aqp-4, Tfrc, Usp2, Gilz, Dbp, and Mapre2 transcripts at ZT2 and ZT12 in whole kidneys of wild-type (white bars) or clock(−/−) triple knockouts (gray bars). Data are means ± SEM of qPCR amplification values that were obtained from seven mice. Statistical significance was calculated using unpaired t-test. *, P = 0.05; **P < 0.05; ***, P < 0.001.
Renal Function in clock(−/−) Mice.
To assess the role of circadian timing system in the kidney, we examined the renal phenotype of clock(−/−) mice. Collection of spot urine and plasma samples was performed at ZT2 and ZT12 from mice maintained on a 12-h light/12-h dark cycle and ad libitum access to food and water (same conditions as those used in microarray and qPCR experiments). As shown in Table 1, the clock(−/−) mice exhibit a significantly decreased urine osmolality at ZT12 and a significantly increased plasma osmolality and hematocrit at ZT12 and ZT2, respectively. In parallel, the knockout mice display an increase in water intake during the active phase (ZT12–ZT24) but a similar to wild-type mice pattern of general motion activity (Figs. S2 A, B, and. C, respectively). Interestingly, when placed in the metabolic cages, a known stressful condition for mice, the capacity of urine concentration in clock(−/−) mice was further impaired (Table 1).
Table 1.
Urine concentrating capacity, plasma electrolytes, and urine excretion rates in wild-type (WT) and clock(−/−) mice
| Spot samples |
Metabolic cages |
|||||
|---|---|---|---|---|---|---|
| ZT2 |
ZT12 |
WT | clock(−/−) | |||
| WT | clock(−/−) | WT | clock(−/−) | |||
| Urine osmolality, mosm/kg H2O) | 1,879 ± 286 | 1,688 ± 380 | 2,097 ± 296 | 1,526 ± 432* | ||
| Plasma Na, mM | 154 ± 5 | 152 ± 3 | 153 ± 4 | 156 ± 8 | ||
| Plasma K, mM | 4.95 ± 0.59 | 4.86 ± 0.47 | 4.50 ± 0.68 | 4.52 ± 0.44 | ||
| Plasma creatinine, mM | 15.0 ± 5.1 | 17.5 ± 5.0 | 16.9 ± 2.4 | 16.8 ± 3.6 | ||
| Plasma osmolality, mosm/kg H2O | 319.9 ± 7.4 | 320.2 ± 3.9 | 316.6 ± 5.1 | 323.0 ± 4.5* | ||
| Hematocrit, %RBC | 46.1 ± 0.5 | 49.6 ± 1.1** | 46.1 ± 0.9 | 48.4 ± 2.8 | ||
| Urine creatinine, mM | 5,365 ± 2483 | 4,109 ± 1591 | 6,375 ± 1108 | 4,165 ± 844** | ||
| FE Na, % | 0.26 ± 0.14 | 0.34 ± 0.12 | 0.16 ± 0.07 | 0.13 ± 0.02** | ||
| FE K, % | 16.8 ± 10.0 | 18.7 ± 6.1 | 20.8 ± 6.3 | 29.1 ± 11.3 | ||
| UNa/UK | 0.62 ± 0.45 | 0.61 ± 0.29 | 0.27 ± 0.09 | 0.20 ± 0.09* | ||
| Urine volume, ml/24 h | 1.27 ± 0.57 | 2.50 ± 1.16* | ||||
| Urine osmolality, mosm/kg H2O | 2,991 ± 690 | 1,332 ± 372*** | ||||
| Body weight, g | 28.26 ± 1.10 | 29.30 ± 2.32 | ||||
Statistical significance was calculated using unpaired t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 6–8. Bold, pairs of values for which the difference is statistically significant.
The clock(−/−) mice exhibit a significant difference in the fractional excretion of sodium (FE Na) between ZT2 and ZT12 (Table 1). The ratio between FE Na at ZT2 and ZT12 reaches ≈2.6 in clock(−/−) mice (P < 0.01), whereas it is only ≈1.6 in wild-type mice (not significant). This indicates that clock(−/−) mice exhibit either a significantly different amplitude or a significantly different kinetic of sodium excretion rhythm. Also, both wild-type and clock(−/−) mice show a marked difference in urinary sodium/potassium ratio (UNa/UK) between ZT2 and ZT12. However, this difference reached a statistical significance only in clock(−/−) mice. Interestingly, the modified pattern of sodium excretion rhythm in clock(−/−) mice correlates with a lower RNA expression levels at ZT2 of epithelial sodium channel, the key regulator of sodium reabsorption in both DCT/CNT and CCD [qPCR assessment of αENaC RNA expression in kidneys of wild-type or clock(−/−) mice (arbitrary units): 100.0 ± 15.0 vs. 76.8 ± 23.8, respectively; P < 0.05, n = 7].
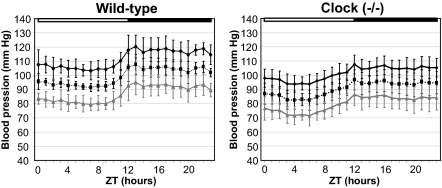
Because, renal sodium handling plays a major role in blood pressure control, we measured blood pressure parameters in conscious unrestrained mice using telemetry. As shown in Fig. 5, both wild-type and clock(−/−) mice exhibit a typical 24-h profile of blood pressure characterized by a dip in systolic and diastolic components during the inactive phase. However, the average mean arterial pressure and mean systolic blood pressure were significantly lower in clock(−/−) mice than in wild-type mice (24-h average mean arterial pressure: 90.3 ± 6.3 mm Hg vs. 99.4 ± 2.9 mm Hg, respectively, P < 0.5, n = 5; 24-h mean systolic blood pressure: 101.2 ± 5.8 mm Hg vs. 111.6 ± 5.6 mm Hg, respectively, P < 0.5, n = 5). The difference in the mean diastolic blood pressure did not reach statistical significance (79.8 ± 6.6 mm Hg vs. 87.1 ± 5.4 mm Hg, respectively, P = 0.11, n = 5).
Fig. 5.
Diurnal profile of blood pressure in wild-type and clock(−/−) mice. The systolic, diastolic, and mean blood pressures are indicates as black, gray, and black dashed lines, respectively. Error bars show SEM (n = 5).
Discussion
Temporal Expression Profiling of DCT/CNT and CCD Transcriptomes.
Our study demonstrates that circadian rhythmicity of renal function correlates with significant changes in expression levels of a large number of DCT/CNT and CCD transcripts. Interestingly, the majority of differentially expressed transcripts from two functional gene families, namely solute carriers (SLC) and enzymes involved in phase I and phase II reactions of metabolism of xenobiotics and other lipophilic compounds, exhibit their maximal expression at ZT12, the time of onset of activity/feeding phase in mice. These results indicate that expression levels of solute carriers and phase I/phase II enzymes are adjusted in anticipation of “programmed” increase in renal load by food components and, concomitantly, by xenobiotics. Because solute carriers are involved in the bulk of organic compounds and ion transport processes, this anticipative mechanism may be of a special functional importance in the kidney. Circadian oscillation in expression of phase I/phase II enzymes has been previously shown in the liver, intestine, and the whole kidney (28). In the kidney, the xenobiotics detoxification is usually attributed to the proximal tubule. However, extensive water removal along the renal tubule may result in a high concentration of xenobiotics and their metabolites in the lumen of the distal nephron and collecting ducts. Thus, the anticipative changes in expression of detoxification enzymes may be beneficial for the protection of DCT/CNT and CCD cells themselves, as well as for the general process of xenobiotics elimination.
Circadian Transcripts in DCT/CNT and CCD.
Statistical analysis revealed that several hundreds of DCT/CNT and CCD transcripts met the formal criteria of circadian transcripts (expression oscillation with ≈24-h period length). It should be noted, however, that cosinor method used for this analysis is applicable only for data sets fitting to cosine function. The formal identification of circadian transcripts exhibiting non-sinusoidal expression profiles will require the temporal profiling of gene expression performed for more than one 24-h period. Importantly, the sets of DCT/CNT and/or CCD circadian transcripts include several key players of renal water and salts homeostasis. Interestingly, circadian genes involved in the same homeostatic function also exhibit a similar temporal expression pattern. For example, vasopressin V2 receptor, aqp-2, and aqp-4 RNA exhibit their maximal expression in the CCD at ZT20, ZT18, and ZT18, respectively. In the DTC/CNT, the expression levels of vitamin D receptor and sodium/calcium exchanger (Slc8a1) transcripts reach their maximum at ZT20 and ZT23, respectively. To what extent this circadian coordination in RNA abundance is also maintained on the level of protein expression or functional activity remains to be established.
Molecular Clock in the Kidney.
The changes in secretion/reabsorption capacities of the distal nephron and collecting ducts are traditionally regarded as a result of changes in hormonal secretion. These latter, in turn, are usually considered as the direct consequences of changes in plasma volume or its composition. Our results demonstrate that DCT/CNT and CCD possess an intrinsic molecular clock, a system allowing self-autonomous regulation of gene expression and, finally, function. Interestingly, recent studies indicate that secretion of hormones controlling salt and water transport in distal nephron and collecting duct is also dependent on the circadian system. Son et al. have shown that adrenal peripheral clock controls circadian rhythm of glucocorticoids synthesis and secretion (29). Because glucocorticoids share a common biosynthesis pathway with aldosterone, a principal hormone regulating sodium reabsorption in the kidney, this indicates that plasma aldosterone levels are also dependent, at least in part, on a molecular clock. The hypothalamic expression of vasopressin is also regulated by the circadian mechanism (30). Collectively, these data indicate that both intrinsic clock and factors depending on extra-renal molecular oscillators may play a major role in renal physiology. To what extent circadian oscillations of gene expression in DCT/CNT and CCD cells depends on the internal clock remains to be established in the tissue-specific knockout models. As of today, the only tissue for which this analysis was performed is the liver. In this organ, more than 90% of circadian transcripts are controlled by the local clock (8).
Renal Function in clock(−/−) Mice.
clock(−/−) mice excrete a diluted urine and exhibit an increased water intake. This correlates with a significant reduction in renal RNA expression of V2R and aqp-2 water channel, two major genes controlling water reabsorption in the distal nephron and collecting ducts. Two possible causes for this phenotype are the primary (psychogenic) polydipsia, a symptom characterized by excessive thirst or, diabetes insipidus, a condition in which the kidney is not concentrating urine properly. The differential diagnostics of these syndromes includes (i) measurement of plasma sodium concentration, plasma osmolality, or plasma hematocrit that are usually decreased in the primary polydipsia but increased in the diabetes insipidus; (ii) water deprivation test; and (iii) test of responsiveness to vasopressin treatment. The two latter tests, however, are of limited significance in the case of a mild phenotype. In clock(−/−) mice, the mild decrease in urine osmolality and an increase in plasma osmolality and hematocrit clearly indicate a mild or partial diabetes insipidus. Interestingly, under a stress condition (metabolic cages), the capacity of clock(−/−) mice to concentrate urine is further impaired. For the moment, we have no explanation for this finding. However, it emphasizes that involvement of circadian timing system in renal function may be different depending on the physiological condition or the pathophysiological state.
The clock(−/−) mice exhibit a modified pattern of sodium excretion with a bigger difference in FE Na between ZT2 and ZT12 comparing to wild-type mice. Also, two major players of sodium reabsorption, namely αENaC and Usp2, have shown significant changes in RNA expression levels. In humans, the amplitude of circadian changes of sodium excretion is a major factor controlling blood pressure (17). In clock(−/−) mice, the arterial blood pressure is significantly lower comparing to wild-type mice. The causative relationship between the modified pattern of sodium excretion and decreased blood pressure remains to be established.
In 1986, Moore-Ede proposed to extend the concept of homeostasis by introducing the term of “predictive homeostasis” or anticipative adaptations of homeostatic reactions to predictable functional challenges (31). At that time, homeostasis was traditionally seen as a corrective reaction to physiological or pathophysiological changes that already occurred (“reactive” homeostasis according to Moore-Ede classification). Although many different lines of evidence indicated the existence of a “predictive” component on the functional level, the molecular basis of this mechanism was missing. The discovery of molecular clock provided this basis for one of the possible “predictive” mechanisms, namely circadian timing system. In our study, we demonstrate that this system controls renal tubular function on both transcriptional and functional levels. This system may provide the kidney with a significant functional advantage through anticipation of changes in requirements for water and solutes reabsorption/secretion.
Methods
Animals.
Male C57BL/6J male mice (Janvier) weighing 25–30 g were used in the microarray experiments. A colony of clock(−/−) mice (C57BL/6J background) was established from breeding pairs of clock(+/-) heterozygous mice originally described by Debruyne et al. (27). The animals were maintained on the standard laboratory chow diet and adapted to 12-h light/12-h dark cycle for 2 weeks before experiments.
Microdissection.
Microdissection of DCT/CNT or CCD was performed from collagenase-treated kidneys as described previously (32).
Microarray.
RNA from microdissected DCT/CNT or CCD was isolated and purified with RNA clean up purification kits from Qiagen. All RNA quantities were assessed by NanoDrop ND-1000 spectrophotometer, and the quality of RNA was controlled on Aligent 2100 bioanalyzer chips. For each sample, 10 ng total RNA were amplified and labeled using the WT-Ovation Pico RNA Amplification System V1, (catalog # 3300–12; NuGen) and labeling with FL-Ovation cDNA Biotin Module V2 (catalog #4200–12; NuGen). Affymetrix Gene Chip Mouse Genome 430 2.0 arrays were hybridized to 5 μg labeled, amplified cDNA, washed, stained, and scanned according to the protocol described in the GeneChip Expression analysis manual (Fluidics protocol EukGeWS2v4_450; Affymetrix).
Data Analysis.
All statistical analysis were performed using the free high-level interpreted statistical language R (R Core, 2004; http://www.R-project.org) and various Bioconductor packages (http://www.Bioconductor.org). Hybridization quality was assessed using Bioconductor “affy” and “affyPLM” packages (33, 34). Log2 normalized expression signals were calculated from Affymetrix CEL files using RMA algorithm (35). Circadian genes were then identified using linear models with a pair of cosine and sine functions as the explanatory variable, with the frequency corresponding to 24-h periodicity as described previously (36). The F-ratio test statistics were converted to P values with the appropriate degrees of freedom. P values were adjusted for multiple testing with Benjamini and Hochberg's method to control the false discovery rate (FDR) (37). For each time point, log2 normalized expression values of the two replicates were averaged (AveExpr), and the amplitude was calculated as the difference between the minimum and the maximum AveExpr.
Analysis of Acrophase Distribution in Different Functional Gene Categories.
The analysis of acrophase distribution was performed using Gene Ontology (GO) annotation on the following gene categories: G protein-coupled receptors; heterotrimeric G proteins; nucleotide cyclases; cyclic-nucleotide phosphodiesterases; serine/threonine kinases; nonreceptor tyrosine kinases; tyrosine kinase receptors; serine/threonine phosphatases; tyrosine phosphatases; dual specificity phosphatases; A kinase anchor proteins; phospholipases; Rab small GTP-binding proteins; Arf small GTP-binding proteins; Rho small GTP-binding proteins; Ras and Ras-related small GTP-binding proteins; SNARE and SNARE-related proteins; coat proteins and clathrin adaptors; actin and actin binding proteins; myosin and myosin-like proteins; microtubule and microtubule-related proteins; intermediate filaments and related proteins; water channels; ion channels and transporters excluding SLC proteins; solute carrier proteins; transcriptional factors; xenobiotics metabolism enzymes (phases I and II).
Metabolic Cages.
The 24-h urine samples were collected in individual metabolic cages (Tecniplast). Urine and plasma osmolarity as well as ionic composition were analyzed in the Laboratoire Central de Chimie Clinique, Centre Hospitalier Universitaire Vaudoise (CHUV) University Hospital (Lausanne, Switzerland).
Characterization of Circadian Drinking and Activity Patterns in wt and clock(−/−) Mice.
The drinking and activity patterns were characterized using Mouse-E-Motion system (Infra-E-Motion Gmbh). This system allows simultaneous online measurements in unstressed conditions of water intake and general motion activity in mice housed in their familiar home cages.
Blood Pressure Measurements.
Blood pressure was measured in conscious unrestrained mice using Data Science International (DSI) telemetry system. Mice were allowed to recover for at least 1 week after the implantation of telemetry device before starting the blood pressure recording.
Supplementary Material
Acknowledgments.
We thank Dr. David Weaver (University of Massachusetts, Amherst, MA) for the generous gift of the clock(−/−) mice; Dr. Ueli Schibler and Dr. Frédéric Gachon (University of Geneva, Switzerland) for sharing with us the RNA samples extracted from kidneys of dbp/hlf/tef triple knockout mice; and Otto Hagenbuchle and Keith Harshman (Lausanne DNA Array Facility) for the microarray profiling studies. This work was supported by the Swiss National Science Foundation Research Grant 3100A0-117824 (to D.F.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904890106/DCSupplemental.
References
- 1.Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 2.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: Time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 3.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stokkan KA, et al. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 5.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 6.Yang S, et al. Genome-wide expression profiling and bioinformatics analysis of diurnally regulated genes in the mouse prefrontal cortex. Genome Biol. 2007;8:R247. doi: 10.1186/gb-2007-8-11-r247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 8.Kornmann B, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minors DS, Waterhouse JM. Circadian rhythms of urinary excretion: The relationship between the amount excreted and the circadian changes. J Physiol. 1982;327:39–51. doi: 10.1113/jphysiol.1982.sp014218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roelfsema F, van der Heide D, Smeenk D. Circadian rhythms of urinary electrolyte excretion in freely moving rats. Life Sci. 1980;27:2303–2309. doi: 10.1016/0024-3205(80)90498-1. [DOI] [PubMed] [Google Scholar]
- 12.Mills JN, Stanbury SW. Intrinsic diurnal rhythm in urinary electrolyte output. J Physiol. 1951;115:18–19. [PubMed] [Google Scholar]
- 13.Moore-Ede MC, Herd JA. Renal electrolyte circadian rhythms: Independence from feeding and activity patterns. Am J Physiol. 1977;232:F128–F135. doi: 10.1152/ajprenal.1977.232.2.F128. [DOI] [PubMed] [Google Scholar]
- 14.Vagnucci AH, Shapiro AP, McDonald RH., Jr Effects of upright posture on renal electrolyte cycles. J Appl Physiol. 1969;26:720–731. doi: 10.1152/jappl.1969.26.6.720. [DOI] [PubMed] [Google Scholar]
- 15.Cohn C, Webb L, Joseph D. Diurnal rhythms in urinary electrolyte excretions by the rat: Influence of feeding habits. Life Sci I. 1970;9:803–809. doi: 10.1016/0024-3205(70)90336-x. [DOI] [PubMed] [Google Scholar]
- 16.Roelfsema F, van der Heide D, Smeenk D. The influence of intravenous infusion of electrolytes on the diurnal excretory rhythms. Life Sci. 1982;30:771–778. doi: 10.1016/0024-3205(82)90612-9. [DOI] [PubMed] [Google Scholar]
- 17.Bankir L, et al. Nighttime blood pressure and nocturnal dipping are associated with daytime urinary sodium excretion in African subjects. Hypertension. 2008;51:891–898. doi: 10.1161/HYPERTENSIONAHA.107.105510. [DOI] [PubMed] [Google Scholar]
- 18.Burnier M, Coltamai L, Maillard M, Bochud M. Renal sodium handling and nighttime blood pressure. Semin Nephrol. 2007;27:565–571. doi: 10.1016/j.semnephrol.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Eastell R, et al. Abnormalities in circadian patterns of bone resorption and renal calcium conservation in type I osteoporosis. J Clin Endocrinol Metab. 1992;74:487–494. doi: 10.1210/jcem.74.3.1740481. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt CP, Homme M, Schaefer F. Structural organization and biological relevance of oscillatory parathyroid hormone secretion. Pediatr Nephrol. 2005;20:346–351. doi: 10.1007/s00467-004-1767-7. [DOI] [PubMed] [Google Scholar]
- 21.De Guchtenaere A, et al. Nocturnal polyuria is related to absent circadian rhythm of glomerular filtration rate. J Urol. 2007;178:2626–2629. doi: 10.1016/j.juro.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 22.Raes A, et al. Abnormal circadian rhythm of diuresis or nocturnal polyuria in a subgroup of children with enuresis and hypercalciuria is related to increased sodium retention during daytime. J Urol. 2006;176:1147–1151. doi: 10.1016/j.juro.2006.04.054. [DOI] [PubMed] [Google Scholar]
- 23.Voogel AJ, et al. Circadian rhythms in systemic hemodynamics and renal function in healthy subjects and patients with nephrotic syndrome. Kidney Int. 2001;59:1873–1880. doi: 10.1046/j.1523-1755.2001.0590051873.x. [DOI] [PubMed] [Google Scholar]
- 24.Buijsen JG, et al. Circadian rhythm of glomerular filtration rate in patients after kidney transplantation. Nephrol Dial Transplant. 1994;9:1330–1333. [PubMed] [Google Scholar]
- 25.Loffing J, Kaissling B. Sodium and calcium transport pathways along the mammalian distal nephron: From rabbit to human. Am J Physiol Renal Physiol. 2003;284:F628–F643. doi: 10.1152/ajprenal.00217.2002. [DOI] [PubMed] [Google Scholar]
- 26.Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA. Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genomics. 2008;32:229–253. doi: 10.1152/physiolgenomics.00201.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debruyne JP, et al. A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 28.Gachon F, et al. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Son GH, et al. Adrenal peripheral clock controls the autonomous circadian rhythm of glucocorticoid by causing rhythmic steroid production. Proc Natl Acad Sci USA. 2008;105:20970–20975. doi: 10.1073/pnas.0806962106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin X, et al. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 31.Moore-Ede MC. Physiology of the circadian timing system: Predictive versus reactive homeostasis. Am J Physiol. 1986;250:R737–R752. doi: 10.1152/ajpregu.1986.250.5.R737. [DOI] [PubMed] [Google Scholar]
- 32.Firsov D, et al. Molecular analysis of vasopressin receptors in the rat nephron. Evidence for alternative splicing of the V2 receptor. Pflugers Arch. 1994;429:79–89. doi: 10.1007/BF02584033. [DOI] [PubMed] [Google Scholar]
- 33.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalized methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 34.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 35.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 36.Wirapati P, et al. Meta-analysis of gene expression profiles in breast cancer: Toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.