Abstract
Drosophila GMP synthetase binds ubiquitin-specific protease 7 (USP7) and is required for its ability to deubiquitylate histone H2B. Previously, we showed that the GMPS/USP7 complex cooperates with the Polycomb silencing system through removal of the active ubiquitin mark from histone H2B (H2Bub). Here, we explored the interplay between GMPS and USP7 further and assessed their role in hormone-regulated gene expression. Genetic analysis established a strong cooperation between GMPS and USP7, which is counteracted by the histone H2B ubiquitin ligase BRE1. Loss of either GMPS or USP7 led to increased levels of histone H2Bub in mutant animals. These in vivo analyses complement our earlier biochemical results, establishing that GMPS/USP7 mediates histone H2B deubiquitylation. We found that GMPS/USP7 binds ecdysone-regulated loci and that mutants display severe misregulation of ecdysone _target genes. Ecdysone receptor (EcR) interacts biochemically and genetically with GMPS/USP7. Genetic and gene expression analyses suggested that GMPS/USP7 acts as a transcriptional corepressor. These results revealed the cooperation between a biosynthetic enzyme and a ubiquitin protease in developmental gene control by hormone receptors.
Proper development requires the coordination of growth and differentiation. Hormones perform essential signaling functions in metazoan organisms, controlling a plethora of processes ranging from homeostasis to key developmental transitions. The steroid hormone 20-hydroxyecdysone (ecdysone) provides critical temporal triggers that direct major developmental transitions in Drosophila (14, 22). A high ecdysone pulse at the end of the third-instar larval stage starts the larval-to-prepupal transition (pupariation). About 10 h later, another ecdysone pulse sets off the prepupal-pupal transition (pupation). The primary mediator of ecdysone signaling is a heterodimer of the Ecdysone Receptor (EcR) and Ultraspiracle (USP), the fly RXR homolog. The EcR/USP heterodimer belongs to the class of nuclear receptors (NRs) that bind their cognate regulatory DNA elements both in the absence and in the presence of hormone (14, 22). Gene regulation by NRs involves the antagonistic activities of transcriptional corepressors and coactivators. Nonliganded NRs recruit transcriptional corepressors to their _target regulatory elements, thus directing gene silencing. Upon hormone binding, there is a conformational transition that causes the replacement of corepressors by coactivators, leading to activation. Typically, coregulators modulate the structure of chromatin and include histone acetyltransferases, deacetylases, methyltransferases, demethylases, and ATP-dependent chromatin-remodeling factors (3, 4, 18, 23).
We previously implicated the biosynthetic enzyme GMP synthetase (GMPS) in transcription regulation via modulation of histone H2B deubiquitylation by ubiquitin-specific protease 7 (USP7) (29). USP7 is an evolutionarily conserved protein which was originally isolated as a binding partner of the herpes simplex virus protein Vmw110/ICP0, hence its alternate name, HAUSP (herpesvirus-associated ubiquitin-specific protease) (11). Several other associated factors and substrates of USP7 have been identified, including p53 (16), Epstein-Barr nuclear antigen 1 (EBNA1) (12), MDM2 (15), DAXX (26), FOXO4 (28), and PTEN (25).
An intriguing feature of GMPS is that it strongly stimulates the activity of USP7 (29). We found that a portion of cellular GMPS was tightly associated with USP7 and was required for H2B deubiquitylation. Histone H2B monoubiquitylation at lysine 120 (H2Bub) by the E3 ligase BRE1 is an active mark linked to transcriptional elongation, whereas H2Aub is associated with silencing (10, 30). In addition, we showed that GMPS and USP7 both act as enhancers of Polycomb-mediated silencing of homeotic genes in vivo (29). These findings suggested that GMPS does double duty: it is required for de novo GMP synthesis but is also involved in transcription control, at least in part, through cooperation with USP7. However, the extent of the involvement of GMPS and USP7 in developmental gene expression control was unclear. Here, we used a combination of genetics, in vivo gene expression analysis, and biochemistry to establish essential roles for GMPS/USP7 in ecdysone signaling. Our results revealed that GMPS/USP7 binds and regulates ecdysone _target loci, implicating a complex of a biosynthetic enzyme and a ubiquitin protease in gene control by hormone receptors.
MATERIALS AND METHODS
Fly strain, genetics, and DNA constructs.
Fly stock maintenance and crosses were performed using standard procedures. UspΔ35 was generated by imprecise excision of the P element in P[SUPor-P]Usp7KG06814 (29). Genomic sequence analysis revealed 24 bp of the P element remaining and loss of a part of the 5′ untranslated region (UTR) and the coding sequence for its first 23 amino acids. P[EP]Bre1kim1 and Bre1RNAi were purchased from GenExel (strain GE22117) and VDRC (strain 15620) (9), respectively. GmpsRNAi was obtained from the National Institute of Genetics Fly Stock Center (strain 9242R-3). To generate UAS-Usp7 and UAS-Usp7C250A transgenic lines, cDNAs encoding full-length USP7, USP7C250A, GMPSS242L, and GMPSC95A were cloned into pUAST and verified by sequencing. P element-mediated germ line transformation was performed according to standard procedures. The Gmpse10 and UAS-Gmps lines were a kind gift of Yong Rao (19). GmpsK07130 has been described previously (29). EcRV559fs and EcRA483T were acquired from the Bloomington stock center. UAS-Usp7#J2 and UAS-Usp7C250A#C4, both integrated on the second chromosome, were each combined with UAS-Gmps#2 (UAS-bur#2 in reference 19) and crossed with a GMR-Gal4 driver line. A stable GMR>Usp7/GMR>Gmps line (#2091) was created by recombination of UAS-Usp7 (line J1, on the third chromosome) and UAS-Gmps (UAS-bur#1 in reference 19) and crossed with a GMR-Gal4 driver line (on the second chromosome). Crosses were performed at 25°C. Details will be provided upon request.
Protein-protein interaction assays.
Recombinant glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21-CodonPlus(DE3)-RIL and purified by glutathione Sepharose 4 fast flow (GE Healthcare) chromatography according to standard procedures (8). 35S-labeled proteins were expressed using TNT coupled rabbit reticulocyte lysates (Promega). Prior to binding reactions, beads were blocked with 3% fetal calf serum in HEMG buffer (25 mM HEPES-KOH [pH 7.6], 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol) containing 1 mM dithiothreitol, 0.2 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin A, 150 mM KCl (making HEMG/150), and 0.1% NP-40. Binding reactions were in the same buffer but lacking serum. Following three washes with HEMG/300/0.1% NP-40 and two with HEMG/150/0.01% NP-40, bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography.
Immunological procedures.
For antibody production, GMPS (amino acids 310 to 563 and 310 to 623) and BRE1 (amino acids 231 to 718) were expressed as GST fusion proteins, purified, and used for immunizations as previously described (8). Anti-USP7, anti-H1, and anti-H2B (PV57/58, raised against purified core histones and mainly recognizing H2B) have been previously published (13, 20, 29). To detect RNA polymerase (Pol) II, we used a mixture of monoclonal antibodies 8WG16, H5, and H14 (Covance). Anti-ubiquityl-histone H2B (clone 56) was from Millipore. Anti-EcR (DDA2.7 and Ag10.2), developed by C. Thummel and D. Hogness, was obtained from the Developmental Studies Hybridoma Bank (Department of Biological Sciences, National Institute of Child Health and Human Development, University of Iowa, Iowa City). Analysis of polytene chromosomes was done as described previously (21). For analysis of polytene chromosomes upon knockdown of GMPS, GmpsRNAi was crossed with an actin driver line. To analyze H2Bub levels, 10 third-instar larvae were lysed in SDS-gel loading buffer using an Eppendorf tube pestle.
Gene expression analyses.
For prepupal analysis, the Usp7kim1, Usp7KG06814, and GmpsK07130 mutant lines were rebalanced with green fluorescent protein (GFP)-marked balancer chromosomes. GFP-negative mutant prepupae and wild-type (WT) prepupae were collected at 2-h intervals from the moment of pupariation (t = 0) for 12 h. RNA was extracted using TRIzol. S2 cells were cultured and treated with double-stranded RNA (dsRNA) as described previously (33). After 3 days, dsRNA was reapplied, and after 5 days, cells were harvested and RNA was extracted using TRIzol. Knockdowns were performed in biological triplicates. Knockdown efficiency was monitored in parallel by immunoblotting. RNA levels in prepupae or S2 cells were analyzed by first-strand cDNA synthesis with Superscript II reverse transcriptase (Invitrogen) and subsequent quantitative PCR (qPCR) with SYBR green I using a MyiQ single-color real-time PCR detection system (Bio-Rad). Analysis of the reverse transcription (RT)-qPCR data was performed using the 2−ΔΔCT method (17). CG11874 was used as an internal control mRNA.
The RT-qPCR primers used were as follows: CG11874, 5′-AGTGTTGCTCTGCCTAAGTGG-3′ and 5′-CGGATGATGGTGCGGATTGG-3′; GAPDH, 5′-TGCTGGAGCCGAGTATGTGG-3′ and 5′-GCCGAGATGATGACCTTCTTGG-3′; ImpL3, 5′-ATCGGCAGCGGCACCAAC-3′ and 5′-CGGCAATGTTCACTCCAGACC-3′; ImpE2, 5′-ACTTCCTGGGCGGCAATCG-3′ and 5′-CTACGGCTGGCTTCTCTGTGG-3′; Ftz-F1, 5′-ATGGAAGGCGAACGAAGGATAC-3′ and 5′-AGTTGGTGGTAGTAGTGATGATGC-3′; E74A, 5′-GATGGTCGTCTTGTTGGAGGTC-3′ and 5′-TGCGGGTTGTTCGGATTGC-3′; E75A, 5′-CCTTTCATTGACTAACTGCCACTC-3′ and 5′-CGAAACGAAACGAACGGAACG-3′; E75C, 5′-CGGCTCGGAAGTTTGTGGTTAG-3′ and 5′-GCTGATGCTGCTGCTGATGC-3′; CG1381, 5′-ACACCCGAACAGGCGAGAATCC-3′ and 5′-TCATCATCGTCGTCGTTGTCATCC-3′.
RESULTS
GMPS and USP7 interact genetically.
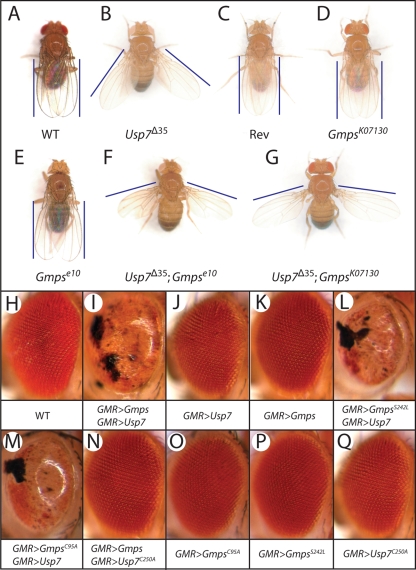
Previously, we presented biochemical evidence revealing that parts of GMPS and USP7 associate in a highly stable complex, mediating epigenetic gene silencing (29). Here, we set out to explore their cooperation in vivo during developmental gene control. To facilitate the genetic analysis of USP7, which is encoded by an essential X-chromosomal gene, we created new alleles by mobilizing the P element in the Usp7KG06814 mutant strain. One of the derived alleles, Usp7Δ35, turned out to be a hemizygous viable hypomorph. Usp7Δ35 lacks part of the USP7 5′ UTR and the coding sequence for its first 23 amino acids and expresses USP7 at a reduced level. Hemizygous Usp7Δ35 males, but not revertants, generated by precise P element excision, displayed a “held-out wing” phenotype (Fig. 1A to C). This phenotype is convenient for genetic interaction assays. Indeed, we found that the “held-out wing” phenotype was strongly enhanced when Usp7Δ35 was combined with either a heterozygous Gmpse10 or GmpsK07130 allele, which by itself did not affect wing positioning (Fig. 1D to G). These results showed that USP7 and GMPS interact not only biochemically but also genetically.
FIG. 1.
Usp7 and Gmps interact genetically. (A to G) Gmps loss-of-function mutations enhance the Usp7Δ35 held-out wing phenotype. The hypomorphic allele Usp7Δ35 was generated by imprecise excision of the P element in Usp7KG06814, whereas the revertant (Rev) resulted from precise excision of the P element. As indicated, the male flies are WT, hemizygous for Usp7Δ35, and/or heterozygous for the indicated Gmps alleles. (H to Q) Ectopic expression of both Usp7 and Gmps driven by the GMR enhancer (GMR>Gmps/GMR>Usp7) causes defective eye development characterized by disorganized or missing ommatidia, loss of bristles, and black necrotic patches. Overexpression of either Usp7 or Gmps by itself did not disrupt eye development. Mutations that abrogate either glutamine hydrolysis (GmpsC95A) or ATP hydrolysis (GmpsS242L) do not affect GMPS cooperation with USP7. However, overexpression of the Usp7C250A catalytic mutant together with GMPS no longer gave an eye phenotype. Single overexpression of GmpsC95A, GmpsS242A, or Usp7C250A has no effect on eye development.
To complement these loss-of-function analyses with an in vivo ectopic expression assay, we employed the GAL4/UAS system (6). We used the glass multimer reporter (GMR) to drive GAL4 expression in the developing eye, resulting in overexpression of USP7 or GMPS under UAS control. Concomitant ectopic expression of USP7 and GMPS (GMR>Usp7/GMR>Gmps) caused clear defects in eye development characterized by disorganized or missing ommatidia, loss of bristles, and black necrotic patches (Fig. 1I). When either only USP7 (Fig. 1J) or only GMPS (Fig. 1K) was expressed ectopically, we observed no eye defects. Thus, the phenotype was strictly dependent on the overexpression of both GMPS and USP7. Next we wondered whether the enzymatic activities of USP7 or GMPS were required for the distorted eye phenotype. The phenotype of ectopic coexpression of USP7 with either a GMPS mutant defective in ATP hydrolysis (GMPSS242L) or a GMPS mutant defective in glutamine hydrolysis (GMPSC95A) was similar to that resulting from the coexpression of WT GMPS (Fig. 1L and M). Thus, in agreement with our earlier in vitro results (29), GMPS's enzymatic activity is not required for stimulation of USP7. In contrast, concomitant overexpression of GMPS and USP7C250A, a deubiquitylation-defective substitution mutant, no longer affected eye development (Fig. 1N). As is the case for the WT proteins, ectopic overexpression of only GMPSC95A, GMPSS242L, or USP7C250A had no effect on eye development. (Fig. 1O, P, and Q). We conclude that GMPS and USP7 cooperation is critically dependent on USP7's deubiquitylating activity but not on de novo GMP synthesis by GMPS.
GMPS/USP7 counteracts BRE1 by deubiquitylation of histone H2B.
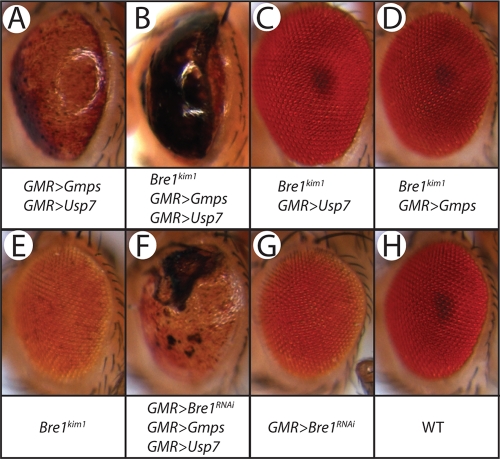
Our earlier in vitro reconstitution experiments suggested that histone H2Bub is a _target for deubiquitylation by GMPS/USP7 (29). Because BRE1 is the E3 ubiquitin ligase that catalyzes H2B ubiquitylation, we wondered whether reduced BRE1 levels would modulate the GMPS/USP7 overexpression phenotype. To test this hypothesis, we combined a heterozygous Bre1kim1 mutant allele with GMR-driven ectopic expression of GMPS and USP7. Note that the recombined GMR>Gmps/GMR>Usp7 line used in these assays has a milder eye phenotype (Fig. 2A) than the one used above (Fig. 1I; see Materials and Methods for details). The heterozygous Bre1kim1 allele, when combined with GMPS/USP7 overexpression, led to strongly (∼50%) reduced viability, whereas the eye phenotype in surviving adults is strongly enhanced (Fig. 2B). The combination of Bre1kim1 with overexpression of only USP7 or only GMPS had no effect on eye development, again emphasizing the dependence on cooperation between GMPS and USP7 (Fig. 2C and D). Development of Bre1kim1 heterozygotes was normal (Fig. 2E). When we used the GMR-GAL4 driver for ectopic GMPS and USP7 expression and to express an interfering RNA _targeting the Bre1 mRNA, we also observed an enhanced phenotype (Fig. 2F). Importantly, the GMR>Bre1RNAi line by itself displayed no defective eye development (Fig. 2G). When GMR>Bre1RNAi was combined with either GMR>Usp7 or GMR>Gmps alone, we observed no phenotype (data not shown). Collectively, these results strongly support the notion that GMPS and USP7 cooperate in vivo to counteract the activity of ubiquitin ligase BRE1.
FIG. 2.
Usp7, Gmps, and Bre1 interact genetically. Concomitant reduction of histone H2B ubiquitin ligase BRE1 levels with GMPS and USP7 overexpression strongly enhances the eye phenotype. (A to E) Heterozygous Bre1kim1 males that overexpress GMPS and USP7 by the GMR-GAL4 driver have a 50% reduction in viability, whereas escapers display an enhanced eye phenotype. Heterozygous Bre1kim1 males which overexpress only USP7, only GMPS, or neither factor have normal eyes. (F) When GMR-GAL4 drives the expression of an RNAi _targeting the Bre1 mRNA, as well as overexpression of USP7 and GMPS, we also observed a clear enhancement of the eye phenotype. (G) GMR>Bre1RNAi by itself does not display any eye defect. WT eyes are shown as a reference.
The ability of GMPS/USP7 to deubiquitylate H2Bub in vitro (29) and the strong genetic interaction between GMPS/USP7 and BRE1 in vivo strongly support the relevance of histone H2Bub as a substrate. To provide further evidence for this notion, we determined H2Bub levels in GmpsK07130 and Usp7KG06814 mutants by immunoblot analysis of protein extracts made from third-instar larvae. Homozygous Usp7KG06814 and GmpsK07130 larvae have strongly reduced levels of USP7 and GMPS, respectively (Fig. 3A). We observed a clear increase in bulk H2Bub levels in homozygous Usp7KG06814 larvae compared to WT or heterozygous larvae (Fig. 3B, compare lanes 5 and 6 with lanes 1 to 4). Note that to show that we are within a quantitative blotting range, we loaded two amounts of protein for each set. In support of the regulatory role of GMPS in the catalytic activity of USP7, we also observed an increase in H2Bub levels in homozygous GmpsK07130 larvae (Fig. 3B, compare lanes 9 and 10 with lanes 1 and 2 and lanes 7 and 8). Collectively, our earlier results (29) and those presented here demonstrate that GMPS/USP7 deubiquitylates histone H2Bub.
FIG. 3.
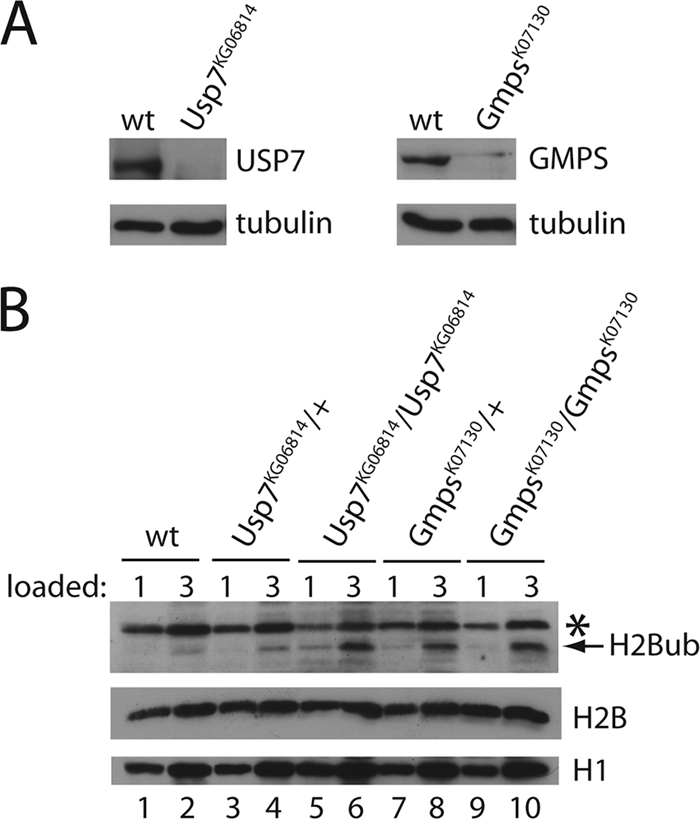
USP7/GMPS _targets H2Bub in vivo. (A) Homozygous Usp7KG06814 and GmpsK07130 mutants have reduced levels of USP7 and GMPS, respectively. Protein extracts of third-instar larvae were analyzed by immunoblotting using the indicated antibodies. (B) Homozygous Usp7KG06814 and GmpsK07130 mutants contain increased levels of histone H2Bub. Immunoblotting on protein extracts of third-instar larvae with the indicated genotypes is shown. A threefold different protein amount of each extract was loaded to ensure that blotting analysis was within a quantitative range. The asterisk indicates a background band that served as an additional loading control. Antisera were directed against the indicated proteins.
GMPS and USP7 are required for ecdysteroid signaling in vivo.
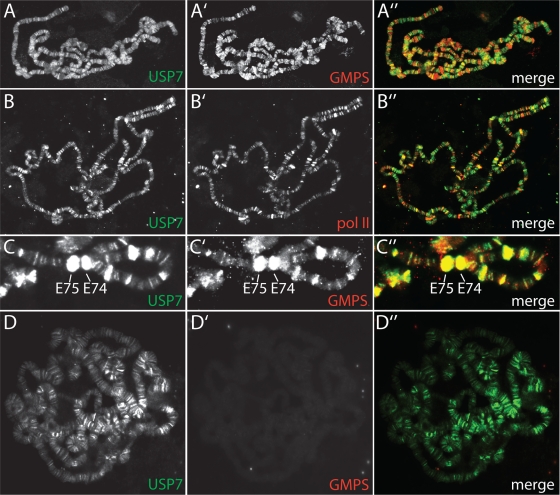
To compare their genome-wide distribution, we used indirect immunofluorescence to determine the binding of GMPS and USP7 on third-instar larval salivary gland polytene chromosomes (Fig. 4A). USP7 and GMPS colocalize on many of their chromosomal binding sites, but each also occupies unique loci. Thus, although USP7 and GMPS can form a tight complex, they can also bind DNA independently. In agreement with this notion, immunodepletion experiments revealed that portions of USP7 and GMPS are not associated with each other (data not shown). Overall, USP7 does not colocalize with RNA Pol II, indicative of a role in gene silencing rather than activation (Fig. 4B). This is consistent with its genetic function as an enhancer of Polycomb silencing (29) and its biochemical activity: removal of the active H2Bub mark. Analysis of their binding pattern revealed that both GMPS and USP7 associate with the E74 and E75 loci harboring ecdysone early-response genes (7, 24) (Fig. 4C). We note that we only observed GMPS and USP7 colocalization in these regions prior to their induction (see below). RNA interference (RNAi)-mediated knockdown of GMPS does not affect USP7 binding to polytene chromosomes (Fig. 4D). These results suggest that the recruitment of USP7 to chromatin is independent of GMPS.
FIG. 4.
GMPS and USP7 bind ecdysone-regulated loci. (A to E) The distribution of USP7, GMPS, and RNA Pol II on Drosophila salivary gland polytene chromosomes was determined by indirect immunofluorescence using the appropriate antibodies. (A) USP7 and GMPS colocalize at many sites but also bind unique loci. (B) USP7 and RNA Pol II distribution is mostly mutually exclusive. (C) Higher magnification of a polytene chromosome showing the binding of USP7 and GMPS at ecdysone-regulated loci E74 and E75 prior to puffing. (D) USP7 remains associated with polytene chromosomes upon RNAi-mediated knockdown of GMPS.
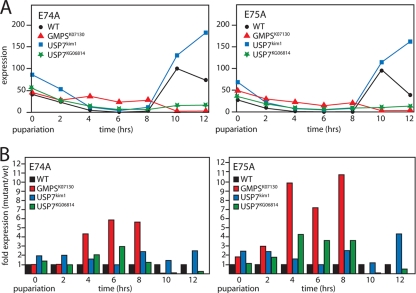
The binding of USP7 and GMPS to the E74 and E75 loci suggested a role in the transcriptional regulation of these gene clusters. Consistent with this notion, strong USP7 and GMPS mutants die at the prepupal or pupal stage, when ecdysone signaling is critical. Furthermore, Gmps and Usp7 mutant pupae frequently lack anterior spiracles, one of the hallmarks of defective ecdysteroid signaling during pupariation (data not shown). To test the role of USP7 and GMPS in ecdysone signaling, we compared the expression profiles of E74A and E75A in homozygous Usp7KG06814, Usp7kim1, and GmpsK07130 prepupae to that in WT animals (Fig. 5A). Prepupae were collected at pupariation (t = 0), and RNA was extracted at 2-h intervals for 12 h and monitored by RT-qPCR. To facilitate comparison, E74A and E75A expression levels in Gmps or Usp7 mutants were also plotted relative to those in WT animals (Fig. 5B). In WT prepupae, E74A and E75A expression reflects the level of ecdysone signaling. Following a peak at pupariation, transcription ceases until, after ∼10 h, another ecdysone pulse strongly activates E74A and E75A transcription. Compared to WT animals, Usp7kim1 prepupae overexpress E74A and E75A at early and late time points. In GmpsK07130 and Usp7KG06814 mutants, there is a striking derepression from t = 4 to 8 h. In contrast to Usp7kim1 mutants, which harbor a weaker allele than Usp7KG06814 and are homozygous viable, the development of GmpsK07130 and Usp7KG06814 mutants arrests prior to pupation and E74A and E75A expression ceases after t = 10 h. In summary, GMPS and USP7 bind ecdysone _target loci and are required for their normal regulation, most likely as transcriptional corepressors.
FIG. 5.
Usp7 and Gmps mutants misexpress EcR _target genes. (A) WT and homozygous Usp7kim1, Usp7KG06814, or GmpsK07130 mutant prepupae were isolated at 2-h intervals from pupariation (t = 0) for 12 h. RNA was extracted, and relative expression levels of E74A (left panel) and E75A (right panel) were determined by RT-qPCR. The highest level of expression in WT prepupae was set at 100. Note that Gmpsk07130 and Usp7KG06814 mutants die during pupation. We omitted the error bars for clarity, but the variation was always less than 5%. (B) E74A and E75A expression in homozygous Usp7kim1, Usp7KG06814, and Gmpsk07130 mutant prepupae plotted relative to that in WT animals.
Binding dynamics at ecdysone _target loci.
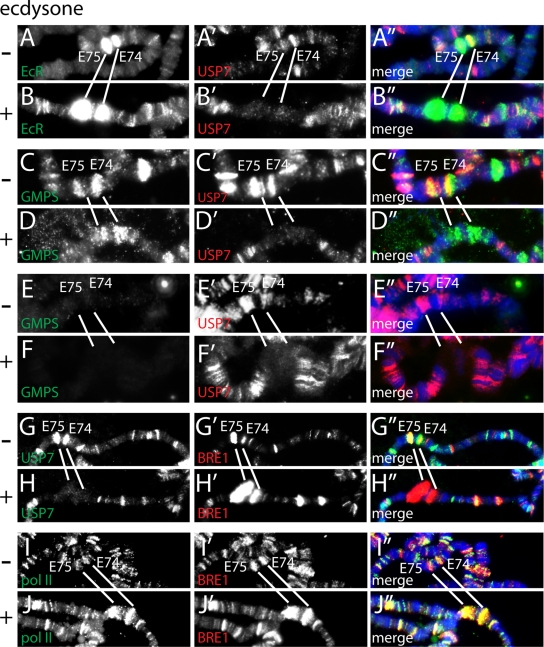
We used indirect immunofluorescence of polytene chromosomes to monitor GMPS and USP7 binding to ecdysone _target loci prior to or after hormone signaling (Fig. 6). An ecdysone pulse at the end of the third-instar larval stage causes activation of the E74 and E75 gene clusters, hallmarked by chromosome “puffing.” EcR and USP7 both bind E74 and E75 prior to induction (Fig. 6A). However, following the ecdysone pulse, additional EcR is recruited but USP7 disappears, as expected of a corepressor (Fig. 6B). GMPS was present at the uninduced E74 and E75 loci (Fig. 6C) but, in contrast to USP7, persisted following activation (Fig. 6D). The dynamics of USP7 binding is independent of GMPS, because it is unaffected by RNAi-mediated depletion of GMPS (Fig. 6E and F). Our genetic analysis indicated that GMPS/USP7 acts antagonistically to BRE1 in vivo (Fig. 2). Therefore, we compared the binding dynamics of BRE1 with those of USP7 and RNA Pol II. In agreement with a low level of transcription prior to ecdysone induction, we detected both BRE1 and RNA Pol II at E74 and E75 (Fig. 6G and I). However, following activation, BRE1 and RNA Pol II levels were dramatically increased, whereas USP7 was removed (Fig. 6H and J). In conclusion, our analysis of binding to ecdysone _target loci supports a role for GMPS/USP7 as a transcriptional corepressor. Moreover, in accordance with their genetic antagonism, and opposing biochemical activities, there is an exchange of the ubiquitin protease USP7 for the ubiquitin ligase BRE1 concomitant with RNA Pol II recruitment.
FIG. 6.
Upon induction, USP7 leaves the E74 and E75 loci, whereas GMPS remains and BRE1 is recruited. Indirect immunofluorescence on third-instar larval polytene chromosomes before (−) and after (+) activation by ecdysone using the indicated antibodies is shown. DNA was visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining (blue). The E74 and E75 loci are indicated. (E and F) RNAi-mediated knockdown of GMPS does not affect the dynamics of USP7 association with E74 and E75.
GMPS/USP7 acts as an EcR corepressor.
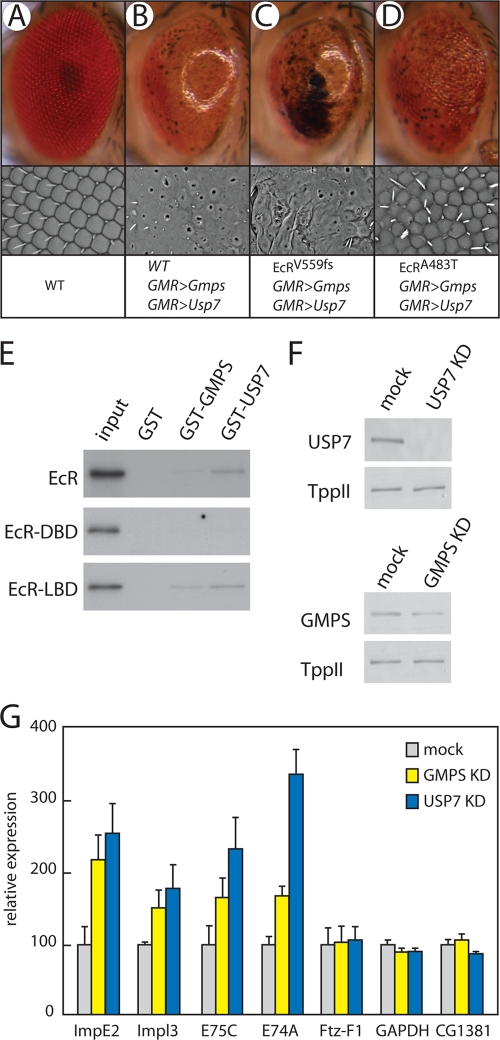
To obtain additional evidence for the role of GMPS/USP7 as an EcR corepressor, we performed genetic interaction assays utilizing two distinct EcR mutants. EcRV559fs is a loss-of-function mutation that has an activation defect (2). In contrast, EcRA483T can still respond to hormone induction but its interaction with the corepressor SMRTER is abrogated, causing enhanced transcription in the absence of hormone (27). When EcRV559fs was combined with GMPS/USP7 ectopic expression in the eye, we observed a clear enhancement of the developmental defect (Fig. 7A to C). In striking contrast, EcRA483T suppressed the GMPS/USP7 overexpression eye phenotype (Fig. 7D). By themselves, the EcRV559fs and EcRA483T mutations result in normal eyes (not shown). Thus, defective activation by EcR enhances the GMPS/USP7 ectopic expression phenotype, whereas loss of repression causes suppression. A plausible molecular explanation is that the failure of EcRA483T to bind the SMRTER corepressor compensates for the overexpression of GMPS/USP7 by lowering the overall EcR-directed transcriptional repression. Alternatively, EcRA483T might be defective for GMPS/USP7 recruitment, possibly through SMRTER. Suggestively, SMRTER was detected by mass spectrometry in our USP7 purifications from Drosophila embryos (J. A. van der Knaap, unpublished results). Whatever the detailed molecular mechanism, these genetic interactions provide further in vivo support for the notion that GMPS/USP7 acts as an EcR corepressor.
FIG. 7.
GMPS/USP7 is an EcR corepressor. (A to D) Genetic analysis suggested that GMPS/USP acts as an EcR corepressor. The effect of ectopic expression of both Usp7 and Gmps (GMR>Gmps/GMR>Usp7) during eye development is enhanced by EcRV559fs, which is activation defective, and suppressed by EcRA483T, which is defective for repression. The upper panels are regular photographs, and the lower panels are scanning electron micrographs. (E) USP7 binds the EcR in vitro. GST, GST-GMPS, or GST-USP7 immobilized on glutathione-Sepharose beads was incubated with 35S-labeled full-length EcR, DBD, or LBD, and after extensive washes, bound proteins were resolved by SDS-PAGE and detected by autoradiography. Eight percent of the input was loaded. (F) RNAi-mediated knockdown of USP7 or GMPS was monitored by immunoblot analysis of whole-cell extracts. Tripeptidyl peptidase II (TppII) served as a loading control. (G) GMPS and USP7 depletion leads to upregulation of ecdysone-regulated genes. RT-qPCR analysis of mock-treated S2 cells or cells treated with dsRNA directed against either USP7 or GMPS. Relative expression levels of the indicated genes were determined by RT-qPCR using the 2−ΔΔCT method. For each gene, the relative level in mock-treated cells was set at 100. Experiments were performed in three independent biological replicates. Error bars represent the standard error of the mean.
To test for direct binding, we expressed and purified GST-tagged GMPS or USP7, which was immobilized on glutathione-Sepharose beads and incubated with radiolabeled EcR. We found that EcR bound efficiently to USP7 but not to GMPS (Fig. 7E). Additional binding studies revealed that USP7 bound the EcR ligand binding domain (LBD) but not the DNA binding domain (DBD). We observed very weak binding of GMPS to the LBD, but considering the lack of binding to full-length EcR, this was most likely background. We conclude that GMPS/USP7 interacts not only genetically with the EcR but also physically, through binding of USP7 to the LBD.
Finally, we decided to test the effect of RNAi-mediated depletion of USP7 or GMPS on a selection of ecdysone-regulated genes in S2 tissue culture cells. Whereas USP7 was efficiently depleted when _targeted by RNAi, only a modest reduction in GMPS levels could be achieved (Fig. 7F). Nevertheless, RT-qPCR revealed clear derepression of the ecdysone-regulated genes ImpE2, ImpL3, E75C, and E74A (1, 32) following USP7 or GMPS depletion. In contrast, reduced USP7 or GMPS levels did not affect the transcription of βFtz-F1, gapdh, or CG1381. We conclude that GMPS/USP7 acts as a gene-selective transcriptional corepressor of ecdysone-inducible genes.
DISCUSSION
Our results demonstrated that a classic biosynthetic enzyme, GMPS, does double duty as a transcription factor involved in gene-specific repression by NRs. Genetic analysis revealed strong cooperation between GMPS and USP7, which was critically dependent on the deubiquitylating activity of USP7 but independent of GMP synthesis by GMPS. A series of observations established that GMPS/USP7 acts as a transcriptional corepressor of the EcR. (i) GMPS and USP7 mutants die at a developmental stage when ecdysone signaling is critical, and they display severe misregulation of endogenous ecdysone _target genes. (ii) GMPS and USP7 bind ecdysone _target genes when these are repressed, but USP7 dissociates upon activation and RNA Pol II recruitment. (iii) USP7 directly binds the EcR. (iv) GMPS/USP7 and the EcR interact genetically. EcR mutants defective for transcriptional activation enhance the GMPS/USP7 ectopic expression phenotype, whereas loss of EcR repression causes suppression. (v) Loss of USP7 or GMPS causes upregulation of ecdysone _target genes, both in vivo and in S2 cells. We conclude that GMPS/USP7 acts as a corepressor during developmental gene control by hormone receptors. Combined with their role in Polycomb-mediated gene silencing (29), these observations revealed surprisingly broad functions in transcription control for the biosynthetic enzyme GMPS and the ubiquitin protease USP7.
Our genetic analysis supports antagonistic roles for GMPS/USP7 and the histone H2B ubiquitin ligase BRE1. These findings dovetail well with our in vitro analysis that showed that GMPS is required for H2B deubiquitylation by USP7 (29). Recently, addition of the repressive histone H2Aub mark has been implicated in silencing of mammalian NR _targets. The H2A ubiquitin ligase 2A-HUB associates with the NR corepressor N-CoR and negatively regulates transcription (34). Conversely, the H2Aub ubiquitin protease 2A-DUB interacts with the transcriptional coactivator p/CAF and stimulates transcriptional activation by the androgen receptor (35). Thus, histone ubiquitylation/deubiquitylation seems to play a key role in gene control by NRs.
Finally, our finding that a key enzyme in the guanine nucleotide biosynthesis pathway is involved in NR _target silencing suggests that GMPS might provide a relay between metabolic state and developmental gene switching. Consistent with this notion, GMPS expression is upregulated in proliferating and tumorigenic cells (5, 31). It will be of interest to take a closer look at other nucleotide biosynthetic enzymes and investigate whether more of them have a second function as transcription factors. They might help provide the critical coupling of cell growth, division, and differentiation that is required for development.
Acknowledgments
We thank Carl Thummel and Yong Rao for providing reagents. We are grateful to Prashanth Kumar Bajpe and Adrie Verhoeven for valuable discussions.
This work was supported by grants from NWO Chemical Sciences (ECHO 700.55.001) and the Dutch government (BSIK 03038, SCDD).
Footnotes
Published ahead of print on 7 December 2009.
REFERENCES
- 1.Beckstead, R. B., G. Lam, and C. S. Thummel. 2005. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 6:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, M., F. B. Imam, W. S. Talbot, B. Ganetzky, and D. S. Hogness. 1997. Drosophila ecdysone receptor mutations reveal functional differences among receptor isoforms. Cell 91:777-788. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447:407-412. [DOI] [PubMed] [Google Scholar]
- 4.Bhaumik, S. R., E. Smith, and A. Shilatifard. 2007. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14:1008-1016. [DOI] [PubMed] [Google Scholar]
- 5.Boritzki, T. J., R. C. Jackson, H. P. Morris, and G. Weber. 1981. Guanosine-5′-phosphate synthetase and guanosine-5′-phosphate kinase in rat hepatomas and kidney tumors. Biochim. Biophys. Acta 658:102-110. [DOI] [PubMed] [Google Scholar]
- 6.Brand, A. H., and N. Perrimon. 1993. _targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 7.Burtis, K. C., C. S. Thummel, C. W. Jones, F. D. Karim, and D. S. Hogness. 1990. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell 61:85-99. [DOI] [PubMed] [Google Scholar]
- 8.Chalkley, G. E., and C. P. Verrijzer. 2004. Immuno-depletion and purification strategies to study chromatin-remodeling factors in vitro. Methods Enzymol. 377:421-442. [DOI] [PubMed] [Google Scholar]
- 9.Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova, M. Fellner, B. Gasser, K. Kinsey, S. Oppel, S. Scheiblauer, A. Couto, V. Marra, K. Keleman, and B. J. Dickson. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151-156. [DOI] [PubMed] [Google Scholar]
- 10.Espinosa, J. M. 2008. Histone H2B ubiquitination: the cancer connection. Genes Dev. 22:2743-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 13.Katsani, K. R., J. J. Arredondo, A. J. Kal, and C. P. Verrijzer. 2001. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 15:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King-Jones, K., and C. S. Thummel. 2005. Nuclear receptors—a perspective from Drosophila. Nat. Rev. Genet. 6:311-323. [DOI] [PubMed] [Google Scholar]
- 15.Li, M., C. L. Brooks, N. Kon, and W. Gu. 2004. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13:879-886. [DOI] [PubMed] [Google Scholar]
- 16.Li, M., D. Chen, A. Shiloh, J. Luo, A. Y. Nikolaev, J. Qin, and W. Gu. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648-653. [DOI] [PubMed] [Google Scholar]
- 17.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 18.Lonard, D. M., and B. W. O'Malley. 2007. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell 27:691-700. [DOI] [PubMed] [Google Scholar]
- 19.Long, H., S. Cameron, L. Yu, and Y. Rao. 2006. De novo GMP synthesis is required for axon guidance in Drosophila. Genetics 172:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohd-Sarip, A., J. A. van der Knaap, C. Wyman, R. Kanaar, P. Schedl, and C. P. Verrijzer. 2006. Architecture of a polycomb nucleoprotein complex. Mol. Cell 24:91-100. [DOI] [PubMed] [Google Scholar]
- 21.Mohrmann, L., K. Langenberg, J. Krijgsveld, A. J. Kal, A. J. R. Heck, and C. P. Verrijzer. 2004. Differential _targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 24:3077-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riddiford, L. M., P. Cherbas, and J. W. Truman. 2000. Ecdysone receptors and their biological actions. Vitam. Horm. 60:1-73. [DOI] [PubMed] [Google Scholar]
- 23.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405-1428. [DOI] [PubMed] [Google Scholar]
- 24.Segraves, W. A., and D. S. Hogness. 1990. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 4:204-219. [DOI] [PubMed] [Google Scholar]
- 25.Song, M. S., L. Salmena, A. Carracedo, A. Egia, F. Lo-Coco, J. Teruya-Feldstein, and P. P. Pandolfi. 2008. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang, J., L. K. Qu, J. Zhang, W. Wang, J. S. Michaelson, Y. Y. Degenhardt, W. S. El-Deiry, and X. Yang. 2006. Critical role for Daxx in regulating Mdm2. Nat. Cell Biol. 8:855-862. [DOI] [PubMed] [Google Scholar]
- 27.Tsai, C. C., H. Y. Kao, T. P. Yao, M. McKeown, and R. M. Evans. 1999. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell 4:175-186. [DOI] [PubMed] [Google Scholar]
- 28.van der Horst, A., A. M. de Vries-Smits, A. B. Brenkman, M. H. van Triest, N. van den Broek, F. Colland, M. M. Maurice, and B. M. Burgering. 2006. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 8:1064-1073. [DOI] [PubMed] [Google Scholar]
- 29.van der Knaap, J. A., B. R. Kumar, Y. M. Moshkin, K. Langenberg, J. Krijgsveld, A. J. Heck, F. Karch, and C. P. Verrijzer. 2005. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol. Cell 17:695-707. [DOI] [PubMed] [Google Scholar]
- 30.Weake, V. M., and J. L. Workman. 2008. Histone ubiquitination: triggering gene activity. Mol. Cell 29:653-663. [DOI] [PubMed] [Google Scholar]
- 31.Weber, G., M. E. Burt, R. C. Jackson, N. Prajda, M. S. Lui, and E. Takeda. 1983. Purine and pyrimidine enzymic programs and nucleotide pattern in sarcoma. Cancer Res. 43:1019-1023. [PubMed] [Google Scholar]
- 32.White, K. P., S. A. Rifkin, P. Hurban, and D. S. Hogness. 1999. Microarray analysis of Drosophila development during metamorphosis. Science 286:2179-2184. [DOI] [PubMed] [Google Scholar]
- 33.Worby, C. A., N. Simonson-Leff, and J. E. Dixon. 2001. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE 2001:pl1. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, W., P. Zhu, J. Wang, G. Pascual, K. A. Ohgi, J. Lozach, C. K. Glass, and M. G. Rosenfeld. 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell 29:69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu, P., W. Zhou, J. Wang, J. Puc, K. A. Ohgi, H. Erdjument-Bromage, P. Tempst, C. K. Glass, and M. G. Rosenfeld. 2007. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell 27:609-621. [DOI] [PMC free article] [PubMed] [Google Scholar]