Abstract
PIWI (P-element-induced wimpy testis) proteins are a subset of the Argonaute proteins and are expressed predominantly in the germlines of a variety of organisms, including Drosophila and mammals. PIWI proteins associate specifically with PIWI-interacting RNAs (piRNAs), small RNAs that are also expressed predominantly in germlines, and silence transposable DNA elements and other genes showing complementarities to the sequences of associated piRNAs. This mechanism helps to maintain the integrity of the genome and the development of gametes. PIWI proteins have been shown recently to contain symmetrical dimethyl arginines (sDMAs), and this modification is mediated by the methyltransferase PRMT5 (also known as Dart5 or Capsuleen). It was then demonstrated that multiple members of the Tudor (Tud) family of proteins, which are necessary for gametogenesis in both flies and mice, associate with PIWI proteins specifically through sDMAs in various but particular combinations. Although Tud domains in Tud family members are known to be sDMA-binding modules, involvement of the Tudor family at the molecular level in the piRNA pathway has only recently come into focus.
Keywords: PIWI, PRMT5, RNA silencing, Tudor, piRNA, sDMA
Argonaute (AGO) proteins associate with small noncoding RNAs of 20–30 nucleotides (nt) to negatively regulate the expression of genes _targeted by the Argonaute–small RNA complexes (Siomi and Siomi 2009). In this mechanism, termed RNA silencing, genes silenced by the catalytic activities of Argonaute proteins are involved in fundamental cellular processes, such as development, differentiation, metabolism, and apoptosis (Kim et al. 2009). Thus, Argonaute proteins are essential for many, if not all, living organisms (Bartel 2009; Malone and Hannon 2009; Voinnet 2009).
The number of Argonaute family members in a species differs; for example, Schizosaccharomyces pombe has only one Argonaute, while Drosophila and humans possess five and eight members, respectively (Hutvagner and Simard 2008). Each Argonaute member falls into one of two subgroups: the AGO and PIWI (P-element-induced wimpy testis) subfamilies (Farazi et al. 2008). Expression of AGO members is ubiquitous, whereas PIWI proteins are detected predominantly in germline cells (Farazi et al. 2008). Depletion of AGO functions often causes developmental defects; for example, Ago2-null mice show embryonic lethality (Liu et al. 2004), while PIWI mutants show defects in gametogenesis, but otherwise develop normally (Cox et al. 1998; Harris and Macdonald 2001; Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007; Li et al. 2009).
AGO proteins associate with microRNAs (miRNAs), ubiquitously expressed small RNAs that function in RNA silencing (Kim et al. 2009). In Drosophila, endogenous siRNAs (endo-siRNAs) are also ubiquitous, and associate with AGO proteins (predominantly with AGO2). Furthermore, exo-siRNAs (siRNAs exogenously introduced into cells to artificially induce RNAi) also associate with AGO in Drosophila and mice (Siomi and Siomi 2009). PIWI proteins associate specifically with PIWI-interacting RNAs (piRNAs) in germline cells, although endo-siRNAs and/or miRNAs are coexpressed with piRNAs in these cells (Kim et al. 2009). Thus, loading of different kinds of small RNAs onto individual Argonaute proteins is considered to be a “molecule-specific” event.
piRNAs have been studied extensively with regard to their biogenesis, characteristics, and functions, especially in Drosophila, fish, nematodes, and mice (in Caenorhabditis elegans, piRNAs are known as 21U RNAs) (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Ruby et al. 2006; Saito et al. 2006; Vagin et al. 2006; Watanabe et al. 2006; Brennecke et al. 2007; Gunawardane et al. 2007; Houwing et al. 2007; Batista et al. 2008; Das et al. 2008). piRNAs are longer than miRNAs and endo-siRNAs by several bases; for example, in Drosophila, piRNAs range between 24 and 30 nt, while miRNAs and endo-siRNAs are ∼20–23 nt long. In addition, piRNAs contain 2′-O-methyl groups at their 3′ ends, unlike miRNAs (except in plants) (Horwich et al. 2007; Houwing et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007; Saito et al. 2007). piRNAs are derived mostly from repetitive intergenic DNA elements, including transposons, and these loci are collectively called “piRNA clusters” (Aravin et al. 2007a). Protein-coding genes such as traffic jam (tj) could also account for piRNA production (Robine et al. 2009; Saito et al. 2009). Because of these features, and considering their limited expression in germlines and their specific associations with PIWI proteins, piRNAs are considered to be a unique set of endogenous small RNAs.
Loss of PIWI proteins in Drosophila and mice causes derepression of transposons and results in severe defects in gametogenesis (Vagin et al. 2006; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008; Li et al. 2009). As a result, the homozygous mutant lines cannot be maintained. Deletion of a Drosophila piRNA cluster, flamenco (flam), located on the X chromosome, also causes derepression of particular transposons, such as gypsy, ZAM, and Idefix (Prud'homme et al. 1995; Desset et al. 2003; Mével-Ninio et al. 2007), because flam gives rise to piRNAs that show strong complementarities to transcripts from these transposons (Brennecke et al. 2007). These studies have made it very clear that both PIWI proteins and piRNAs are required for transposon silencing. _targets of PIWI–piRNA complexes are not limited to transposons. In fact, a subset of piRNAs in Drosophila has been shown to function in silencing protein-coding genes. The best examples are piRNAs derived from suppressor of stellate [su(ste)] and tj, which down-regulate protein-coding stellate (ste) and fasciclin III (fas III) genes, respectively (Livak 1984; Aravin et al. 2001, 2004; Vagin et al. 2006; Nishida et al. 2007; Saito et al. 2009).
In Drosophila, AGO3 (Argonaute3), Piwi , and Aub (Aubergine) belong to the PIWI protein family, while in mice, MILI, MIWI, and MIWI2 belong to the mouse PIWI family (Siomi and Siomi 2009). Bioinformatic analyses of piRNAs that associate with PIWI proteins in both Drosophila and mice germlines have led to two models for piRNA biogenesis: the amplification loop pathway (also termed the Ping-Pong pathway) and the primary processing pathway (Aravin et al. 2007a; Siomi and Siomi 2009). Observations in Drosophila support the concepts that, in the amplification loop pathway, Aub (mainly associated with antisense piRNAs, which show a preference for a uracil [U] at the 5′ end) and AGO3 (mainly associated with sense piRNAs, which show a preference for an adenine [A] at position 10) reciprocally cleave _target RNAs in sense and antisense orientations, respectively, and that this reciprocal _target RNA cleavage by Aub and AGO3 constantly gives rise to abundant piRNAs in germline cells (Brennecke et al. 2007; Gunawardane et al. 2007). The vast majority of their RNA _targets are transposon transcripts; thus, transposon silencing occurs in germlines in parallel with piRNA production through the amplification loop. In mice, of the three PIWI proteins, MILI and MIWI2 function in the amplification loop system (Aravin et al. 2007b).
Involvement of Aub and AGO3 in the primary processing pathway, however, is unlikely—at least in somatic cells of Drosophila ovaries. Evidence supporting this notion was obtained from studies on ovarian somatic cells (OSCs) and ovarian somatic sheets (OSSs). In Drosophila OSCs/OSSs, levels of Aub and AGO3 are below detection, but piRNAs are abundantly expressed and loaded onto Piwi (Lau et al. 2009; Saito et al. 2009). Deep sequencing analyses of small RNAs in piRNA-related mutants also provided strong evidence for an Aub–AGO3-independent piRNA pathway in somatic cells in ovaries (Li et al. 2009; Malone et al. 2009). The requirement for MILI and MIWI2 in the mouse somatic primary processing pathway remains undetermined. Factors necessary for primary piRNA processing may include Zucchini (Zuc), a putative cytoplasmic nuclease, because loss of Zuc function caused a severe reduction in the expression levels of primary piRNAs, such as flam-originating piRNAs (in ovaries) (Malone et al. 2009) and tj-originating piRNAs (in OSCs) (Saito et al. 2009). dicer mutant ovaries accumulate piRNAs, similarly to wild-type ovaries (Vagin et al. 2006). Dicer-independent piRNA production was also observed in zebrafish (Houwing et al. 2007). Thus, it seems that both the amplification loop and primary processing pathways for piRNA production do not require Dicer. Other genes—such as Armitage, Spindle E (Spn-E), Maelstrom, Krimper, Vasa, and Squash—might be involved in piRNA biogenesis (Vagin et al. 2006; Lim and Kai 2007; Pane et al. 2007). However, the molecular details of the requirement of these genes in piRNA biogenesis remain unclear.
A recent study has shown that PIWI proteins in Drosophila, mice, and Xenopus contain sDMAs (symmetrical dimethyl arginines), and that the factor mediating this post-translational modification is PRMT5 (Kirino et al. 2009). sDMA is one of various methyl group modifications found on specific arginines in protein molecules (Bedford and Clarke 2009). sDMA is known to modify the ability of a protein to perform its biological activities. For example, Sm proteins, factors needed for splicing machinery, contain sDMAs in their arginine–glycine-rich (RG) domains and associate, through their sDMAs, with a Tud domain-containing protein, SMN (Survival motor neuron) protein (Brahms et al. 2001; Friesen et al. 2001). This Sm–SMN association recruits U snRNA, leading to the efficient assembly of U snRNP (Meister et al. 2002). This whole system serves as a “gatekeeper” that prevents the misassembly of Sm proteins to non_target RNA and also prevents Sm protein aggregation (Pellizzoni et al. 2002; Chari et al. 2008). Other Tud family members interact with particular proteins through methylated arginines in the _target proteins and to regulate their functions (Côté and Richard 2005). Genetic studies in Drosophila and mice had indicated that Tud family members are linked to gametogenesis, even before the discovery of piRNAs and the characterization of PIWI proteins (Boswell and Mahowald 1985; Pan et al. 2005; Chuma et al. 2006). Based on these observations, it was inferred that the biological functions of PIWI proteins would be regulated by Tud proteins via specific associations with PIWI-sDMA modifications. These predictions gradually have been shown to be correct through recent studies conducted mainly in Drosophila and mice.
PIWI proteins are sDMA-modified by PRMT5
The first evidence to show that PIWI proteins contain sDMAs was provided by Kirino et al. (2009). Kirino et al. (2009) demonstrated that a protein pool obtained by immunoprecipitation from mouse testes using an sDMA-specific antibody, Y12, contained both MILI and MIWI. PIWI proteins in Drosophila ovaries and Xenopus oocytes (Xiwi and Xili are the Xenopus PIWI) were also shown to contain sDMAs by Western blot analyses using Y12 or SYM11, another sDMA-specific antbody (Kirino et al. 2009; Nishida et al. 2009). Y12 coimmunoprecipitated piRNAs with PIWI proteins (Kirino et al. 2009), indicating that sDMA modification does not abolish the PIWI–piRNA association.
The involvement of PRMT5 in the PIWI-sDMA modification was investigated for two major reasons: (1) Loss of PRMT5 caused complete loss of Sm-sDMA modification in Drosophila (Gonsalvez et al. 2006; Anne et al. 2007), and (2) prmt5 mutants phenocopy aub mutants (Harris and Macdonald 2001). No PIWI-sDMA modification was detectable in prmt5 mutants (Kirino et al. 2009). Thus, it was concluded that PRMT5 is the factor responsible for the sDMA modification.
The binding capacity of PIWI proteins for piRNAs seems unaltered by prmt5 mutations (Kirino et al. 2009). However, derepression of a transposon, Het-A, was observed in prmt5 mutant ovaries (Kirino et al. 2009). This result made it clear for the first time that prmt5 is tightly connected to transposon silencing. PRMT5-mediated sDMA modification appears to control the subcellular localization of PIWI proteins in ovaries because, in prmt5 mutants, Aub does not concentrate at the nuage (Nishida et al. 2009), an electron-dense material associated with nurse cell nuclei. This suggests that nuage localization of Aub is important for silencing. Whether Aub localization at the posterior pole of oocytes (Kirino et al. 2009), where materials required for primordial germ cell (PGC) specification are accumulated, is affected by loss of PRMT5 remains controversial (Kirino et al. 2009; Nishida et al. 2009).
Studies describing mouse prmt5 mutants have not yet been published. In transgenic mice expressing individual mouse PIWI proteins, the PRMT5 complex containing WDR77 (also known as MEP50), a cofactor of PRMT5, was able to associate with all three PIWI proteins (Vagin et al. 2009). MIWI2 and MILI association with the PRMT5–WDR77 complex, termed the methylosome, was observed in embryonic testes, while MIWI (and also MILI) association with the methylosome was observed in adult testes. This was simply due to the stage-specific expression of individual PIWI proteins during germline development. Whether other members of the PRMT family associate with mouse PIWI proteins remains undetermined.
The Drosophila homolog of WDR77/MEP50 is Valois (Vls) (Anne and Mechler 2005; Cavey et al. 2005). Genetic disruption of vls results in phenocopies of tud mutants; both mutants cause a grandchild-less phenotype (Boswell and Mahowald 1985; Schupbach and Wieschaus 1986). Vls, like Aub and Tud, is a component of the nuage and pole plasm (Bardsley et al. 1993; Harris and Macdonald 2001). Vls interacts with not only PRMT5, but also Tud, and localization of Tud at the nuage and at the posterior pole of oocytes depends on Vls expression (Anne and Mechler 2005). These observations clearly indicate a direct relationship of Vls with Aub and Tud in Drosophila.
sDMA modification status of PIWI proteins in mice and Drosophila
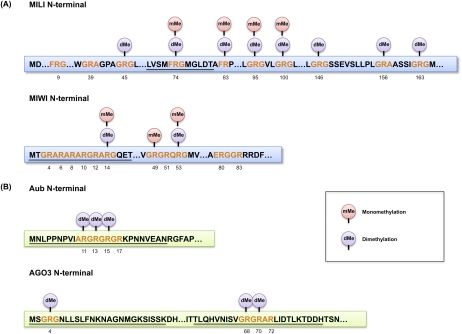
Putative “sDMA motifs” GRG and ARG/GRA (G, glycine; R, arginine; A, alanine), present in animal PIWI proteins, were suggested by Kirino et al. (2009). The precise positions of sDMAs in mouse and Drosophila PIWI proteins were determined later by mass spectrometry (MS). The first R residue identified to have a symmetric dimethyl group was R74 in MILI (Reuter et al. 2009). Interestingly, R74 resides in the sequence FRG (F, phenylalanine), which was not one of the predicted sDMA motifs. More comprehensive mapping of methylated arginines, including monomethylation, in MILI and MIWI was performed later (Chen et al. 2009; Vagin et al. 2009). Vagin et al. (2009) found that R100, R146, R163, and R549 in MILI are dimethylated, whereas R95 in MILI and R49 and R371 in MIWI are monomethylated. R74 in MILI and R14 in MIWI were detected to be both mono- and dimethylated. Chen et al. (2009) used endogenous MILI and MIWI isolated from adult testes and found that R53 in MIWI can be both mono- and dimethylated. R74, R83, R95, and R100 in MILI appear to be both mono- and dimethylated, whereas R45, R146, R156, and R163 are only dimethylated. All dimethylation was most likely symmetrical (sDMAs). There is some discrepancy between the two analyses that might reflect the methylation status from distinct sources. This may suggest that the PIWI methylation can be regulated dynamically through spermatogenesis. The current methylation status of mouse PIWI proteins, as determined by MS, is summarized in Figure 1A. The methylation status of MIWI2 remains undetermined.
Figure 1.
Arginine methylation status of PIWI proteins. Mouse PIWI (A; MILI and MIWI) and fly PIWI (B; Aub and AGO3) N-terminal sequences are shown with putative sDMA motifs (red). Identified methylation sites (dimethylation [dMe] or monomethylation [mMe]) are shown above the relevant arginine, with the residue numbers below. Underlined sequences indicate the synthetic peptides that have been used in studies for pull-down assays. The MILI peptide corresponds to L69–T80 and R74 is an sDMA (Reuter et al. 2009). The MIWI peptide corresponds to M1–T18, and R4, R6, R8, R10, R12, and R14 are sDMAs (Kirino et al. 2010). The Aub peptides correspond to M1–N20 (Kirino et al. 2010) and M1–N25 (Nishida et al. 2009), and R11, R13, and R15 are sDMAs. The AGO3 peptides correspond to M1–K25 and T58–H82, and R4 in the M1–K25 peptide and R68 and R70 in the T58–H82 peptide are sDMAs (Nishida et al. 2009).
In Drosophila, endogenous Aub and AGO3 were subjected to liquid chromatography (LC)-MS/MS analysis. R11, R13, and R15 in Aub and R4, R68, and R70 in AGO3 were determined to be sDMAs (Fig. 1B; Nishida et al. 2009). R17 in Aub and R72 in AGO3 were predicted to be dimethylated in silico; however, MS as performed by Nishida et al. (2009) did not reveal these modifications. Monomethylated Rs were not detected in either Aub or AGO3 (MC Siomi, unpubl.). The methylation status of Piwi in ovaries remains undetermined.
Association of members of the Tud family with PIWI proteins
The tud gene was first discovered in Drosophila as a gene required for the assembly of the germ plasm, a specialized cytoplasm containing electron-dense polar granules (Boswell and Mahowald 1985). The tud gene encodes an ∼280-kDa protein with 11 repeat motifs (Thomson and Lasko 2004, 2005; Arkov et al. 2006). These domains are termed Tud domains (Ponting 1997). Tud domains are commonly found in proteins other than Tud in a wide range of organisms (Ponting 1997; Talbot et al. 1998; Maurer-Stroh et al. 2003).
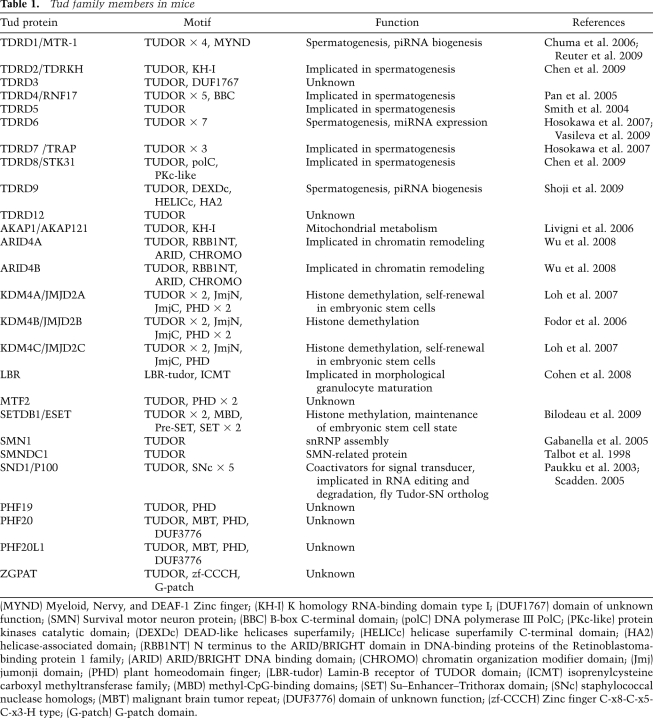
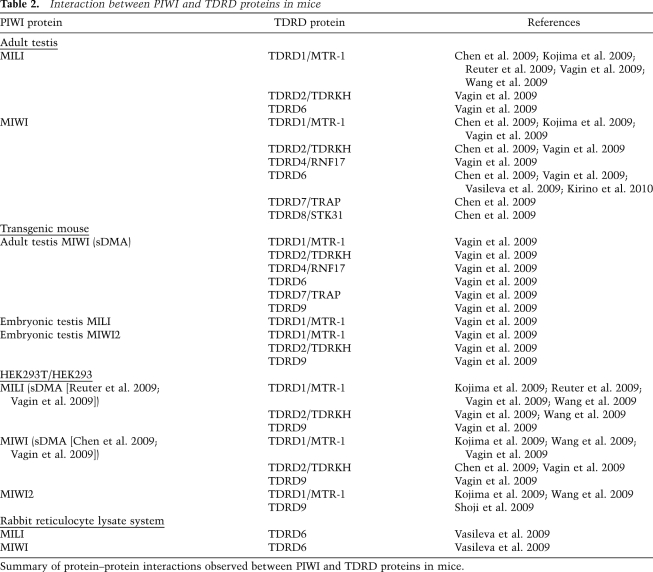
Based on data in the NCBI HomoloGene database, mice express 26 members of the Tud family (Table 1). Of these, 10 proteins are considered to be members of the Tud domain-containing (TDRD) family (Table 1). TDRD1, which contains four Tud domains along with a MYND (myeloid–nervy–DEAF-1) domain, is necessary for spermatogenesis (Chuma et al. 2006) and interacts with MILI (Table 2; Reuter et al. 2009; Vagin et al. 2009; Wang et al. 2009). MIWI also interacts with TDRD1 (Table 2; Chen et al. 2009; Kojima et al. 2009; Reuter et al. 2009; Vagin et al. 2009; Wang et al. 2009), although whether TDRD1 shows a tight association with MIWI2 remains controversial (Kojima et al. 2009; Reuter et al. 2009; Vagin et al. 2009; Wang et al. 2009). This type of discrepancy can be caused by different groups using different conditions to analyze protein–protein interactions.
Table 1.
Tud family members in mice
(MYND) Myeloid, Nervy, and DEAF-1 Zinc finger; (KH-I) K homology RNA-binding domain type I; (DUF1767) domain of unknown function; (SMN) Survival motor neuron protein; (BBC) B-box C-terminal domain; (polC) DNA polymerase III PolC; (PKc-like) protein kinases catalytic domain; (DEXDc) DEAD-like helicases superfamily; (HELICc) helicase superfamily C-terminal domain; (HA2) helicase-associated domain; (RBB1NT) N terminus to the ARID/BRIGHT domain in DNA-binding proteins of the Retinoblastoma-binding protein 1 family; (ARID) ARID/BRIGHT DNA binding domain; (CHROMO) chromatin organization modifier domain; (Jmj) jumonji domain; (PHD) plant homeodomain finger; (LBR-tudor) Lamin-B receptor of TUDOR domain; (ICMT) isoprenylcysteine carboxyl methyltransferase family; (MBD) methyl-CpG-binding domains; (SET) Su–Enhancer–Trithorax domain; (SNc) staphylococcal nuclease homologs; (MBT) malignant brain tumor repeat; (DUF3776) domain of unknown function; (zf-CCCH) Zinc finger C-x8-C-x5-C-x3-H type; (G-patch) G-patch domain.
Table 2.
Interaction between PIWI and TDRD proteins in mice
Summary of protein–protein interactions observed between PIWI and TDRD proteins in mice.
In vivo interaction assays with various MILI deletion mutants determined a domain necessary for TDRD1 interaction (Reuter et al. 2009). This domain was in the N-terminal region of MILI and contained dimethylated R74 (Reuter et al. 2009). Treatment with MTA [5′-deoxy-5′-(methylthio)-adenosine], a methyltransferase inhibitor, disrupted MILI-sDMA modification and abolished the association of MILI with TDRD1 (Reuter et al. 2009; Vagin et al. 2009). However, unlike full-length MILI, the N-terminal region was not detected by Y12 (Reuter et al. 2009); this implies that sDMA may not be absolutely necessary for the interaction between MILI and TDRD1. Loss of the MYND domain in TDRD1 does not affect the MILI–TDRD1 interaction (Reuter et al. 2009), indicating that the association occurs through Tud domains.
Tud domain-containing proteins that can associate with PIWI proteins in mouse testes are not limited to TDRD1. Seven Tud domain-containing proteins—TDRD1 to TDRD9, except TDRD3 and TDRD5—were found to be associated with PIWI proteins by various means, both in vivo and in vitro (Table 2; Chen et al. 2009; Kojima et al. 2009; Shoji et al. 2009; Vagin et al. 2009; Vasileva et al. 2009; Wang et al. 2009; Kirino et al. 2010). sDMA dependency of some TDRD proteins to interact with PIWI proteins has been determined. For example, TDRD6—considered the mouse homolog to Drosophila Tud, although TDRD6 contains seven Tud domains (Hosokawa et al. 2007), whereas Tud contains 11 Tud domains (Thomson and Lasko 2004, 2005; Arkov et al. 2006)—was able to associate with MILI and MIWI (Chen et al. 2009; Vagin et al. 2009; Vasileva et al. 2009; Kirino et al. 2010), and the interaction most likely occurred in an sDMA-dependent manner (Kirino et al. 2010). TDRD9 interacts specifically with MIWI2 (Table 2; Shoji et al. 2009; Vagin et al. 2009). TDRD1 may also associate with MIWI2 (Vagin et al. 2009; Wang et al. 2009). Whether the TDRD1–MIWI2 and TDRD9–MIWI2 interactions require sDMAs remains unknown.
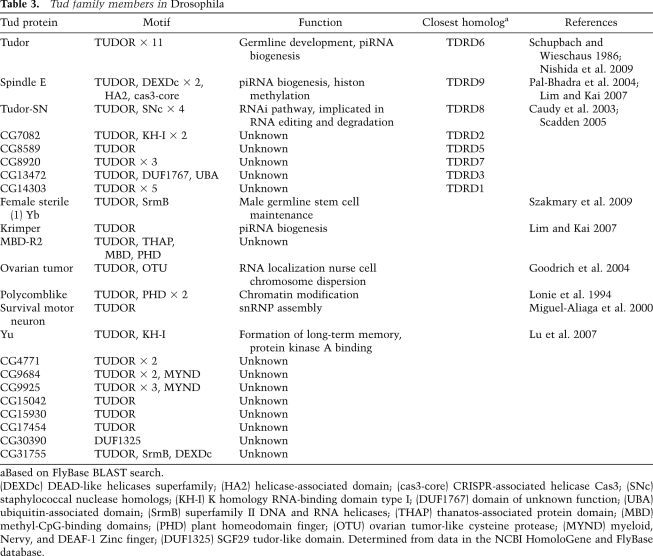
sDMA-mediated binding of PIWI proteins with Tud family members is evolutionarily conserved. Pull-down assays from Drosophila ovary lysates using synthetic Aub peptides, with and without sDMAs, show that a protein of ∼280 kDa associates only with the Aub-sDMA peptide (Kirino et al. 2009; Nishida et al. 2009). This protein was identified as Tudor by MS, and the association was confirmed by Western blot analysis using anti-Tud antibodies. Tud members in Drosophila are listed in Table 3. Of these, CG14303 is the closest homolog of mouse TDRD1; however, CG14303 does not show an sDMA-dependent association with Aub peptides (Nishida et al. 2009). Also, Spn-E, the Drosophila homolog of mouse TDRD9 (Shoji et al. 2009), shows no detectable association with the peptides (Nishida et al. 2009). Thus, association of Aub with Tud is rather specific. Similar assays were performed for AGO3 using peptides that correspond to two different sDMA-modified regions of the protein (Nishida et al. 2009). The sDMA peptide corresponding to the N-terminal end of AGO3 (Fig. 1B) showed no association with Tud, but the second sDMA peptide, spanning amino acids 58–82 of AGO3, interacted strongly with Tud. These data indicate clearly that sDMA modifications alone are not sufficient to promote the PIWI–Tud association, and that the association occurs in a peptide sequence-specific manner. None of the AGO3 peptides associate with Spn-E (Nishida et al. 2009). AGO3 is considered the Drosophila homolog of MIWI2, in the sense that both proteins interact specifically with secondary piRNAs produced by the amplification loop, which is operated by Aub and AGO3 in Drosophila and by MILI and MIWI2 in mice (Aravin et al. 2008). Here, a species difference is obvious with regard to the PIWI–Tud association.
Table 3.
Tud family members in Drosophila
aBased on FlyBase BLAST search.
(DEXDc) DEAD-like helicases superfamily; (HA2) helicase-associated domain; (cas3-core) CRISPR-associated helicase Cas3; (SNc) staphylococcal nuclease homologs; (KH-I) K homology RNA-binding domain type I; (DUF1767) domain of unknown function; (UBA) ubiquitin-associated domain; (SrmB) superfamily II DNA and RNA helicases; (THAP) thanatos-associated protein domain; (MBD) methyl-CpG-binding domains; (PHD) plant homeodomain finger; (OTU) ovarian tumor-like cysteine protease; (MYND) myeloid, Nervy, and DEAF-1 Zinc finger; (DUF1325) SGF29 tudor-like domain. Determined from data in the NCBI HomoloGene and FlyBase database.
Significance of Tud–PIWI associations for PIWI–piRNA interactions
Many Tud family members interact with PIWI proteins. Thus, it can be speculated that this might be the most prominent pathway for Tud biology. Many groups have looked into how Tud members affect the association between PIWI and piRNAs. Immunoprecipitation from adult mouse testes using an anti-MILI antibody copurified MILI with piRNAs (Kirino et al. 2009; Reuter et al. 2009). In addition, immunoprecipitation using Y12 copurified MILI with piRNAs (Kirino et al. 2009; Reuter et al. 2009). Y12 (antibody) and TDRD1 (protein) recognize the same sDMA region at the N terminus of MILI; this implies that a fraction of MILI that is not associated with TDRD1 is recognized by Y12. This suggests that, even without association with TDRD1, Mili is able to maintain binding with piRNAs. MILI has additional sDMAs (Fig. 1; Chen et al. 2009; Vagin et al. 2009); however, the MILI mutant (termed m1), in which R9, R39, R45, and R74 were mutated to lysines (Ks), showed no association with TDRD1 or TDRD9 (Reuter et al. 2009). Thus, TDRD1 is not likely to be required for MILI to maintain the association with piRNAs once the small RNAs have been loaded onto MILI. Whether Y12 copurifies TDRD proteins with MILI remains undetermined.
Loss of TDRD1 function does not change the ability of MILI to associate with piRNAs in embryonic testes (Reuter et al. 2009), although the total amounts of piRNAs are severely decreased in Tdrd1 mutants (Vagin et al. 2009). Deep sequencing analysis revealed that loss of TDRD1 function leads to overrepresentation of small RNAs derived from protein-coding transcripts (from both exons and introns) and of 5S ribosomal RNAs (rRNAs) associated with MILI (Reuter et al. 2009). Representation of transposon-derived small RNAs is not altered by tdrd1 mutations (Reuter et al. 2009). The molecular mechanisms that create this discrepancy observed between wild-type and tdrd1 mutant testes remain unclear. The overall piRNA profiles in spermatocytes, in contrast, do not seem to be changed by loss of TDRD6 function (Vagin et al. 2009). TDRD6 may also be required for regulation of miRNAs (Vagin et al. 2009; Vasileva et al. 2009); however, the effect may be indirect, because changes in developmental timing in tdrd6 mutant mice may result in altered miRNA profiles, as has been reported by Vagin et al. (2009).
Loss of Tud function in Drosophila ovaries causes Aub to be associated with a greater abundance of piRNAs compared with Aub in wild-type ovaries (Nishida et al. 2009). Deep sequencing for piRNAs associated with Aub in both wild-type and tud ovaries revealed that overrepresentation of any kind of Aub-associated small RNAs did not occur in tud; however, the population of transposon-derived piRNAs was altered significantly by loss of Tud function. An obvious change in the strand bias of transposon-derived piRNAs was not seen in the mutants. How this alteration is caused by Tud loss remains unclear; however, these results suggest that Tud is required for the quality control of transposon-derived piRNAs. Obvious derepression of transposons in tud ovaries has not been reported. This might be because, although the population was altered, Aub is still associated with transposon-derived piRNAs, even in the absence of Tud function, and thus the Aub–piRNA association state is not “perfect” but is somewhat “manageable” to silence transposons, at least in ovaries. However, offspring of tud mutants are devoid of germ cells. One explanation for this might be that the phenomenon is simply independent of the transposon derepression issue.
Depletion of tdrd9 in mice testes causes a massive increase of LINE1-derived piRNAs, while other piRNAs, such as the ones derived from IAP and Sine B1 transposons, show a decrease in Tdrd9 mutants (Shoji et al. 2009); hence, the population of transposon-derived piRNAs is changed by TDRD9. In this sense, Tud in Drosophila and TDRD9 in mice are functionally similar to each other, although their similarity at the peptide sequence level is not particularly high.
Further significant aspects of Tud association with PIWI proteins
PIWI proteins in Drosophila germlines were originally reported to be destabilized by loss of sDMA modification (Kirino et al. 2009). However, this interpretation was revised later (Nishida et al. 2009; Kirino et al. 2010). There now seems to be a consensus that the stability of PIWI proteins is not changed by the loss of sDMA modification. In tud mutants as well as in prmt5 mutants, the total amounts of Aub and AGO3 were not reduced, indicating that Tud association with the PIWI proteins, regardless of the presence of sDMAs, is not required for their stabilization in vivo. However, the localization of PIWI proteins in ovaries was greatly affected by Tud depletion. In tud mutants, Aub is not detected at the nuage, although the protein is detected at the posterior pole of oocytes (Nishida et al. 2009). Kirino et al. (2010) reported that Aub localization to the posterior pole is markedly reduced in tud mutants. Thus, it remains controversial whether the posterior pole localization of Aub is dependent on its association with Tud.
The protein levels of MILI and its cytoplasmic localization in mouse testes were barely affected by the loss of TDRD1 expression. However, MIWI2 localization was largely affected. In wild-type testes, MIWI2 is found predominantly in the nucleus and at the nuage. Without TDRD1, MIWI2 becomes mislocalized, being evenly distributed in the cytoplasm (Reuter et al. 2009; Shoji et al. 2009; Vagin et al. 2009). The expression level of MIWI2 was not changed by the loss of TDRD1. Similar mislocalization of MIWI2 was observed previously in MILI mutants, where piRNAs were found not to be loaded onto MIWI2 (Aravin et al. 2008). This was because piRNAs associated with MIWI2 require MILI–Slicer for their production. This suggested that, without piRNA loading, MIWI2 is not imported into the nucleus. A possible scenario might then be that, in Tdrd1 mutants, the amplification loop for producing MIWI2-associated piRNAs would be somehow unavailable or not functional, as in mili mutants, and, consequently, this defect freed MIWI2 from piRNA association, and this absence of piRNAs caused MIWI2 to be localized in the cytoplasm. Indeed, it was found that the levels of piRNAs in the antisense orientation are lowered selectively by TDRD1 loss (Vagin et al. 2009). Antisense piRNAs are thought to be loaded predominantly onto MIWI2 through the amplification loop pathway in mice. Therefore, the decreased amount of piRNAs associated with MIWI2 might indeed be the main cause for MIWI2 mislocalization. Loss of TDRD9 does not cause the mislocalization of MILI, MIWI2, or TDRD1 (Shoji et al. 2009); this is likely to be because piRNAs are loaded onto MIWI2, even in this mutant. It seems that the requirements of TDRD1 and TDRD9 in the piRNA production system differ from each other. In tdrd6 mutants, MIWI was found to be dispersed in the cytoplasm (Vasileva et al. 2009), although the cause of this observation remains unclear.
Earlier studies have revealed that the loss of MILI and MIWI2 causes the expression levels of both LINE1 and IAP transposons to be much higher compared with those in wild-type testes (Aravin et al. 2007a; Kuramochi-Miyagawa et al. 2008). Loss of MILI and MIWI2 impairs DNA methylation of transposons (Aravin et al. 2007b, 2008; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008). Interestingly, loss of both TDRD1 and TDRD9 derepresses LINE1 (Reuter et al. 2009; Shoji et al. 2009; Vagin et al. 2009); however, silencing of IAP remains as in wild type. The DNA methylation state of LINE1, but not of IAP, was also found to be decreased in tdrd1 and tdrd9 mutants (Reuter et al. 2009; Shoji et al. 2009). It is thus obvious that both TDRD1 and TDRD9 have important roles, at least in LINE1 silencing in mouse testes.
The current body of evidence shows clearly that sDMA modifications are made on PIWI proteins by PRMT5. Also, PIWI proteins associate, via their sDMA modifications, with Tud family members. These mechanisms are highly evolutionarily conserved and are indispensable for gametogenesis. In fact, genes encoding protein factors required for this sequential molecular flow are classified mostly as grandchild-less genes. This simply indicates that the system may be germline-specific. Tud family members have impacts on piRNA production and on piRNA association with PIWI proteins; however, no impact on ubiquitously expressed miRNAs or endo-siRNAs has so far been reported (except for the possible involvement of TDRD6 in miRNA regulation). Indeed, none of the ubiquitous AGO proteins that function in RNA silencing by associating with miRNAs and endo-siRNAs have so far been shown to be sDMA-modified, in contrast to PIWI proteins. Thus, regulation by PRMT5-directed sDMA modification and sDMA-specific association with Tud might be a specialized event for the PIWI–piRNA pathway. Why such an elaborative system exists specifically for the piRNA pathway remains unknown. It can be speculated that germlines need to selectively use PIWI proteins containing sDMA and PIWI without sDMAs at certain stages during gametogenesis. To clarify the possibility, it would be interesting to study a dynamic event for PIWI-sDMA modification in germlines.
How and with what factors are piRNA production and piRNA association with PIWI proteins regulated? Until recently, this question was unanswered; however, we now know that particular members of the Royal Family of Tudor—TDRD proteins in mice and Tudor in Drosophila—have roles in piRNA pathways. It is tempting to postulate that Tud members act as the sovereigns of the whole piRNA process. Further investigation should help clarify if this interesting scenario reflects reality, and resolve other related questions regarding the Royal Family of Tudor and the piRNA pathways occurring in germlines.
Acknowledgments
We thank Satomi Kuramochi-Miyagawa and Yoichiro Chuma for useful discussions. H.S. is supported by MEXT (Ministry of Education, Culture, Sports, Science, and Technology). M.C.S. is supported by NEDO (New Energy and Industrial Technology Development Organization) and CREST (Core Research for Evolutional Science and Technology) from JST (Japanese Science and Technology Agency).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1899210.
References
- Anne J, Mechler BM. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuléen. Development. 2005;132:2167–2177. doi: 10.1242/dev.01809. [DOI] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuléen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D-melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Wang JYS, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- Bardsley A, McDonald K, Boswell RE. Distribution of tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development. 1993;119:207–219. doi: 10.1242/dev.119.1.207. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: _target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu WF, Vasale JJ, Duan SH, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes & Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- Brahms H, Meheus L, De Brabandere V, Fischer U, Lührmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Cavey M, Hijal S, Zhang XL, Suter B. Drosophila valois encodes a divergent WD protein that is required for Vasa localization and Oskar protein accumulation. Development. 2005;132:459–468. doi: 10.1242/dev.01590. [DOI] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhäger M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal snRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo YH, Tenaglia E, Xu C, Gish G, Min JR, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TV, Klarmann KD, Sakchaisri K, Cooper JP, Kuhns D, Anver M, Johnson PF, Williams SC, Keller JR, Stewart CL. The lamin B receptor under transcriptional control of C/EBPɛ is required for morphological but not functional maturation of neutrophils. Hum Mol Genet. 2008;17:2921–2933. doi: 10.1093/hmg/ddn191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. doi: 10.1093/genetics/164.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes & Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein _targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum Mol Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian LP, Matera AG. The Sm-protein methyltransferase, Dart5, is essential for germ-cell specification and maintenance. Curr Biol. 2006;16:1077–1089. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Goodrich JS, Clouse KN, Schüpbach T. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–1958. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin HF. A novel class of small RNAs in mouse spermatogenic cells. Genes & Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li CJ, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: Domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: Key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K, Kuramochi-Miyagawa S, Chuma S, Tanaka T, Nakatsuji N, Kimura T, Nakano T. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells. 2009;14:1155–1165. doi: 10.1111/j.1365-2443.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Vagin VV, Lee SH, Xu J, Ma SM, Xi HL, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JD, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livigni A, Scorziello A, Agnese S, Adornetto A, Carlucci A, Garbi C, Castaldo I, Annunziato L, Avvedimento EV, Feliciello A. Mitochondrial AKAP121 links cAMP and src signaling to oxidative metabolism. Mol Biol Cell. 2006;17:263–271. doi: 10.1091/mbc.E05-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes & Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonie A, D'Andrea R, Paro R, Saint R. Molecular characterisation of the Polycomblike gene of Drosophila melanogaster, a trans-acting negative regulator of homeotic gene expression. Development. 1994;120:2629–2636. doi: 10.1242/dev.120.9.2629. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lu YS, Shuai Y, Feng C, Tully T, Xie Z, Zhong Y, Zhou HM. The AKAP Yu is required for olfactory long-term memory formation in Drosophila. Proc Natl Acad Sci. 2007;104:13792–13797. doi: 10.1073/pnas.0700439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: A complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- Mével-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Chan YB, Davies KE, van den Heuvel M. Disruption of SMN function by ectopic expression of the human SMN gene in Drosophila. FEBS Lett. 2000;486:99–102. doi: 10.1016/s0014-5793(00)02243-2. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine–piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Nagami-Okada T, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Pan JY, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development. 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol Endocrinol. 2003;17:1805–1814. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A. flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol. 2009;19:1–11. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C-elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of PIWI-interacting RNAs at their 3′ ends. Genes & Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113:443–448. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9–MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Smith JM, Bowles J, Wilson M, Teasdale RD, Koopman P. Expression of the tudor-related gene Tdrd5 during development of the male germline in mice. Gene Expr Patterns. 2004;4:701–705. doi: 10.1016/j.modgep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Szakmary A, Reedy M, Qi H, Lin H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J Cell Biol. 2009;185:613–627. doi: 10.1083/jcb.200903034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Miguel-Aliaga I, Mohaghegh P, Ponting CP, Davies KE. Characterization of a gene encoding Survival motor neuron (SMN)-related protein, a constituent of the spliceosome complex. Hum Mol Genet. 1998;7:2149–2156. doi: 10.1093/hmg/7.13.2149. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Tudor and its domains: Germ cell formation from a Tudor perspective. Cell Res. 2005;15:281–291. doi: 10.1038/sj.cr.7290297. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li CJ, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang XH, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes & Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang JQ, Saxe JP, Tanaka T, Chuma S, Lin HF. Mili interacts with Tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes & Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Eldin KW, Beaudet AL. Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst. 2008;100:1247–1259. doi: 10.1093/jnci/djn253. [DOI] [PMC free article] [PubMed] [Google Scholar]