Summary
Deubiquitinating enzymes (DUBs) regulate diverse cellular functions by cleaving ubiquitin from specific protein substrates. How their activities are modulated in various cellular contexts remains poorly understood. The yeast deubiquitinase Ubp8 protein is recruited and activated by the SAGA complex and, together with Sgf11, Sus1, and Sgf73, forms a DUB module responsible for deubiquitinating histone H2B during gene expression. Here we report the crystal structure of the complete SAGA DUB module, which features two functional lobes structurally coupled by Sgf73. In the “assembly lobe”, a long Sgf11 N-terminal helix is clamped onto the Ubp8 ZnF-UBP domain by Sus1. In the “catalytic lobe”, an Sgf11 C-terminal zinc finger domain binds to the Ubp8 catalytic domain next to its active site. Our structural and functional analyses reveal a central role of Sgf11 and Sgf73 in activating Ubp8 for deubiquitinating histone H2B and demonstrate how a DUB can be allosterically regulated by its non-substrate partners.
Introduction
Protein ubiquitination controls a broad range of cellular functions including gene transcription, signal transduction, cell cycle progression, protein trafficking, DNA repair, and DNA replication (Hershko and Ciechanover, 1998). Ubiquitin E3 ligases mediate the ubiquitin conjugation reaction at the end of a three-enzyme (E1-E2-E3) cascade, whereas deubiquitinating enzymes (DUBs) cleave ubiquitin from the substrate polypeptides or polyubiquitin chains. To achieve temporal control, the activities of both ubiquitin ligases and DUBs must be tightly regulated. This is attained at the level of substrate recognition, catalytic competency, sub-cellular localization, and enzyme abundance. In contrast to the much studied ubiquitin ligases, how DUBs are regulated remains poorly understood.
The yeast and human genomes encode close to 20 and 100 DUBs, respectively (Amerik et al., 2000; Komander et al., 2009; Nijman et al., 2005; Reyes-Turcu et al., 2009). A recent survey of 75 human DUBs has revealed that most DUBs form protein complexes with multiple interacting partners in the cell (Sowa et al., 2009). The ubiquitin-specific processing proteases (UBPs in yeast and USPs in humans) represent the largest sub-family of DUBs. With a conserved catalytic domain, UBP/USPs are characterized by highly variable regions flanking or inserted in the catalytic core (Komander et al., 2009). Sequence diversities in these regions are believed to dictate UBP/USPs binding to substrates, recruitment of regulatory factors, and integration into unique cellular machinery. Recent structural studies focusing on individual domains of several USPs have revealed a common fold adopted by their homologous catalytic domains, many of which show a considerable degree of structural plasticity (Avvakumov et al., 2006; Hu et al., 2006; Hu et al., 2002; Hu et al., 2005; Komander et al., 2008; Renatus et al., 2006). Although the conformational changes reported so far are associated with the binding of the ubiquitin moiety of the substrate, they raise immediate questions as to whether the catalytic domains of UBP/USPs can be allosterically regulated by non-substrate partners, and how full-length multi-domain UBPs/USPs, together with their binding proteins, function in a physiologically relevant form.
The yeast Ubp8 protein is an evolutionarily conserved DUB with a functional role in gene expression. Transcription of eukaryotic genes requires dynamic regulation of chromatin structure through coordinated posttranslational modifications of histones. The yeast Spt-Ada-Gcn5-Acetyl transferase (SAGA) complex is a chromatin-modifying transcription coactivator, which is best characterized for its histone acetyltransferase activity (HAT) encoded by the central subunit Gcn5 (Daniel and Grant, 2007; Lee and Workman, 2007). Recent studies have identified Ubp8 as yet a second enzymatic subunit of SAGA, responsible for catalyzing histone H2B deubiquitination (Daniel et al., 2004; Henry et al., 2003). H2B Lys123 undergoes dynamic mono-ubiquitination, which is sequentially mediated by the Rad6-Bre1 E2-E3 pair and the SAGA-bound Ubp8 (Hwang et al., 2003; Robzyk et al., 2000; Wood et al., 2003). H2B mono-ubiquitination serves as a transient signal that orchestrates a diverse set of events during transcription (Fleming et al., 2008; Shilatifard, 2006; Weake and Workman, 2008). Further investigations have uncovered three novel proteins, Sgf11, Sus1, and Sgf73, which are required for tethering Ubp8 to SAGA (Ingvarsdottir et al., 2005; Köhler et al., 2006; Köhler et al., 2008; Lee et al., 2009; Powell et al., 2004; Rodriguez-Navarro et al., 2004). Importantly, Ubp8 alone does not possess H2B deubiquitinating activity unless it is integrated into SAGA (Lee et al., 2005). Purified recombinant Ubp8 can only hydrolyze Ub-AMC, a model ubiquitin conjugate, when Sgf11, Sus1, and Sgf73 are all present (Köhler et al., 2008). Remarkably, USP22, the human ortholog of Ubp8, forms a similar SAGA DUB module and deubiquitinates both H2A and H2B (Zhang et al., 2008; Zhao et al., 2008). Abnormalities the human SAGA DUB module are potentially linked to cancer and the neurological disorder spinocerebellar ataxia type 7 (Glinsky, 2006; Helmlinger et al., 2006; Palhan et al., 2005; Zhang et al., 2008). Here we report the crystal structure of the entire yeast SAGA DUB module complex, which reveals the mechanisms by which the DUB module assembles and activates Ubp8 for H2B deubiquitination.
Results
Overall structure of the SAGA DUB module
To assemble a recombinant SAGA DUB module, we co-expressed, purified, and crystallized the full-length Ubp8, Sgf11, Sus1 and the N-terminal fragment of Sgf73 (amino acid 1–104) as a quaternary complex from E. coli. The complex structure was determined at 2.70 Å resolution by a combination of zinc single-wavelength anomalous dispersion (SAD) and molecular replacement (MR) methods (Table S1).
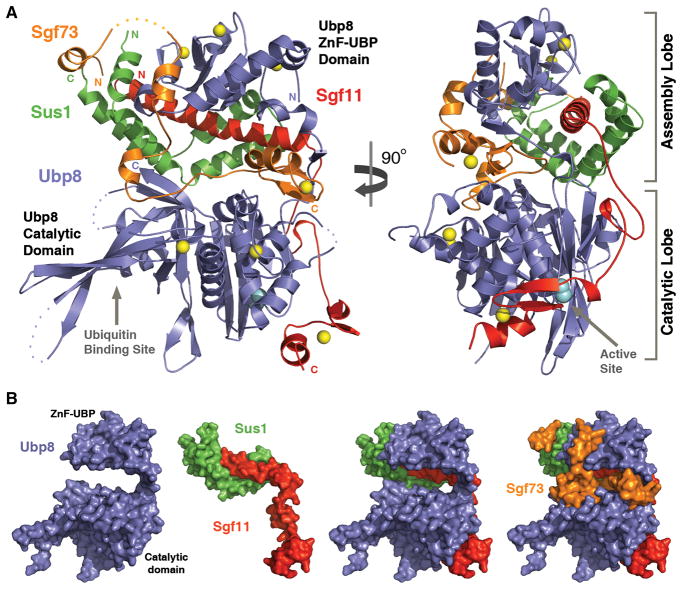
The Ubp8-Sgf11-Sus1-Sgf73 complex adopts an overall compact and highly organized structure, which involves extensive protein interaction interfaces (Figure 1A and 1B). Each subunit of the hetero-tetrameric complex makes physical contacts with all three other subunits. Overall, the DUB module has two distinct functional lobes, which are organized by two separate domains of the core subunit Ubp8. The N-terminal ZnF-UBP domain of Ubp8 nucleates the other three subunits, forming the “assembly lobe” of the module, whereas the C-terminal catalytic domain of Ubp8 together with Sgf11 C-terminal ZnF domain constitutes the “catalytic lobe” (Figure 1A). The two functional lobes of the module are joined together by the Sgf73 N-terminal fragment, which inserts itself in between the two Ubp8 domains.
Figure 1. Overall view of the yeast SAGA DUB module.
(A) Two orthogonal views of the complex. Ubp8 (blue), Sgf11(red), Sus1 (green), and Sgf73 (orange) are shown in ribbon representation with disordered regions in dashed lines. The zinc ions are shown as yellow spheres. The active site cysteine is shown in cyan CPK model.
(B) Individual or different combination of the DUB module subunits are shown in surface representation to demonstrate their spatial relationships. The orientation of all subunits is the same as shown in (A, left).
See Table S1 for crystallographic information.
The “assembly lobe” of the DUB module mediates tetrameric complex formation and is characterized by tightly inter-locked subunits. At the center of this lobe is a long Sgf11 N-terminal helix, which is completely enclosed by Sus1 and the Ubp8 ZnF-UBP domain. The “catalytic lobe” of the DUB module features a catalytically competent form of the Ubp8 catalytic domain, which orients its active center and ubiquitin-binding site away from the “assembly lobe” (Figure 1A). The striking highlight of the “catalytic lobe” is an Sgf11 C-terminal sequence, which contains a small zinc finger domain and a linker sequence connected to the long N-terminal helix. Anchored to the DUB module via the “assembly lobe”, Sgf11 uses this C-terminal sequence to interact with the Ubp8 catalytic domain in the close vicinity of the catalytic triad of the deubiquitinase.
The Ubp8 ZnF-UBP domain as a scaffold for complex assembly
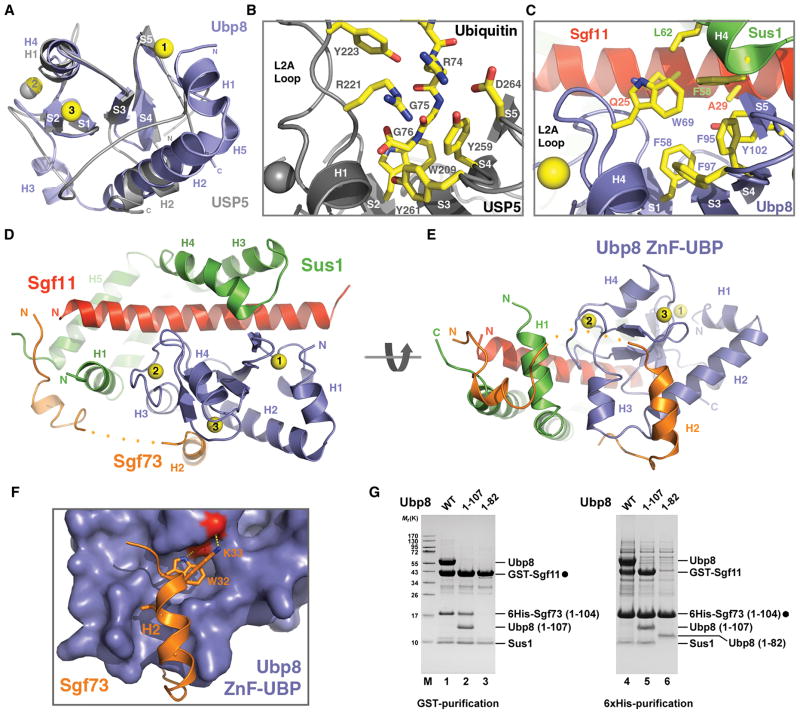
The yeast Ubp8 protein belongs to a sub-family of UBP/USPs that share a common ZnF-UBP domain N-terminal to the catalytic domain (Bonnet et al., 2008). The Ubp8 ZnF-UBP domain adopts the typical UBP-type zinc finger fold as reported for three mammalian USP proteins, USP5, USP16, and USP33 (Allen and Bycroft, 2007; Pai et al., 2007; Reyes-Turcu et al., 2006). Superposition analysis of the ZnF-UBP domains of Ubp8 and USP5 shows that their central sequence regions have the same core structure, which consists of a five-stranded β-sheet with a nearby α-helix (S1–S5 and H4 in Ubp8) (Figure 2A). Their flanking sequences, however, form distinct structural elements (Figure 2A, see also Figure S1).
Figure 2. The Ubp8 ZnF-UBP domain mediates complex assembly of the DUB module.
(A) Superposition analysis of the ZnF-UBP domains in Ubp8 (blue) and USP5 (grey). The three zinc ions in the Ubp8 ZnF-UBP domain are numbered (yellow spheres). Secondary structure elements of both proteins are labeled (H for α-helix and S for βstrand).
(B) The ubiquitin tail-binding pocket of the USP5 ZnF-UBP domain. The side chains of key ubiquitin-contacting residues in USP5 and the last three ubiquitin residues are shown in sticks.
(C) The corresponding region of Ubp8 as shown for USP5 in (B). The side chains of key residues from Ubp8, Sus1, and Sgf11 for forming a three-protein junction are shown in sticks.
(D) and (E) Two views of the entire “assembly lobe” of the DUB module.
(F) The interface between the Ubp8 ZnF-UBP domain and the second helix of Sgf73 featuring the conserved WK motif. The side chains of the Sgf73 WK motif are shown in sticks. The Ubp8 ZnF-UBP domain is shown in surface representation. The two backbone carbonyl groups of Ubp8, which accept hydrogen bonds from the Sgf73 WK motif are colored in red.
(G) The indicated DUB module components were polycistronically expressed in E. coli. Protein lysates were split and subjected in parallel to either GST- or 6His-affinity purifications. Lanes 1 and 4, 2 and 5, 3 and 6 correspond to each other. Purified complexes were eluted with either GSH or imidazole and analyzed by SDS-PAGE and Coomassie staining. Filled circles indicate bait proteins. Labeled proteins were identified by mass spectrometry.
See also Figure S1.
The Ubp8 ZnF-UBP domain does not have the ubiquitin tail-binding pocket found in USP5 and USP16. The USP5 ZnF-UBP domain binds the extended C-terminal tail of ubiquitin through a deep pocket formed between the central β-sheet and the long L2A loop (Figure 2B). Most ubiquitin-interacting residues inside the pocket, including Arg221 of the L2A loop, are conserved in USP5 and USP16. In contrast, the ZnF-UBP domain of Ubp8 features a much shorter L2A loop and lacks the conserved residues for recognizing ubiquitin. Although a hydrophobic pocket is formed between the L2A loop and the twisted central β-sheet, it is mostly occupied by the bulky side chain of the Trp69 residue, which replaces the ubiquitin-interacting arginine found in USP5 and USP16 (Figure 2C). Instead of binding ubiquitin, this site of the Ubp8 ZnF-UBP domain exposes a hydrophobic surface area that participates in a three-protein junction with residues from Sus1 and Sgf11.
The Ubp8 ZnF-UBP domain organizes the “assembly lobe” of the DUB module by packing against the central two-thirds of the 40 amino acid long Sgf11 N-terminal helix, contacting two separate regions of Sus1 which wraps around the Sgf11 helix, and interacting with an Sgf73 helix and its following loop (Figure 2D). Joined together as one structural unit, Sus1 and the Sgf11 helix surround half of the Ubp8 ZnF-UBP domain, opposite to the Sgf73 binding site. The interactions between the Sgf11 helix and the Ubp8 ZnF-UBP domain mainly involve polar residues and polypeptide backbone groups from both sides. The middle region of the interface, however, is nucleated by hydrophobic interactions predominantly through two aromatic residues of Ubp8, Trp69 and Tyr102 (Figure 2C). At this site, two hydrophobic amino acids, Phe58 and Leu62, on the Sus1 H4 helix also participate in the inter-molecular interactions, which all together give rise to a tight three-protein junction. The association among Sus1, Sgf11, and the Ubp8 ZnF-UBP domain is further reinforced by a second Sus1-Ubp8 interface, which involves the Sus1 H1 helix (Figure 2D).
The Sgf73 H2 helix docks to the Ubp8 ZnF-UBP domain from the opposite side of the zinc finger fold (Figure 2D, 2E). The N-terminal end of the Sgf73 helix is embedded in a surface groove formed between the Ubp8 H2 helix and a loop coordinating the third zinc ion (Figure 2E, 2F). The hallmark of this Sgf73-Ubp8 interface is the completely buried Sgf73 residue Trp32 and the residue Lys33 covering it (Figure 2F). Each donating a hydrogen bond to a backbone carbonyl group of Ubp8, these two Sgf73 amino acids are strictly conserved in yeast species, suggesting a key role in anchoring Sgf73 to the Ubp8 ZnF-UBP domain. Overall, the Ubp8 ZnF-UBP domain serves as an assembly scaffold, holding all subunits of the module together.
To verify our structural analysis, we constructed two Ubp8 truncation mutants to test whether the Ubp8 ZnF-UBP domain itself is indeed sufficient for associating with the other three subunits of the DUB module. As shown in Figure 2G, a Ubp8 fragment representing the minimal Ubp8 ZnF-UBP domain (Ubp8 1–107), but not one lacking the last two β-strands and the C-terminal sequence (Ubp8 1–82), can form a stable complex with Sgf11, Sus1, and Sgf73. Furthermore, the ability of the truncated Ubp8 ZnF-UBP domain (Ubp8 1–82) to interact with Sgf73 but not Sus1 is consistent with two separate subunit interfaces as revealed by the crystal structure (compare lanes 3 and 6).
Sus1 as an elastic clamp for fastening the Sgf11 helix to Ubp8
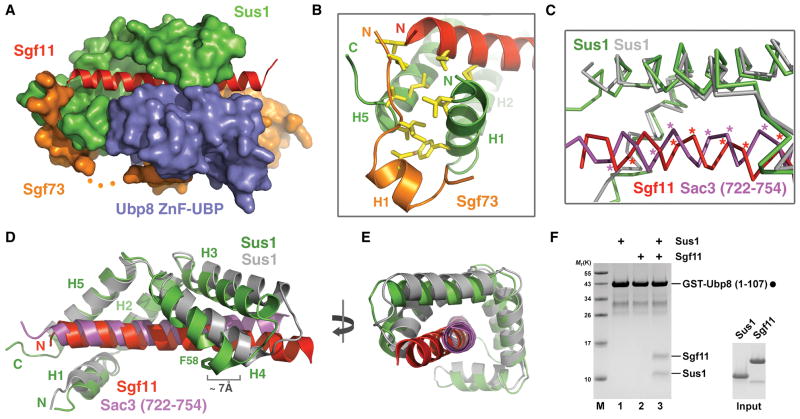
Sus1 is a bifunctional protein serving as a critical subunit of the SAGA DUB module and the TREX-2 complex, an mRNA export machinery formed by three additional subunits, Sac3, Cdc31, and Thp1 (Rodriguez-Navarro et al., 2004). Based on the physical association of the two complexes mediated by Sgf73 (amino acids 227–402) and a role of Sus1 in TREX-2 _targeting, both proteins have been proposed to participate in connecting transcription and mRNA export at the nuclear pore, a process known as “gene gating” (Köhler et al., 2008). Previous crystallographic studies of Sus1 in complex with a Sac3 C-terminal domain and Cdc31 have shown that two Sus1 molecules, each adopting a common hinged helical hairpin fold, interact with Sac3 by sequentially wrapping around a long continuous Sac3 helix (Jani et al., 2009). In the “assembly lobe” of the DUB module, Sus1 folds into essentially the same spiral clamp conformation and binds to the long Sgf11 N-terminal helix in a similar fashion. However, distinct from its simple role of gripping long helical structures in the partial TREX-2 complex, Sgf11-bound Sus1 in the DUB module uses its H1 and H4 helices to form two additional protein interfaces with the Ubp8 ZnF-UBP domain (Figure 2D). As a result, the Sgf11 helix becomes entirely enclosed by Sus1 and the ZnF-UBP domain of Ubp8 (Figure 3A), losing its freedom to dissociate without disrupting the three-protein complex.
Figure 3. Sus1 clamps down the Sgf11 N-terminal helix onto the Ubp8 ZnF-UBP domain.
(A) Overall view of the “assembly lobe” in the DUB module.
(B) A three-protein junction formed among the two terminal helices of Sus1 (green) and the N-terminal end of Sgf11 (red) and Sgf73 (orange).
(C) Superposition analysis of Sus1 in complexes with Sgf11 (red) and Sac3 (magenta). The Sus1 molecule in the Sus1-Sgf11 and Sus1-Sac3 complexes is colored in green and grey, respectively. The Cα of Sus1-contacting residues in Sgf11 and Sac3 are labeled with asterisks.
(D) and (E) Two orthogonal views of the superimposed Sus1-Sgf11 and Sus1-Sac3 complex structures. H2 and H5 helices of Sus1 in the two structures were superimposed to demonstrate the structural plasticity of Sus1.
(F) Reconstitution assay with recombinant proteins. GST-Ubp8 (1–107) was immobilized on GSH beads (filled circle) and incubated with either Sus1 or Sgf11 alone or with equal molar amounts of both proteins (input).
Intriguingly, an Sgf73 N-terminal sequence further secures the Sgf11-Sus1 complex by capping the elongated complex at one end. Connected to the Ubp8-interacting H2 helix via a disordered loop, the Sgf73 H1 helix and its N-terminal sequence interact with the N-terminal H1 and C-terminal H5 helices of Sus1 while overarching to the N-terminal end of the nearby Sgf11 helix (Figure 2D). A number of hydrophobic residues from Sgf73, Sus1, and Sgf11 congregate and form a hydrophobic core of yet another three-protein junction interface (Figure 3B).
A comparison of the Sus1-Sgf11 complex of the DUB module and a Sus1-Sac3 sub-complex readily reveals that Sus1 is an adaptable α-helix clamp with a considerable degree of structural plasticity. When the H2 and H5 helices of Sus1 in the two complexes are superimposed, it becomes clear that Sus1 recognizes the overall shape and hydrophobic surface pattern of the _target helix instead of specific residues. In fact, the helical polypeptide backbones of the Sgf11 and Sac3 helices interfaced with the same H2 and H5 helices of Sus1 are significantly shifted in the superimposed structure (Figure 3C). Moreover, the remaining three helices of Sus1 have to undergo substantial movements around the hinge regions in between helices to transit from one structural state to the other in the two complexes (Figure 3D). For example, the Ubp8-interacting residue at the tip of the Sus1 H4 helix in the Sus1-Sgf11 complex (Figure 2C) is separated from its position in the Sus1-Sac3 complex by nearly 7 Å (Figure 3D). Overall, the two distal ends of the Sus1 clamp, i.e. the N-terminal tips of the H1 and H4 helices of Sus1, appear to be closer to each other in the DUB module. This feature is also associated with a slight bending of the Sgf11 helix in the middle (Figure 3D, 3E). Interestingly, both distal ends of the Sus1 molecule and the middle region of the Sgf11 helix make direct contacts with the Ubp8 ZnF-UBP domain in the DUB module (Figure 2D), implicating that an induced fit mechanism might be involved in the cooperative assembly of the protein complex. Consistent with the spatial arrangement and a multiple inter-subunit cross-talk, the minimal Ubp8 ZnF-UBP domain can efficiently pull down Sus1 and Sgf11 only when both are present in an in vitro binding assay (Figure 3F).
The “catalytic lobe” of the SAGA DUB module
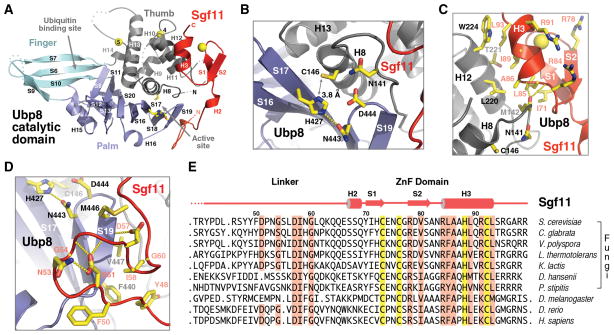
Tightened together in the “assembly lobe” via their N-terminal regions, Ubp8 and Sgf11 join their C-terminal regions to constitute the “catalytic lobe” of the DUB module. Following the published nomenclature (Hu et al., 2002), the structure of the Ubp8 catalytic domain can be divided into three sub-domains, Fingers, Palm, and Thumb (Figure 4A). The curved Fingers domain presents a large surface pocket, which is expected to cradle the globular domain of ubiquitin with support from the other two subdomains (Figure 1, 4A). In between the Palm and Thumb domain is the predicted cysteine protease active site, which connects to the ubiquitin-binding site through a narrow channel that accommodates the extended C-terminal tail of ubiquitin. Two unique zinc-coordinating sites are found in the Ubp8 Thumb domain, one facing the predicted ubiquitin-binding surface and the other packing against it from behind (Figure 4A and S2).
Figure 4. The Sgf11 ZnF domain binds to the Ubp8 catalytic domain next to the active site.
(A) Overall view of the “catalytic lobe” of the SAGA DUB module. The Ubp8 catalytic domain is differentially colored to show the three sub-domains, Thumb (grey), Palm (blue), and Fingers (cyan). The catalytic triad of Ubp8 are shown in sticks.
(B) A close-up view of the Ubp8 active site with the catalytic triad (C146, H427, and N443) and two amino acids (N141 and D444) that might help stabilize the oxyanion hole.
(C) Interface between the Sgf11 ZnF domain and the Ubp8 Thumb domain. The three conserved arginine residues exposed at the outer surface of the Sgf11 ZnF domain are also shown.
(D) Interface between the Sgf11 linker sequence and the Palm domain within the Ubp8 catalytic domain. The Ubp8 catalytic triad, the two Sgf11 DxxG motifs, and their surrounding residues are shown in sticks.
(E) Sequence alignment of Sgf11 orthologs with secondary structure assignment. The zinc-coordinating residues are highlighted in yellow. Other highly conserved residues are highlighted in salmon.
See also Figure S2.
In the crystal, Ubp8 is in a catalytically competent conformation. As shown in Figure 4B, the candidate catalytic triad of Ubp8 formed among Cys146, His427, and Asn443 is arranged in a nearly optimal geometry for catalysis. Specifically, the Nδ1 atom in the side chain of His427 is ready to deprotonate the Sγ atom of Cys146, which is 3.8 Å away. Meanwhile, the adjacent Asn443 residue accepts a hydrogen bond from His427 at a distance of 2.9 Å, fulfilling the role of stabilizing the catalytic histidine residue. Finally, two additional nearby amino acids, Asp141 and Asp444, are poised to form the necessary hydrogen bond network to support the oxyanion hole involved in the catalytic reaction.
The Sgf11 C-terminal region represents the most striking component of the “catalytic lobe” by forming an extended interface with the Ubp8 catalytic domain. It comprises a small CCHC-type zinc finger (ZnF) domain at the very C-terminal end and a winding linker sequence leading to the long N-terminal helix in the “assembly lobe” (Figure 1A, 4A). The Sgf11 ZnF domain forms a C2H2-like zinc finger fold, which features a β-hairpin, an α-helix, and a zinc ion bound in the middle (Figure 4C). Through a hydrophobic interface, the Sgf11 ZnF domain uses one side of its helix to pack against the H12 helix in the Thumb domain of Ubp8. The functional importance of this interface is underscored by the interacting hydrophobic residues, most of which are strictly conserved in the orthologs of Sgf11 and Ubp8 from yeast to humans (Figure 4E and S1). Importantly, the N-terminal end of the Sgf11 H3 helix also interacts with a critical loop in the Ubp8 Thumb domain, which is formed by the signature Cys box motif conserved in all UBP/USP family members (Wilkinson, 1997). This Cys box loop precedes the H8 helix and holds two essential catalytic site residues, Cys146 and Asn141, of Ubp8 (Figure 4B, 4C). Although no single amino acid of the Sgf11 ZnF domain directly interacts with these Ubp8 catalytic residues, the highly complementary inter-molecular interface formed with their polypeptide backbone implicates a plausible role of the Sgf11 ZnF domain in sculpting the enzymatic center of the deubiquitinase. On the opposite side of the zinc finger fold, the Sgf11 ZnF domain harbors a cluster of basic residues that are completely exposed to the solvent. Three of these residues, Arg78, Arg84, and Arg91, are conserved among Sgf11 orthologs, suggesting yet another functional role of the Sgf11 ZnF domain in the DUB module (Figure 4E).
The Sgf11 linker sequence also participates in regulating Ubp8 by clinging to the Palm domain near the active site from the other side of the catalytic cleft (Figure 4A). In a highly sinuous structure, the Sgf11 linker region is characterized by two tandem DxxG motifs (Figure 4D), one providing an intra-molecular hydrogen bond network supporting the formation of a large loop, the other adopting a β-turn structure and forming two intermolecular hydrogen bonds with the Ubp8 S19 β-strand adjacent to the catalytic site residue Asn443 (Figure 4D). Similar to the ZnF domain, the Sgf11 linker sequence helps shape the Ubp8 active site without directly interacting with the residues in the catalytic triad. Overall, the Sgf11 C-terminal region embraces the Ubp8 catalytic core like a cuff, gripping on both its Thumb and Palm domains (Figure 4A).
Activation of Ubp8 by Sgf11
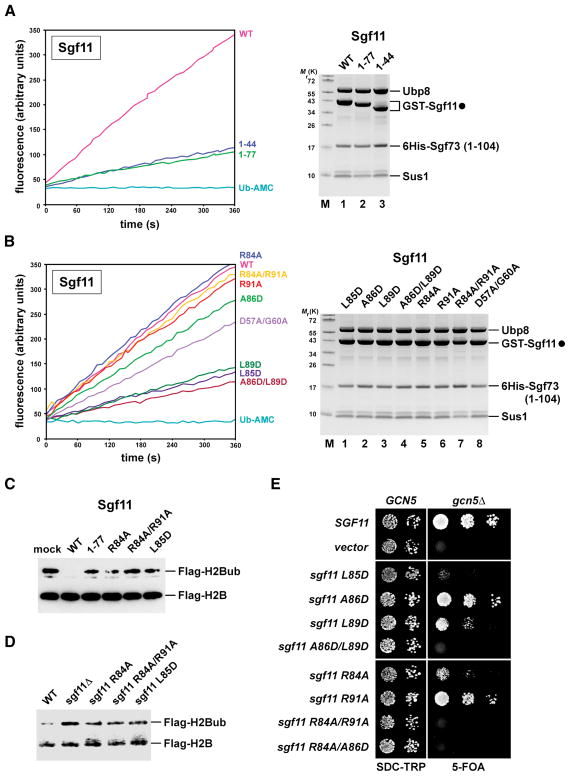
Prior to solving the DUB module structure, we generated a comprehensive set of mutations to determine whether its ZnF domain could be required to regulate Ubp8 activity. Notably, C-terminal truncations of the 99-amino acid protein Sgf11, which disrupt (Sgf11 1–77) or completely remove (Sgf11 1–44) its ZnF domain, caused a pronounced decrease of Ubp8 activity towards Ub-AMC, without affecting DUB assembly (Figure 5A).
Figure 5. The Sgf11 ZnF domain activates Ubp8 and mediates Ub-H2B deubiquitination.
(A) and (B) Ub-AMC hydrolysis assay. Coomassie staining shows equalized amount of Ubp8 and overall composition of the complexes.
(C) In vitro H2B deubiquitination assay. Hyperubiquitinated Flag-tagged H2B purified from ubp8Δ cells was incubated with equal amounts of the indicated wild-type and mutant DUB tetramers as depicted in (A) and (B). Ubiquitinated and unmodified H2B were detected by anti-Flag western blotting.
(D) Anti-Flag immunoprecipitates derived from cells expressing Flag–tagged H2B. sgf11Δ cells were transformed with plasmids carrying the indicated wild-type, mutant sgf11 alleles or an empty vector. Recovered proteins were analyzed by SDS–PAGE and anti-Flag western blotting.
(E) A GCN5/sgf11Δ shuffle strain containing a GCN5 cover plasmid (URA3) was transformed with wild-type SGF11, empty vector or the indicated sgf11 mutant alleles (TRP1 plasmids). Growth was followed on SDC-Trp and on SDC+5-fluoroorotic acid (5-FOA) plates. Cell density was normalized, cells were spotted onto plates in 10-fold serial dilution steps and incubated at 30°C for 2 days (SDC-TRP) and 4 days (SDC+5-FOA).
See also Figure S3 and Table S2 for yeast strains and plasmids used.
To precisely dissect the contribution of specific Sgf11 ZnF residues to catalysis, we mutated the conserved hydrophobic amino acids Leu85, Ala86, Leu89 and the basic residues Arg84 and Arg91 (Figure 5B). Although Ub-AMC hydrolysis by the Sgf11 A86D mutant was only moderately impaired, the single mutants L85D, L89D and the double mutant A86D/L89D exhibited a strongly reduced activity, comparable in extent to the effect of removing the entire ZnF (i.e. Sgf11 1–44). In contrast, R84A and R91A alone and the combined Sgf11 R84A/R91A mutation did not have a noticeable effect.
To explore a potential role of these basic residues in substrate recognition, we assessed whether mutating Arg84 and Arg91 would impair hydrolysis of mono-ubiquitinated histone H2B, the natural Ubp8 substrate. This in vitro assay uncovered a failure to efficiently deubiquitinate H2B by the single R84A and the double R84A/R91A mutant (Figure 5C). Since Sgf11 1–77 and L85D mutants were already impaired in Ub-AMC hydrolysis, they predictably failed to properly hydrolyze Ub-H2B (Figure 5C). As a result, the global level of Ub-H2B was increased in cells harboring the sgf11 R84A, R84A/R91A or L85D ZnF mutations (Figure 5D).
The DUB module structure provides a powerful rationale for interpreting the effects of the mutants described above. Introducing a repulsive negative charge at the hydrophobic Sgf11 ZnF-Ubp8 interface (e.g. L85D) is expected to disrupt the intimate packing of the Sgf11 H3 helix against the Ubp8 H12 helix and thus to destabilize the adjacent catalytic triad (Figure 4C). In contrast, mutating the solvent-exposed basic residues on the opposite side of the Sgf11 H3 helix would leave the intermolecular hydrophobic interactions with Ubp8 H12 intact and Ub-AMC hydrolysis unaffected. Moreover, neither Arg84 nor Arg91 is expected to be directly involved in binding ubiquitin. Importantly, the outward orientation of the basic patch on the Sgf11 ZnF domain and its close proximity to the catalytic center suggest a role in recognizing some part of histone H2B or a larger nucleosomal surface and may explain the specific defect in hydrolyzing Ub-H2B but not Ub-AMC. Based on the observed structure of the DIxG motif and its role in configuring the Sgf11 linker, a D57A/G60A mutant also lowered Ub-AMC hydrolysis (Figure 5B).
Finally, we analyzed the in vivo significance of each mutation by genetic methods (Table S2). Yeast cells can compensate the loss of either SAGA histone acetyltransferase (gcn5Δ) or histone deubiquitinating activity (sgf11Δ). However, combining gcn5Δ with sgf11Δ was found to cause synthetic lethality, indicating a synergism between SAGA HAT and DUB function (Figure 5E) (Lee et al., 2005). Accordingly, the L85D and L89D alleles, which could displace the Sgf11 ZnF domain from the Ubp8 surface, conferred a strong synthetic enhancement phenotype when combined with gcn5Δ. In line with its modest defect in Ub-AMC hydrolysis (Figure 5B), the isolated A86D mutation exhibited a weak synthetic growth phenotype, whereas the sgf11 A86D/L89D double mutant was inviable. Consistent with a key role of the solvent exposed surface of the Sgf11 ZnF domain in mediating H2B deubiquitination, the isolated R84A allele displayed a robust synthetic enhancement with gcn5Δ. Although the sgf11 R91A mutant conferred only subtle synthetic growth defects, it caused lethality when mutated together with R84A (R84A/R91A). The observed genetic phenotypes do not result from Sgf11 protein instability, as all mutants were expressed essentially at wild-type levels (Figure S3). In essence, the structural and functional data support a key role of the Sgf11 ZnF domain in activating Ubp8 by structurally configuring its active site for catalysis. These results also hint at an important role of the solvent-exposed basic surface of the same Sgf11 domain in H2B deubiquitination, possibly by dictating substrate specificity.
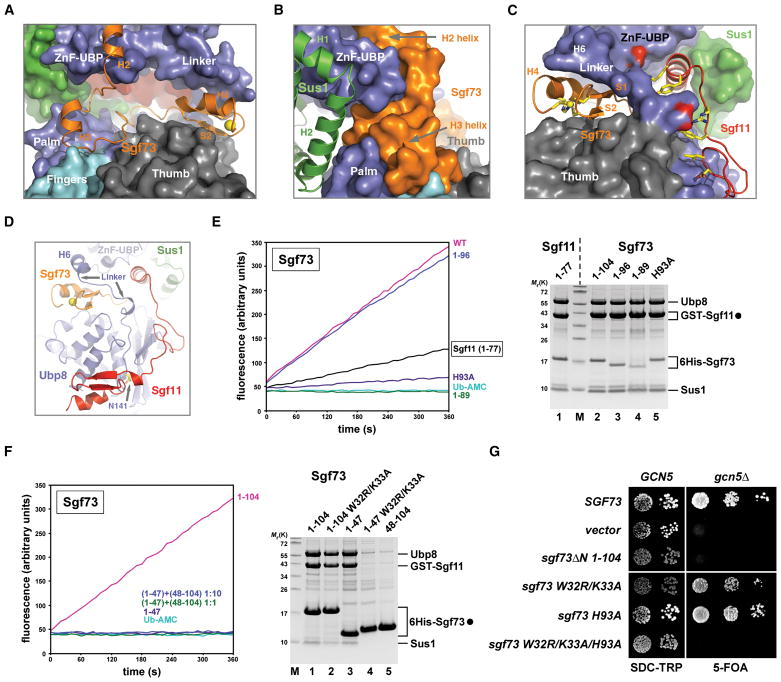
Sgf73 activates Ubp8 by coupling the two functional lobes of the DUB module
The N-terminal 104 amino acid fragment of Sgf73 is necessary for DUB module activation and recruitment to SAGA (Köhler et al., 2008). The crystal structure reveals that Sgf73 adopts a largely extended conformation with three short N-terminal helices and a C-terminal ZnF fold similar to the one found in Sgf11 (Figure 1A and 6A, see also Figure S4A). Following the WK motif-bearing H2 helix (Figure 2F), the Sgf73 polypeptide forms a continuous and curvy interface with the backside of the Ubp8 catalytic domain in the “catalytic lobe” (Figure 6A). The H2-loop-H3 region of Sgf73 docks onto the backside of the Ubp8 Palm and Fingers domains, supporting one end of the “assembly lobe” while wrapping around the Ubp8 ZnF-UBP domain (Figure 6B). At the opposite C-terminal end, the Sgf73 ZnF domain acts like a wedge, inserting itself between the Ubp8 Thumb domain and the N-terminal portion of the Ubp8 linker sequence (Figure 6C, 6D). The H6 helix in the Ubp8 linker sequence stacks on top of the Sgf73 ZnF domain and buttresses the Ubp8 ZnF-UBP domain. Overall, the Sgf73 N-terminal fragment enables the “catalytic lobe” of the DUB complex to piggyback the entire “assembly lobe” so that the two act together as one structurally integrated module (Figure 1B).
Figure 6. Sgf73 couples the two functional lobes of the DUB module and potentiates Ubp8.
(A) A close-up view of the space between the two Ubp8 domains occupied by Sgf73. The structure is depicted in the same orientation as seen in Figure 1B. Sgf73 is shown in ribbon with the rest of the DUB complex shown in surface representation.
(B) Left side view of (A) showing that the H2-loop-H3 region of Sgf73 holds together the two domains of Ubp8 at one end of the complex.
(C) Right side view of (A) showing that the Sgf73 ZnF domain functions as a wedge to fix the conformation of the Ubp8 linker sequence, which is also in contact with the Sgf11 linker region. Key residues of the Sgf11 linker are shown in sticks. Two Ubp8 residues forming salt bridges with two positively charged Sgf11 residues are colored in red.
(D) A focused view of the DUB module showing the spatial relationships among the Sgf73 ZnF domain, the Ubp8 linker sequence, the Sgf11 C-terminal region, and the Ubp8 active site.
(E) Ub-AMC hydrolysis assay. Coomassie staining shows equalized amount of Ubp8 and overall composition of the complexes (filled circle marks bait proteins).
(F) A tetrameric complex containing Sgf73 amino acids 1–47 (lane 3) was preincubated with recombinant Sgf73 amino acids 48–104 at a 1:1 or 1:10 stoichiometry (30° for 15min) and subsequently assayed for Ub-AMC hydrolytic activity. Coomassie-stained gel shows imidazole eluates of affinity-purified 6His-Sgf73 complexes (filled circle).
(G) A GCN5/sgf73Δ shuffle strain was transformed with wild-type SGF73, empty vector or the indicated sgf73 mutant alleles (TRP1 plasmids). Growth was followed on SDC-Trp and on SDC+5-fluoroorotic acid (5-FOA) plates.
See also Figure S4.
To dissect the function of Sgf73, we first assessed the importance of its ZnF domain for enzymatic activity. A truncation of the 4th zinc-coordinating residue Cys98 (Sgf73 1–96) had no significant effect on Ub-AMC hydrolysis as compared to an intact ZnF (Sgf73 1–104) (Figure 6E). However, a point mutation of the 3rd zinc-coordinating residue (Sgf73 H93A) or removal of the entire H4 helix (Sgf73 1–89) essentially abrogated DUB activity (Figure 6E). In comparison, disrupting the Sgf11 ZnF (Sgf11 1–77 or Sgf11 1–44) retained a low basal level of enzymatic activity. These data indicate that Sgf73 plays an essential role in priming Ubp8 for activation. Ubp8 activation, therefore, involves a two-fold mechanism, in which the Sgf73 and Sgf11 ZnF domains operate in synergy.
Given that the Sgf11 ZnF domain is topologically positioned via a distant molecular tether at the “assembly lobe” (i.e. Sus1), we reasoned that the Sgf73 ZnF domain might also require an analogous anchoring mechanism. We constructed a DUB module devoid of enzymatic activity, which was assembled but contained only a short N-terminal fragment of Sgf73 whose cut-off point lies at the junction between the two lobes (Sgf73 1–47) (Figure 6F lane 3). Intriguingly, adding back the complementary Sgf73 polypeptide (Sgf73 48–104), even at 10 folds excess, could not form a full module and restore Ub-AMC hydrolysis (Figure 6F). This result suggests that the Sgf73 ZnF domain requires a physical connection to the “assembly lobe” to perform its function and cannot act as an independent allosteric activator of Ubp8.
Next, we tested the importance of the Sgf73 WK motif in mediating Sgf73 tethering within the “assembly lobe”. Indeed, an Sgf73 1–104 W32R/K33A double mutant co-purified reduced amounts of Ubp8, Sgf11 and Sus1 (Figure 6F, lane 2). The same mutation in the context of the small Sgf73 fragment described above (Sgf73 1–47 W32R/K33A) completely disrupted the Sgf73-Ubp8 interaction with a concomitant loss of Sgf11 and Sus1 (Figure 6F, lane 4). On the other hand, the Sgf73 ZnF fragment (amino acids 48–104) alone lacked sufficient intrinsic affinity towards Ubp8 when co-expressed with the other three DUB module components (Figure 6F lane 5). Thus, the WK motif is a prime Sgf73 attachment site within the “assembly lobe”, which aids in the stable positioning of the Sgf73 ZnF domain to the backside of the Ubp8 catalytic domain.
Finally, we examined the in vivo significance of these findings in the context of the full-length Sgf73 protein (658 residues). In analogy to the genetic interaction between SGF11 and GCN5, a synthetic lethal phenotype was apparent in sgf73Δgcn5Δ cells. Truncating the SGF73 N-terminus (sgf73ΔN 1–104) also caused synthetic lethality with gcn5Δ, consistent with a failure to recruit the DUB module to SAGA (Köhler et al., 2008). The sgf73 W32R/K33A mutation or the H93A mutant alone displayed only modest synthetic enhancement phenotypes (note a smaller colony size as compared to wild-type). In contrast, the combined triple mutation (W32R/K33A/H93A) resulted in synthetic lethality just like the complete deletion of SGF73. Accordingly, Ubp8 was no longer incorporated into SAGA, as assessed by a purification of the entire SAGA complex from yeast cells (Figure S4B). Taken together, these findings help define Sgf73 as a second enzyme-activating subunit in the DUB module.
Discussion
An increasing number of studies have demonstrated that the enzymatic activities of many DUBs are dictated by cellular context. For example, association with the proteasome is known to activate at least three DUBs of different types, USP14, UCH37, and Rpn11 (Reyes-Turcu et al., 2009). Recognition of the free C-terminus of the proximal ubiquitin in a poly-ubiquitin chain has been shown to stimulate the activity of USP5 (Reyes-Turcu et al., 2006). Importantly, a variety of cellular proteins unrelated to the ubiquitin-proteasome system can either activate or inhibit specific DUBs through direct interactions, oftentimes by modulating their intrinsic catalytic activity (Borodovsky et al., 2001; Cohen et al., 2003; Cohn et al., 2007; Soncini et al., 2001; van der Knaap et al., 2005; Yao et al., 2008). Although previous crystallographic studies have revealed significant structural reorganization of the active center or the ubiquitin-binding site of various DUBs, the detected conformational changes are mostly associated with substrate (ubiquitin)-induced fit, instead of allosteric (in)activation by a regulatory protein (Avvakumov et al., 2006; Hu et al., 2002).
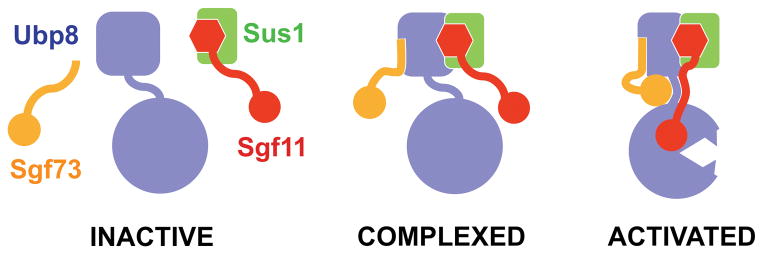
The crystal structure of the yeast SAGA DUB module unravels the structural basis of how a DUB is integrated into multi-subunit protein machinery and becomes activated by its non-substrate binding partners. We present a simple model deciphering the assembly and activation codes of the SAGA DUB module. As shown in Figure 7, formation of the module can be dissected into two functional, but not necessarily sequential, steps — assembly and activation. The formation of the module complex is primarily mediated by the ZnF-UBP domain of Ubp8, which simultaneously binds the N-terminal helix of Sgf11 and the Sgf73 N-terminal region. Sus1 stabilizes the Sgf11-Ubp8 interaction as a molecular clamp. Sgf11 and Sgf73 in turn use their ZnF domains to activate the Ubp8 catalytic domain via two separate interfaces. Sgf11 binds to the Ubp8 catalytic domain near the active site, supporting the construction of a competent catalytic center of the DUB enzyme, whereas Sgf73 potentiates Ubp8 by structurally coupling the two domains of Ubp8 and fixing the conformation of its linker sequence. Noticeably, Ubp8 activation unlikely involves the two “blocking loops” of the UBP/USP catalytic domain previously proposed for the activation mechanism of USP14 (Figure S5) (Hu et al., 2005). In addition to activating the Ubp8 catalytic domain, our studies reveal yet a second potential role of Sgf11 in substrate recognition. Future work will address the details of the DUB module-nucleosome interaction and the inactive forms of Ubp8.
Figure 7.
Model for the assembly and activation of the SAGA DUB module. Free Ubp8 (blue) has a ZnF-UBP domain (square) and an inactive catalytic domain (full circle). Independent from other subunits, Sus1 (green) can form a complex with Sgf11 (red) by wrapping around its N-terminal helix (hexagon). The correctly positioned ZnF domains of Sgf11 (red circle) and Sgf73 (orange circle) are both required to fully activate Ubp8 (sharp teeth).
See also Figure S5.
Finally, our study highlights the remarkable evolutionary plasticity of Sus1, which can operate as a co-factor of a histone DUB and as a nuclear pore _targeting element of the TREX-2 mRNA exporter. The role of Sus1 as an elastic clamp in the DUB module raises the possibility that Sus1 might similarly fasten the Sac3 α-helix onto a component of the nuclear pore as a mechanism for tethering the gene expression machinery to the nuclear envelope.
Experimental Procedures
Protein Overexpression and Purification
The full-length S. cerevisiae SGF11, SUS1, and codon-optimized UBP8 genes were co-expressed with the SGF73 amino acid 1–104 gene fragment in BL21-CodonPlus(DE3)-RIL E. coli cells on the polycistronic expression vector pST44 (Tan et al., 2005). SGF11 contains a TEV-cleavable N-terminal GST-tag and SGF73 aa1–104 a cleavable N-terminal hexahistidine (6His) tag. The tetrameric complex was isolated by glutathione (GSH) affinity and anion exchange chromatography, followed by TEV cleavage and gel filtration chromatography. Proteins were concentrated to 5 mg/ml in a final solution of 20 mM Tris-HCl (pH = 8.0), 200 mM NaCl, and 5 mM dithiothreitol (DTT).
Crystallization and Data Collection
Crystals were grown at 4 °C by the hanging-drop vapor-diffusion method with 1.5 μl protein samples mixed with an equal volume of reservoir solution containing 0.2 M ammonium citrate pH 5.8 and 18% w/v PEG 3,350. The crystals form in space group P21, with a = 64.4 Å, b = 103.5 Å, c = 70.3 Å, and β = 108.1°, and contain one complex in the asymmetric unit. All data sets were collected on BL8.2.1 at the Advanced Light Source in Berkeley, using crystals flash-frozen in the crystallization buffers supplemented with 20% glycerol at −170 °C. Reflection data were indexed, integrated, and scaled (Table S1) using the HKL2000 package (Otwinowski and Minor, 1997).
Structure Determination and Refinement
The nature complex structure was determined by a combination of molecular replacement (MR) and zinc single anomalous dispersion (SAD) methods. The Ubp8 catalytic domain was located by MR using a poly-alanine chain derived from the human USP2 structure (PDB accession #2HD5) as a search model. After seven zinc atoms were located via the model phases, a zinc SAD dataset was collected. The PHENIX package (Adams et al., 2002) was used to combine phases from MR and SAD and to calculate an interpretable map, which allowed structural model building. Iterative cycles of model building and refinement were performed using O, CNS, and the CCP4 programs (Brunger et al., 1998; CCP4, 1994; Jones et al., 1991). The final model was refined to 2.70 Å. All structure figures were prepared with PyMOL (DeLano, 2002).
Affinity Purification Analysis
GST-affinity purification was performed in buffer containing 100 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1.5 mM MgCl2, 0.15% NP-40, 1 mM DTT and eluted with GSH. 6xHis purifications were performed with Talon cobalt-based affinity resin (Clontech) in the same buffer without DTT and eluted with 150 mM imidazole. To purify reconstituted complexes, recombinant proteins were mixed, immunoprecipitated and, after washing and elution, detected by SDS-PAGE and Coomassie staining. Tandem Affinity Purifications from yeast were performed according to a published procedure (Rigaut et al., 1999).
Yeast strains and plasmid constructions
Genes in yeast were deleted by a standard one-step PCR-base technique. Strains and plasmids are listed in Supplementary Table S2.
Ub-AMC hydrolysis assay
Assays were conducted as described previously (Dang et al., 1998). Measurements were made in parallel using a Gemini XPS microplate spectrofluorometer. For complex reconstitution, recombinant proteins were pre-incubated at 30 °C for 15 min prior to the assay.
Histone H2B ubiquitination/deubiquitination assays
Global H2B ubiquitination was examined as described previously (Kao and Osley, 2003). In vitro H2B deubiquitination assays were performed as described in Köhler et al., 2008.
Supplementary Material
Acknowledgments
We thank ALS beamline staff for assistance and all members of the Zheng lab and Andrea Barta (MFPL) for invaluable discussions. This study is supported by Howard Hughes Medical Institute and NIH grant CA107134 to N.Z. and grants of the Deutsche Forschungsgemeinschaft (SFB638/B3) to A.K and E.H.
Footnotes
The Protein Data Bank accession numbers for the Ubp8-Sgf11-Sus1-Sgf73 complex structure is 3M99.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Allen MD, Bycroft M. The solution structure of the ZnF UBP domain of USP33/VDU1. Protein Sci. 2007;16:2072–2075. doi: 10.1110/ps.072967807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, Dhe-Paganon S. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Romier C, Tora L, Devys D. Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem Sci. 2008;33:369–375. doi: 10.1016/j.tibs.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol. 2003;5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D’Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- Daniel JA, Grant PA. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res. 2007;618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR, 3rd, Grant PA. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem. 2004;279:1867–1871. doi: 10.1074/jbc.C300494200. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User’s Manual. San Carlos: DeLano Scientific; 2002. [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, Emili A, Hughes TR, Greenblatt JF, Berger SL. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani D, Lutz S, Marshall NJ, Fischer T, Köhler A, Ellisdon AM, Hurt E, Stewart M. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell. 2009;33:727–737. doi: 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kao CF, Osley MA. In vivo assays to study histone ubiquitylation. Methods. 2003;31:59–66. doi: 10.1016/s1046-2023(03)00088-4. [DOI] [PubMed] [Google Scholar]
- Köhler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Lee KK, Florens L, Swanson SK, Washburn MP, Workman JL. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- Pai MT, Tzeng SR, Kovacs JJ, Keaton MA, Li SS, Yao TP, Zhou P. Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin. J Mol Biol. 2007;370:290–302. doi: 10.1016/j.jmb.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DW, Weaver CM, Jennings JL, McAfee KJ, He Y, Weil PA, Link AJ. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259. doi: 10.1128/MCB.24.16.7249-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M, Parrado SG, D’Arcy A, Eidhoff U, Gerhartz B, Hassiepen U, Pierrat B, Riedl R, Vinzenz D, Worpenberg S, et al. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14:1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Soncini C, Berdo I, Draetta G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene. 2001;20:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. Faseb J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Jin J, Cai Y, Takahashi H, Swanson SK, Washburn MP, Florens L, Conaway RC, Cohen RE, et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–917. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.