Abstract
Par-1 is an evolutionarily conserved protein kinase required for polarity in worms, flies, frogs, and mammals. The mammalian Par-1 family consists of four members. Knockout studies of mice implicate Par-1b/MARK2/EMK in regulating fertility, immune homeostasis, learning, and memory as well as adiposity, insulin hypersensitivity, and glucose metabolism. Here, we report phenotypes of mice null for a second family member (Par-1a/MARK3/C-TAK1) that exhibit increased energy expenditure, reduced adiposity with unaltered glucose handling, and normal insulin sensitivity. Knockout mice were protected against high-fat diet-induced obesity and displayed attenuated weight gain, complete resistance to hepatic steatosis, and improved glucose handling with decreased insulin secretion. Overnight starvation led to complete hepatic glycogen depletion, associated hypoketotic hypoglycemia, increased hepatocellular autophagy, and increased glycogen synthase levels in Par-1a−/− but not in control or Par-1b−/− mice. The intercrossing of Par-1a−/− with Par-1b−/− mice revealed that at least one of the four alleles is necessary for embryonic survival. The severity of phenotypes followed a rank order, whereby the loss of one Par-1b allele in Par-1a−/− mice conveyed milder phenotypes than the loss of one Par-1a allele in Par-1b−/− mice. Thus, although Par-1a and Par-1b can compensate for one another during embryogenesis, their individual disruption gives rise to distinct metabolic phenotypes in adult mice.
Cellular polarity is a fundamental principle in biology (6, 36, 62). The prototypical protein kinase originally identified as a regulator of polarity was termed partitioning defective (Par-1) due to early embryonic defects in Caenorhabditis elegans (52). Subsequent studies revealed that Par-1 is required for cellular polarity in worms, flies, frogs, and mammals (4, 17, 58, 63, 65, 71, 89). An integral role for Par-1 kinases in multiple signaling pathways has also been established, and although not formally addressed, multifunctionality for individual Par-1 family members is implied in reviews of the list of recognized upstream regulators and downstream substrates (Table 1). Interestingly, for many Par-1 substrates the phosphorylated residues generate 14-3-3 binding sites (25, 28, 37, 50, 59, 61, 68, 69, 78, 95, 101, 103). 14-3-3 binding in turn modulates both nuclear/cytoplasmic as well as cytoplasmic/membrane shuttling of _target proteins, thus allowing Par-1 activity to establish intracellular spatial organization (15, 101). The phosphorylation of Par-1 itself promotes 14-3-3 binding, thereby regulating its subcellular localization (37, 59, 101).
TABLE 1.
Multifunctionality of Par-1 polarity kinase pathwaysa
| Regulator or substrate | Function | Reference(s) |
|---|---|---|
| Regulators (upstream function) | ||
| LKB1 | Wnt signaling, Peutz-Jeghers syndrome, insulin signal transduction, pattern formation | 2, 63, 93 |
| TAO1 | MEK3/p38 stress-responsive mitogen-activated protein kinase (MAPK) pathway | 46 |
| MARKK | Nerve growth factor signaling in neurite development and differentiation | 98 |
| aPKC | Ca2+/DAG-independent signal transduction, cell polarity, glucose metabolism | 14, 37, 40, 45, 59, 75, 95 |
| nPKC/PKD | DAG-dependent, Ca2+-independent signal transduction (GPCR) | 101 |
| PAR-3/PAR-6/aPKC | (−); regulates Par-1, assembly of microtubules, axon-dendrite specification | 19 |
| GSK3β | (−); tau phosphorylation, Alzheimer's dementia, energy metabolism, body patterning | 54, 97 |
| Pim-1 oncogene | (−); G2/M checkpoint, effector of cytokine signaling and Jak/STAT(3/5) | 5 |
| CaMKI | (−); Ca2+-dependent signal transduction, neuronal differentiation | 99 |
| Substrates (downstream function) | ||
| Cdc25C | Regulation of mitotic entry by activation of the cdc2-cyclin B complex | 25, 72, 78, 103 |
| Class II HDAC | Control of gene expression and master regulator of subcellular trafficking | 28, 50 |
| CRTC2/TORC2 | Gluconeogenesis regulator via LKB1/AMPK/TORC2 signaling, PPARγ1a coactivator | 49 |
| Dlg/PSD-95 | Synaptogenesis and neuromuscular junction, tumor suppressor (102) | 104 |
| Disheveled | Wnt signaling, translocation of Dsh from cytoplasmic vesicles to cortex | 73, 94 |
| KSR1 | Regulation of the Ras-MAPK pathway | 68, 69 |
| MAP2/4/TAU | Dynamic instability (67, 83) of microtubules, Alzheimer's dementia (30) | 11, 31-33, 47, 70, 96 |
| Mib/Notch | Mind bomb (Mib degradation and repression of Notch signaling results in neurogenesis) | 57, 74, 81 |
| Par3/OSKAR/Lgl | Cytoplasmic protein segregation, cell polarity, and asymmetric cell division | 7, 10 |
| Pkp2 | Desmosome assembly and organization; nuclear shuttling | 68, 69 |
| PTPH1 | Linkage between Ser/Thr and Tyr phosphorylation-dependent signaling | 103 |
| Rab11-FIP | Regulation of endocytosis (23), trafficking of E-cadherin (64) | 34 |
LKB1 also is known as Par-4; MARKK also is known as Ste20-like; (−), inhibitory/negative regulation has been shown; GPCR, G protein-coupled receptors. MARKK is highly homologous to TAO-1 (thousand-and-one amino acid kinase) (46).
The mammalian Par-1 family contains four members (Table 2). Physiological functions of the Par-1b kinase have been studied using _targeted gene knockout approaches in mice (9, 44). Two independently derived mouse lines null for Par-1b have implicated this protein kinase in diverse physiological processes, including fertility (9), immune system homeostasis (44), learning and memory (86), the positioning of nuclei in pancreatic beta cells (35, 38), and growth and metabolism (43).
TABLE 2.
Terminology and localization of mammalian Par-1 family members
| Synonymsa | Subcellular localization |
|---|---|
| Par-1a, MARK3, C-TAK1, p78/KP78, 1600015G02Rik, A430080F22Rik, Emk2, ETK-1, KIAA4230, mKIAA1860, mKIAA4230, M80359 | Basolateralb/apicalc |
| Par-1b, EMK, MARK2, AU024026, mKIAA4207 | Basolateral |
| Par1c, MARK1 | Basolateral |
| Par1d, MARK4, MARKL1 | Not asymmetricd |
Par should not to be confused with protease-activated receptor 1 (PAR1 [29]); C-TAK1, Cdc twenty-five C-associated kinase 1; MARK, microtubule affinity regulating kinase; MARKL, MAP/microtubule affinity-regulating kinase-like 1.
Basolateral to a lesser degree than Par-1b (37).
Human KP78 is asymmetrically localized to the apical surface of epithelial cells (76).
Variant that does not show asymmetric localization in epithelial cells when overexpressed (95).
Beyond Par-1b, most information regarding the cell biological functions of the Par-1 kinases comes from studies of Par-1a. Specifically, Par-1a has been implicated in pancreatic (76) and hepatocarcinogenesis (51), as well as colorectal tumors (77), hippocampal function (100), CagA (Helicobacter pylori)-associated epithelial cell polarity disruption (82), and Peutz-Jeghers syndrome (48), although the latter association has been excluded recently (27). As a first step toward determining unique and redundant functions of Par-1 family members, mice disrupted for a second member of the family (Par-1a/MARK3/C-TAK1) were generated. We report that Par-1a−/− mice are viable and develop normally, and adult mice are hypermetabolic, have decreased white and brown adipose tissue mass, and unaltered glucose/insulin handling. However, when challenged by a high-fat diet (HFD), Par-1a−/− mice exhibit resistance to hepatic steatosis, resistance to glucose intolerance, and the delayed onset of obesity relative to that of control littermates. Strikingly, overnight starvation results in a complete depletion of glycogen and lipid stores along with an increase in autophagic vacuoles in the liver of Par-1a−/− but not Par-1b−/− mice. Correspondingly, Par-1a−/− mice develop hypoketotic hypoglycemia. These findings reveal unique metabolic functions of two Par-1 family members.
MATERIALS AND METHODS
Animal procedures.
The Washington University School of Medicine Animal Studies Committee approved all animal procedures. Unless stated otherwise, animals were fed a standard chow diet (Lab Diet 5063 containing 4.5% fat, 55% carbohydrate, 20% protein, and 4.7% fiber; Purina Mills, St. Louis, MO) and were housed with free access to food and water under a 12-h light/dark cycle. Unless indicated, only male mice were analyzed throughout this study.
Construction of the Par-1a _targeting construct.
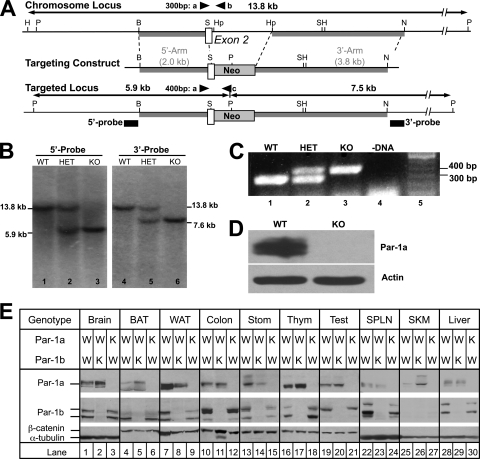
A Par-1a genomic clone was isolated from a 129X1/SvJ mouse embryonic stem (ES) cell genomic library by hybridization with an isogenic 0.9-kb Par-1a-specific cDNA probe. The genomic organization of the mouse Par-1a gene was disrupted by replacing a 1.9-kb SacI/HpaI fragment with a 1.2-kb neomycin phosphotransferase cassette (Fig. 1 A). The replacement disrupts exon 2 encoding the ATP binding domain of Par-1a and also destroys the 5′ splice site between introns 2 and 3. The insertion of the neomycin selection cassette alters the PstI fragment from a single 13.8-kb fragment to two fragments of 7.5 and 5.9 kb, and this change was used to identify _targeted ES cell clones and knockout (KO) animals by Southern blotting (Fig. 1B) using the 5′ and 3′ probes indicated in Fig. 1A. Using mouse genomic DNA as the template, the 425-bp 5′ probe was amplified by PCR with primers of 5′-TGGTGGTACACGCCTGTATTC and 5′-CTGATCTACACAATTCCAGGAC. The 204 bp of 3′ probe was amplified by PCR with primers of 5′-GCATCTTGGGCTGTAAGTGATG and 5′-GGCTTACAACCATCTGTACAG.
FIG. 1.
_targeted disruption of Par-1a in mice and tissue distribution of Par-1a and Par-1b. (A) Par-1a locus (top), _targeting vector (middle), and chromosomal organization after recombination (bottom). The Par-1a gene was disrupted by the insertion of a neomycin (neo) phosphotransferase cDNA as a selectable marker. Sizes of upstream (2 kb) and downstream (3.8 kb) homologous arms are indicated (gray boxes), as is exon 2 (open box). Black boxes indicate positions of the probes for genomic Southern blotting; triangles depict locations of PCR primers for genotyping. Abbreviations: P, PstI; B, BglII; S, SacI; Hp, HpaI; N, NcoI; H, HindIII. (B) Southern blot analysis demonstrating homologous recombination at the Par-1a locus. Genomic DNA was digested with PstI and subjected to Southern blotting using the 5′ and 3′ probes indicated in panel A. Genotypes are indicated (WT, wild-type mouse; HET, heterozygous; KO, homozygous). (C) PCR analysis of mouse DNA (tail), amplified with three PCR primers (a, b, and c) shown in panel A. The WT allele produces a 300-bp PCR product, and null alleles produce a 400-bp PCR product. (D) Western blot analysis of Par-1a protein in brain lysates prepared from C57BL/6-mice. (E) Tissues from wild-type, Par-1a−/−, and Par-1b−/− mice were isolated. Protein lysates from the indicated tissues were resolved by SDS-PAGE and subjected to Western blotting for the indicated proteins: brain (0.2 mg), brown adipose tissue (BAT; 1.1 mg), white adipose tissue (WAT; 1.1 mg), colon (0.4 mg), stomach (Stom; 0.4 mg), thymus (Thym; 0.4 mg), testis (Test; 0.4 mg), spleen (SPLN; 0.4 mg), skeletal muscle (SKM; 0.4 mg), and liver (0.13 mg). W, wild type; K, knockout.
PCR analysis.
PCR of tail DNA/tissues was achieved using a three-primer PCR with one 5′ primer from exon 2 (5′-GGAGACGGGAGGCAAGAAGTCA), a 3′ primer from the intronic sequence between exons 2 and 3 (5′-GGCTCATTCCCTTGGTGTACTT), and a 3′ primer from the neomycin cassette (5′GAGCAGCCGATTGTCTGTTGT), resulting in 400-bp (KO) or 300-bp (wild-type [WT]) bands, respectively (Fig. 1C).
Generation of mice harboring the Par-1a mutation.
RW4 ES cells were electroporated with linearized _targeting vector and selected with Geneticin (G418; Invitrogen) using established protocols developed in the Murine Embryonic Stem Cell Core of the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital (details are available at http://escore.im.wustl.edu). A total of 96 G418-resistant ES cell clones were analyzed for homologous recombination by Southern blotting, and three clones were found to be positive. Positive ES clones were karyotyped and microinjected into 3.5-day postcoitus (dpc) C57BL/6 blastocysts, which subsequently were implanted into the uteri of pseudopregnant C57BL/6 × C3HF1 foster mothers. Male chimeras selected by the percentage of agouti color were mated to C57BL/6 females. Germ line transmission was determined by agouti coat color. F1 animals were tested for the _targeted Par-1a allele by Southern blotting and the PCR analysis of tail DNA. Heterozygous (HET) F1 males/females were interbred to generate the F2 littermates used for subsequent breeding and analysis and are referred to as wild-type, heterozygous, and null pups/mice.
Western blotting.
Tissues from wild-type and knockout animals were homogenized in 800 μl of mammalian cell lysis buffer (MCLB; 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 0.1 M NaCl, 0.5% NP-40, 2 mM dithiothreitol [DTT]) containing 1 μM microcystin, 1 mM sodium orthovanadate, 10 mM β-glycerol phosphate, 1 mM sodium fluoride, 2 μM phenylmethylsulfonyl fluoride (PMSF), and protease (Sigma, St. Louis, MO) and phosphatase inhibitor cocktails (Calbiochem, Gibbstown, NJ). Homogenates were rocked for 15 min at 4°C and then clarified twice by centrifugation. Clarified lysates were resolved by SDS-PAGE on a 10% SDS gel, and Western blotting was carried out with antibodies specific to actin (1:4,000; Sigma Chemical Co.), tubulin (1:1,000; Sigma), β-catenin (1:10,000; BD Biosciences/Pharmagen), Par-1a ascitic fluid (1:10,000) (45), glycogen synthase (1:1,000; Cell Signaling), phosphoglycogen synthase (1:1,000; Cell Signaling), and LC-3 (1:1,000; Cell Signaling). Par-1b was detected with ascites generated using a monoclonal antibody produced against keyhole limpet hemocyanin (KLH)-conjugated peptide corresponding to amino acids 288 to 307 (PDYKDPRRTELMVSMGYTRE) of human Par-1b. Bound primary antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Jackson, West Grove, PA), and proteins were visualized by chemiluminescence.
Tissue distribution.
The distribution of Par-1a and Par-1b in various tissues was examined using a modified Western blot protocol. In brief, mouse tissues harvested from ∼6-month-old mice were homogenized in 0.8 ml of mammalian cell lysis buffer (as described above). Lysates were rocked for 15 min at 4°C, followed by centrifugation for 10 min at 20,800 × g. Clarified supernatants were transferred to prechilled tubes. Proteins were resolved by SDS-PAGE on 7.5% gels until the 50-kDa molecular mass marker was at the bottom of the gel. Proteins were transferred to nitrocellulose membranes in CAPS buffer [0.1 M 3-(cyclohexylamino)-1-propanesulfonic acid and 10% methanol (C2632; Sigma, St. Louis, MO)] at 1.2 mA for 1.5 h. Membranes were blocked in 1× TBST (100 mM Tris, pH 8.0, 1.5 M sodium chloride, 2% Tween 20) containing 5% milk for 1 h. Membranes were probed with Par-1a ascites (1:10,000), Par-1b ascites (1:1,000), actin (1:4,000; Sigma Chemical Co.), or β-catenin antibody (1:30,000) by being rocked overnight at 4°C. Membranes were washed four times for 13 min each in 1× TBST. Membranes then were incubated with a goat anti-mouse antibody (dilution, 1:30,000; Jackson, West Grove, PA) for 1 h at room temperature. Membranes then were washed four times in 1× TBST for 13 min each. Blots were developed using ECL detection reagent (GE Healthcare, Piscataway, NJ).
Breeding, animal weights, and growth measurements.
Embryos and pups generated from heterozygous crosses were weighed and genotyped by PCR analysis. Plug observation was recorded as 0.5 dpc; embryos were harvested, rinsed, and weighed. Mean body weights at each time point were compiled from at least 6 mice per genotype. Breeding schemes followed standard protocols, and for double-knockout (DKO) experiments genotypes are abbreviated by a two-letter code (the first letter indicates Par-1b status, and the second letter indicates Par-1a status: W, +/+; H, +/−; K, −/−; e.g., HK). Binning and comparison within and between genotypes via univariate and multivariate analyses tested trends over time. We assessed the number and genotype of litters in 10 superovulated timed pregnancies at 8.5 and 10.5 dpc. Offspring ratios for Par-1a−/−, Par-1b−/−, and all genotypes in DKO experiments were recorded as pups/litter and total numbers per genotype. The significance of deviations from expected Mendelian ratios were tested using chi-square statistics with 2 (WT, HET, and KO) or 8 degrees of freedom (KK, WW, WK, KW, WH, HW, KH, HK, and HH) in Par-1a null and DKO experiments, respectively (two-tailed P values are provided).
Metabolic measurements (indirect calorimetry).
Metabolic measurements were determined using previously established protocols (8, 43). Briefly, metabolic characterization included metabolic rate, energy expenditure, activity, food consumption (in g/day), and body temperature. During data analysis, energy expenditure and metabolic rate were normalized with respect to body weight. Energy expenditure and respiratory quotient (RQ) were calculated from the gas exchange data. RQ was the ratio of the rate of the elimination of carbon dioxide (VCO2) to VO2 and the equation for energy expenditure (or heat) was (3.815 + 1.232 × RQ) × VO2. Activity was measured on x and z axes using infrared photobeams to count the number of beam breaks.
HFD studies.
HFD studies followed previously published protocols (43) and consisted of a total of 55 mice. In 35 3-week-old mice (WT, Par-1a−/−, Par-1b−/−), the regular rodent chow diet (CD) was replaced with an adjusted-calories diet (42% fat) from Harlan Teklad (88137; Western diet; Madison, WI). The weight of the HFD chow was recorded, and food consumption as well as mouse weight were determined for individual mice on a weekly basis. Although resistance to weight gain on HFD has been reported previously for Par-1b null mice (43), for the direct comparison and determination of specific organ uptake values, the HFD experiment included a new group of nine Par-1b null mice and therefore consisted of six different trial groups: WT-CD, WT-HFD, Par-1a−/−-CD, Par-1a−/−-HFD, Par-1b−/−-CD, and Par-1b−/−-HFD. Metabolic profiling after a total diet time of 16 weeks consisted of (i) fasting (n = 18 CD, 35 HFD) and blood glucose levels after feeding (n = 6 per genotype, randomly selected), (ii) glucose tolerance testing (GTT) of 18 CD mice (6 per genotype, randomly selected) and all HFD mice, (iii) fludeoxyglucose ([18F]FDG) biodistribution experiments (n = 19 CD, 31 HFD), (iv) organ weights, and (v) histologic examination. The HFD trial was designed to allow at least 24 h between GTT and [18F]FDG biodistribution studies, during which animals were kept on HFD.
Serum factor quantification.
Serum levels of insulin, triglycerides, adiponectin, leptin, and cholesterol were determined by Ani Lytics Incorporated (Gaithersburg, MD; n ≥ 11 animals per genotype and sex); glucagon and free fatty acids were determined according to previously established protocols (18).
Glucose and insulin measurements and tolerance tests.
Randomly fed or fasted mice were analyzed, and blood glucose levels were determined using a β-glucose photometer and β-glucose cuvettes (HemoCue AB, Angelholm, Sweden) or Accucheck advantage glucometer (Roche Diagnostics Corp., Indianapolis, IN). Normoglycemia was defined as blood glucose values between 90 to 130 mg/dl. Intraperitoneal (i.p.) glucose tolerance tests were performed on fasted (12 h) animals on CD and all HFD trial mice. Mice were injected i.p. with d-glucose (20% solution; 1 g/kg of body weight), and blood glucose levels were determined at 15, 30, 60, 90, 120 min, and >24 h postinjection. In addition, insulin levels were determined during GTT immediately before and 15, 30, and 60 min after glucose injection. Therefore, ∼20 to 30 μl of blood was collected using the Microvette 200 capillary blood collection system (Sarstedt, Newton, NC) and centrifuged at 30,000 × g for 5 min. Fasting serum insulin levels were determined for 50 male mice on CD (n = 24 WT, 8 Par-1a−/−, 18 Par-1b−/−) and 18 randomly selected mice on HFD (n = 6 per genotype) using rat insulin as a standard and enzyme-linked immunosorbent assays (ELISA) as specified by the manufacturer (Crystal Chem, Inc., IL). Insulin tolerance tests (ITT) were performed on 6-h-fasted male mice (n = 9 Par-1a+/+, 9 Par-1a−/−). Insulin sensitivity in Par-1b null mice has been reported (43) and was not repeated. Blood glucose values were measured immediately before and at 15-min intervals for a total of 90 min after the i.p. injection of insulin (0.30 IU/kg HumulinR; Eli Lilly and Company, Indianapolis, IN).
MicroPET studies.
Micro-positron emission tomography (microPET) studies entailed the imaging of cohorts of male WT and Par-1a null mice that were repetitively imaged once a week for four consecutive weeks, each time under a different metabolic condition. Brown adipose tissue (BAT), skeletal muscle (SM), heart, liver, and brain were examined under the following four conditions: (i) fasted with saline, (ii) fasted with insulin, (iii) nonfasted with saline, or (iv) nonfasted with insulin. The next morning, either 0.5 U/kg insulin (Humulin N; Eli Lilly, Indianapolis, IN) or saline was administered to mice by i.p. injection. Thirty minutes after the injection, mice were lightly anesthetized with isoflurane, followed by a tail vein injection of [18F]FDG (∼200 to 500 μCi in saline). Immediately after the injection of radiotracer, mice were placed supine in the microPET scanner (Focus120 or Focus220 scanner; Concorde MicroSystems, Knoxville, TN) and imaged (acquisition time, ∼10 min; one bed position; ordered set expectation maximum reconstruction). Mice were allowed to recover and then were anesthetized and imaged again at 1 and 2 h after the injection of radiotracer. The Par-1b null and WT mice used for comparison were age-matched males and treated as described above, except that they were imaged on an R4 scanner (43). MicroPET images were corrected for decay, but not attenuation or scatter, and then stacked regions of interest (ROI) of relevant tissues and organs were analyzed with AnalyzePC 6.0 software. ROI counts were converted to counts per gram of tissue (nCi/g), assuming a tissue density of 1 g/ml. Data for the accumulation of [18F]FDG on microPET images were expressed as standard uptake values (SUV), representing counts per gram of tissue divided by the injected dose of radioactivity per gram of animal weight (87, 88). After statistical analysis using a one-way analysis of variance (ANOVA) and Student's t tests, comparison between genotypes and experiments was enabled by the normalization of the SUV to the corresponding WT littermates, with data for Par-1a- and Par-1b-disrupted mice against their littermate WT as the normalized average SUV ± standard errors of the means (SEM); errors were propagated (see below).
[18F]FDG biodistribution experiments.
As previously reported, white adipose tissue (WAT) cannot be analyzed by noninvasive microPET imaging (43). As an alternative, we examined the glucose uptake of WAT and other organs by traditional [18F]FDG biodistribution (12, 20, 53, 88). Mice were anesthetized with isoflurane (2% vaporized in oxygen), followed by the tail vein injection of [18F]FDG (20 μCi in 100 μl saline). Sixty minutes after [18F]FDG injections, mice were sacrificed by cervical dislocation (under anesthesia). Blood, liver, heart, brain, muscle, WAT, and BAT were rapidly removed and weighed, and their radioactivity was measured using a Beckman 8000 gamma counter. The percent injected dose per gram of tissue (% ID/g) was calculated [(tissue μCi) (injected Ci)−1 (g tissue)−1 × 100] and subsequently compared among tissues, genotypes, and diets. Biodistribution analysis was performed on 31 HFD mice (17 WT, 8 Par-1a−/−, 6 Par-1b−/−) and 19 mice from the CD arm (age-matched control group; total n = 50). Tissue samples from these mice were fresh-frozen and stored at −80°C. Subsequent cryostat sections confirmed tissues and allowed histomorphological/histochemical comparison to tissues obtained from animals that did not undergo [18F]FDG uptake (n = 5; no differences observed) or HFD (see Results). For these HFD trial biodistribution experiments, the 50 mice were prospectively assigned to one of nine experimental days with an average of six animals per experiment (range, 3 to 10 mice/day). Excluding the 16 weeks on HFD (as described above), these experiments were performed during a 10-month period.
Starvation experiments.
Starvation experiments consisted of the determination of blood glucose as well as ketone levels using a MediSense Precision Xtra diabetes glucose and ketone monitor system (Abbott, Alameda, CA). After overnight fasting (20 h), measurements were repeated; one group of mice was subjected to extended fasting (an additional 4 h), and the other group of mice was given an i.p. injection of 2 mg/g glucose for 2 h. Tissues were collected at all time points and samples separated for Western blotting, routine histology, electron microscopy (EM), and glycogen determination.
Dissection and histology.
Organs were excised, weighed, rinsed in phosphate-buffered saline (PBS), fixed in 10% neutral-buffered formalin, rinsed in PBS, and stored in 70% ethanol. Formalin-fixed tissues were paraffin embedded (FFPE) and sectioned (∼5 μm) using standard procedures. For morphological evaluation and fat content, hematoxylin-and-eosin (H&E) and oil red O staining was performed on FFPE and fresh-frozen sections (stored at −80°C), respectively. Morphological analysis was performed using multiple independent samples per site/organ (13 to 26 samples per genotype) as well as all 55 animals in the HFD trial. We quantified BAT and WAT samples taken from the interscapular and gonadal fat pads, respectively. Quantification was performed using digital image processing via ImageJ (v. 1.37; http://rsb.info.nih.gov/ij/) and Photoshop CS3 (Adobe Systems) according to previously established semiautomatic protocols (16, 39). Specifically, oil red O positivity was quantified from ≥20 high-power fields, cell numbers or lipid vacuoles were quantified in 6 to 20 randomly chosen images per sample, and periodic acid Schiff (PAS) staining was quantified using integrated density measurements (IntDen; equivalent to the product of the area and mean gray value, with gray value being the luminescence value between 0 [black] and 255 [white]).
Glycogen determination.
Fifty milligrams of liver and skeletal muscle samples were extracted with 0.9 N perchloric acid and 99% ethanol to precipitate glycogen. The glycogen from the pellet was dialyzed and digested to free glucose with amyloglucosidase. Glucose concentrations were measured by the glucose oxidase method (glucose analyzer II; Beckman Instruments, Fullerton, CA).
EM.
EM was performed according to established protocols (85). Tissues were fixed overnight at 4°C in modified Karnovsky's fixative containing 3% glutaraldehyde and 1% paraformaldehyde in sodium cacodylate buffer, pH 7.4. After being rinsed in sodium cacodylate buffer, samples were postfixed in phosphate cacodylate-buffered 21% OsO4 for 1 h, dehydrated in graded ethanols with a final dehydration in propylene oxide, and embedded in Embed-812 (Electron Microscopy Sciences, Hatfield, PA). One-micron-thick plastic sections were examined by light microscopy after being stained with toluidine blue. Ultrathin sections (90 nm thick) of individual samples were cut onto formvar-coated slot grids, which permit the visualization of larger cross-sections. Sections were poststained with uranyl acetate and Venable's lead citrate and viewed with a JEOL model 1200EX electron microscope (JEOL, Tokyo, Japan). Digital images were acquired using the AMT advantage HR (Advanced Microscopy Techniques, Danvers, MA). Plastic sections as well as an ultrastructural review were performed blinded to the genotype.
Digitization and image analysis.
Analyses were performed using an Olympus DP70 digital camera (Olympus, Tokyo, Japan) connected to an Olympus BX51 light microscope or a Scanscope XT whole-slide scanner/ImageScope v10.0.36.1805 (Aperio, Vista, CA). Image and pixel quantification employed established semiautomatic threshold algorithms (60) using the software package ImageJ (http://rsb.info.nih.gov/ij) (1). Statistical testing between groups was performed using Student's t test and the Wilcoxon two-sample test.
Statistics.
Chi-squared t tests (two-tailed), the nonparametric alternative Mann-Whitney U test, one-way and two-way ANOVA, and Bonferroni posttests to compare replicate means were used when appropriate. In experiments with small numbers of samples (defined as n ≤ 12), the D'Agostino-Pearson omnibus K2 normality test (24) was used to approximate the Gaussian distribution of values. P values of <0.05 indicate non-Gaussian distribution, and we assessed these experiments with both parametric and nonparametric tests; both P values are provided when applicable. In all studies, the accepted level of significance was P < 0.05, and data are reported as means ± SEM. In selected experiments and for comparison between strains and experiments, values were normalized to the corresponding WT and errors were propagated according to the formula σx̄WT/x̄WT)2 + (σx̄KO/x̄KO)2] 1/2 × x̄NKO, where σ̄ is the SEM, x̄ is the average, and N is the normalized average. N̄KO was adapted from reference 66.
Error propagation and statistical tests in general were chosen assuming the most pessimistic situation (80). All data were analyzed using Prism 5.0b (GraphPad Software Inc., La Jolla, CA) or Microsoft Excel 2008 (version 12.1.9; Microsoft Corporation, Redmond WA).
RESULTS
Gene _targeting was employed to determine the consequences of Par-1a loss in mice (Fig. 1A). The _targeted Par-1a allele was introduced by electroporation into RW4 embryonic stem (ES) cells derived from the mouse strain 129X1/SvJ. Southern blot analysis identified correct _targeting events (Fig. 1B). Southern blotting and PCR analysis demonstrated that the agouti offspring produced by the chimeric males were heterozygous for the Par-1a locus (data not shown). F1 heterozygous offspring were intercrossed, and F2 offspring were genotyped by PCR (Fig. 1C). Western blotting demonstrated that the _targeted disruption of Par-1a produced a null allele of the locus (Fig. 1D), and multitissue Western blotting confirmed the ablation of Par-1a/b in all tissues of the corresponding null-mice (Fig. 1E).
Higher levels of Par-1a were noted in several tissues of Par-1b null mice, including brain, BAT, thymus, and muscle (Fig. 1E, lanes 2, 5, 17, and 26), which may provide an explanation for Par-1a compensation of Par-1b loss (see below). However, increased Par-1b levels were not observed in Par-1a null tissues. Also observed in Fig. 1E is the differential expression of Par-1a splice variants in various mouse tissues. The NCBI database contains five Par-1a splice variants with their respective protein products, ranging from 659 to 753 amino acids. It is not known whether functions differ among the various splice variants.
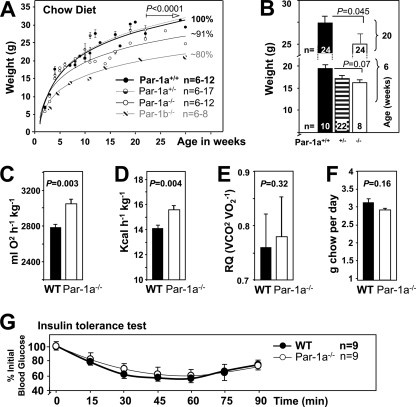
Par-1a−/− mice are hypermetabolic and show 9% reduction in body weight.
When Par-1a heterozygotes were intercrossed, the offspring contained WT, HET, and KO pups, although genotype proportions deviated significantly from expected Mendelian ratios with the loss of homozygous KO mice (n = 750; WT, 30%; HET, 52%; KO, 17%; χ2 = 27.864, P < 0.0001). All pups developed without any visible dysmorphisms or obvious external pathology, and animals survived to adulthood. Par-1a null mice exhibited significant weight differences compared to their wild-type littermates (e.g., at 32 weeks, WT mice [n = 6] were 33.4 ± 0.94 g and Par-1a−/− mice [n = 6] were 25.5 ± 0.42 g; P < 0.0001, t test). In contrast to Par-1b null mice that show reduced weights at 6 weeks of age (9, 42), in Par-1a null mice the difference was not discernible this early (Fig. 2 A and B).
FIG. 2.
Par-1a−/− mice are growth retarded and hypermetabolic. (A) Body weights of mice from birth to 30 weeks of age (n = 4 to 14 per genotype per time point). (B) Body weights at 6 and 20 weeks. Body weights of Par-1a+/+, Par-1a+/−, and Par-1a −/− mice at 6 weeks of age are shown (number of animals per sex indicated). Also shown are metabolic rate (C), energy expenditure (D), respiratory quotients (RQ = VCO2/VO2) (E), and food intake (F) in WT and Par-1a null mice. (G) Insulin tolerance tests were performed by the i.p. injection of 0.30 U/kg insulin into WT and Par-1a−/− mice; tail bleeds were obtained and glucose levels were monitored at the indicated time points. Glucose levels were plotted as percent blood glucose at time zero before injection.
Body weight represents a net balance of food intake and energy expenditure. Based on the decreased weight in Par-1a null mice, we measured total energy expenditure (EE), total metabolic rate (MR), and respiratory quotients of male mice on a regular chow diet. Par-1a null mice exhibited higher O2 consumption (MR showed an ∼9% increase) (Fig. 2C) and higher EE (10% increase) (Fig. 2D). We also analyzed the respiratory quotient (RQ = VCO2/VO2) as a measure of fuel-partitioning patterns, and RQ ranged from 0.76 to 0.78 (P = 0.32, t test) (Fig. 2E). Food intake in Par-1a null mice was slightly less than that of WT littermates, although this difference was not statistically significant (WT, 3.15 ± 0.09; Par-1a−/−, 2.94 ± 0.04; P = 0.16, t test) (Fig. 2F). Thus, unlike Par-1b null mice, Par-1a null mice are not hyperphagic (43). The lack of effect on RQ suggests that there was no difference in fuel selection between carbohydrates and lipids, leading us to explore whether the increase in energy expenditure occurs secondarily to increased thermogenesis. However, body temperatures of Par-1a null mice were similar to those of WT littermates (WT, 34.3 ± 0.5; Par-1a−/−, 34.1 ± 0.8; P = 0.8, t test), and the expression of uncoupling proteins in selected tissues showed no significant difference (not shown). Lastly, Par-1a null mice showed similar levels of activity (compared to that of WT mice; P = 0.96), and basic sensorimotor testing as well as open-field tests showed no significant difference (P = 0.64 [not shown]).
Par-1a−/− mice are normoglycemic and normoinsulinemic and show no insulin sensitivity.
Serum levels of insulin, glucagon, leptin, adiponectin, triglycerides, cholesterol, and free fatty acids in Par-1a null mice were measured and compared to values from WT and Par-1b null mice (20-week-old mice on regular chow diet) (Table 3). Male Par-1a null mice exhibited levels of adiponectin that were 1.6-fold higher than those of WT littermates. Par-1b null mice showed a significant decrease in female adiponectin values. In contrast to Par-1b null mice, Par-1a null mice showed no difference in serum insulin levels (Table 3), and blood glucose was normal (WT, 152 ± 18 mg/dl; Par-1a−/−, 147 ± 5 mg/dl; P = 0.216, t test). Par-1a null mice on CD exhibited ITT curves identical to those of WT littermates (Fig. 2G; P = 0.87, one-way ANOVA). On CD, Par-1a null mice showed no significant difference in GTT relative to that of WT littermates (Fig. 3 C) (P = 0.15, t test; and P = 0.181, two-way ANOVA for the entire time course; Par-1b−/− data not shown).
TABLE 3.
Metabolic characteristics of WT, Par-1a−/−, and Par-1b−/− animalsa
| Animal typed | Insulin (ng/ml) | Glucagon (pg/ml) | Leptin (ng/ml) | Adiponectin (μg/ml) |
Triglyceride (mg/dl) | Cholesterol (mg/dl) | NEFAc (μmol/liter) | |
|---|---|---|---|---|---|---|---|---|
| Male | Female | |||||||
| WT | 0.65 ± 0.10 | 45.5 ± 11.7 | 3.68 ± 0.26 | 9.8 ± 0.66b | 17.9 ± 1.7 | 74.9 ± 3.46 | 83.27 ± 2.4 | 1,303 ± 65.8 |
| Par-1a−/− | 0.61 ± 0.08 | 31 ± 4.33 | 3.64 ± 0.16 | 16.5 ± 1.6 | 18.4 ± 2.2 | 73.1 ± 3.77 | 74.95 ± 2.87 | 1,329 ± 107 |
| Par-1b−/− | 0.37 ± 0.04 | 34 ± 2.25 | 3.02 ± 0.16 | 11.4 ± 1.3 | 11.7 ± 0.84 | 62.2 ± 3.39 | 73.45 ± 2.81 | 1,149 ± 66.3 |
Twenty-week-old mice on a regular chow diet were fasted overnight (n ≥ 11 per genotype, all male unless otherwise indicated); serum values are expressed as the means ± SEM of measurements obtained.
Male mice had significantly lower adiponectin levels (P = 0.0003). Note that this gender difference is lost in both Par-1 mice.
NEFA, nonesterified fatty acids.
P values were determined by Student's t tests comparing Par-1 mice to the WT. P values for WT and Par-1a−/− mice were the following: insulin, 0.74; glucagon, 0.27; leptin, 0.79; adiponectin (male), 0.0008; adiponectin (female), 0.87; triglyceride, 0.74; cholesterol, 0.03; NEFA, 0.84. Values for adiponectin (male) and cholesterol were significant. P values for Par-1b−/− mice were the following: insulin, 0.04; glucagon, 0.39; leptin, 0.16; adiponectin (male), 0.28; adiponectin (female), 0.005; triglyceride, 0.013; cholesterol, 0.011; NEFA, 0.11. Values for adiponectin (female), triglyceride, and cholesterol were significant.
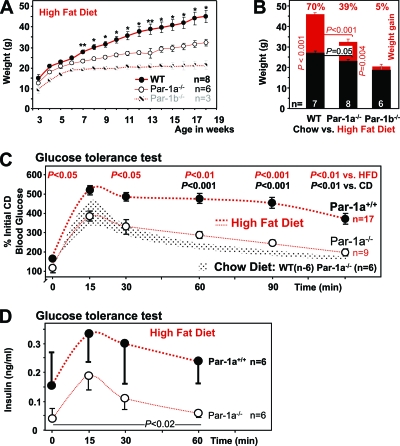
FIG. 3.
Resistance of Par-1a−/− mice to high-fat diet-induced metabolic changes. (A) Body weights during 15 weeks of HFD starting at 3 weeks of age in male mice (*, P < 0.001; **, P < 0.005). (B) Body weight gain (as percentages) indicated above weight comparisons of the control group (black/white/gray) and the HFD arm of the trial (red) in 18-week-old male mice. (C) Glucose tolerance testing was performed by the i.p. injection of d-glucose at 1 mg/g body weight into all genotypes on chow and high-fat diet. Glucose levels were monitored as indicated and expressed in percentages of initial glucose normalized to their littermates on chow; for comparison, a dotted band shows the time course for WT and Par-1a−/− mice on chow diet. Statistical testing was performed using original values, and P values are provided for testing between genotypes on HFD and between WT-HFD and WT-CD, when significant. (D) Serum insulin levels as determined by ELISA for the first four time points of the glucose tolerance test after HFD.
Par-1a−/− mice show delayed weight gain on high-fat diet.
The striking resistance to HFD-induced weight gain in Par-1b null mice (43) prompted us to challenge Par-1a null mice with a similar HFD for 16 weeks (Fig. 3A and B). The main finding was the reduced weight gain of Par-1a null mice compared to that of WT littermates on HFD (Fig. 3A). Specifically, while Par-1b null mice on HFD showed a statistically significant lack of weight gain after 7 weeks [designated t(7w); t(7w) = 3.297; P < 0.05; t(17w) = 7.234; P < 0.001, two-way ANOVA with Bonferroni posttests; weight gain, +5%; weight on CD, 19 ± 0.75 g; weight on HFD, 20.3 ± 1.34; P = 0.4097, t test], Par-1a null mice on HFD continued to gain weight during the entire experiment (Par-1a−/− weight gain, +39%; CD weight, 23.47 ± 1.6 g; HFD weight, 32.4 ± 1.47; P = 0.004, t test), albeit at a reduced rate and significantly different from that of WT littermates (+70% weight gain; CD weight, 27.12 ± 0.5 g; HFD weight, 46 ± 0.9; P = 0.001, t test). HFD food intake of Par-1a null mice was similar to that of WT littermates (P = 0.39, t test). While the Par-1a phenotype with a reduced rate of weight gain is, at first glance, less dramatic than that observed in Par-1b null mice [Par-1a−/− versus WT: t(all) = 2.779, P < 0.05, one-way ANOVA with Bonferroni's multiple comparison test], it is a highly significant finding (P = 0.001) (Fig. 3B). Par-1a null mice required 7 additional weeks of HFD to reach the original weight of their WT littermates.
Par-1a−/− mice show resistance to glucose intolerance on high-fat diet.
Par-1a null mice fed an HFD were subjected to glucose tolerance testing (Fig. 3C). As expected, WT mice on HFD exhibited glucose intolerance relative to that of WT mice on CD, with a prolonged elevation of blood glucose of ∼400% from 15 min onwards (t value = 3.645, P < 0.01, two-way ANOVA). Strikingly, Par-1a null mice exhibited improved glucose tolerance on HFD relative to that of WT controls on the same diet (P = 0.041, t test [two genotypes]; t value = 3.38, P < 0.01, one-way ANOVA [all genotypes]) (Fig. 3D). GTT response in Par-1a null mice under HFD conditions was essentially identical to that of WT and Par-1a null mice on CD (P = 0.4781, t test [two genotypes]; t value = 0.346, P > 0.05, one-way ANOVA [all genotypes]) (Fig. 3D and data not shown). We conclude that Par-1a disruption leads to the maintenance of normal glucose tolerance even under HFD conditions; accordingly, Par-1a null mice on HFD were normoglycemic (P = 0.3, t test). We next examined serum insulin levels in HFD-fed mice during the first 60 min of the GTT (Fig. 3D) and found significantly lower levels in Par-1a null mice (t value = 3.16, P = 0.0194, t test [two genotypes]; F [mean square between groups over the mean square within groups] = 6.454, P = 0.018, one-way ANOVA). Although we did not formally assess insulin tolerance in Par-1a null mice on HFD, the combination of improved glucose tolerance and reduced insulin secretion during the GTT is indicative of peripheral insulin hypersensitivity. Thus, Par-1a null mice demonstrate normal insulin and glucose sensitivity unless challenged by HFD. In contrast, Par-1b null mice exhibited insulin hypersensitivity even on CD (43). The direct assessment of pancreatic islets has proven difficult with Par-1 mice (38); however, morphological and morphometric differences were not observed in the islet organ of Par-1a null mice (not shown).
Par-1a−/− mice show diet-dependent reduced adiposity.
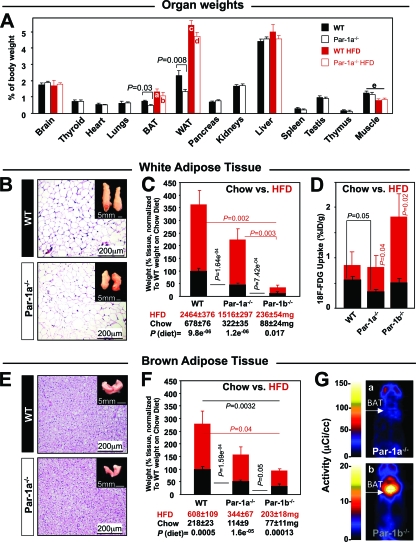
We determined whether the reduced body weight of Par-1a null mice was reflected disproportionately in different tissues (Fig. 4 A). Measurements of tissue weights in Par-1a null mice indicated that decreased weights of most organs were proportional to total body weight and consistent with a decrease in overall body mass rather than in specific organs. However, two tissues did not follow this rule and were disproportionately smaller: white adipose tissue (WAT from gonadal fat pads) and brown adipose tissue (BAT). We also performed the dissection of tissues in the mice on HFD and, as expected, the energy excess in WT mice led to a disproportionate increase in overall adiposity (BAT-CD versus BAT-HFD, P = 0.001; WAT-CD versus WAT-HFD, P = 2.5e−5; t tests) (Fig. 4A). Par-1a null mice showed a similar disproportionate increase in overall adiposity (BAT-CD versus BAT-HFD, P = 0.007; WAT-CD versus WAT-HFD, P = 1.4e−9; t tests) with a weight increase in BAT and gonadal WAT that eliminated the reduced adiposity observed under chow diet (Fig. 4A, columns a versus b and c versus d, both P = 0.08, t tests).
FIG. 4.
Reduced adiposity and decreased glucose uptake in Par1a−/− mice. (A) Tissue weights normalized to total body weight in the indicated tissues from 18-week-old male WT and Par-1a−/− mice fed chow diet (black) or HFD (red; selected tissues); n ≥ 7 samples per tissue. For an explanation of labels a to e, see Results. (B) Hematoxylin and eosin (H&E) stain of WAT from Par-1a−/− and Par-1a+/+ mice; insets, gonadal fat pads. (C) Weight comparisons of WAT between WT, Par-1a−/−, and Par-1b−/− mice on chow (black) and after 16 weeks of high-fat diet (red); weights were normalized to that of the WT on a chow diet. Statistical tests and indicated P values derived from original weights (provided below bars). (D) Invasive biodistribution experiments following the injection of [18F]FDG and the dissection of gonadal WAT 1 h postinjection; values are expressed as the percentage of injected dose per gram of tissue (%ID/g). (E) H&E stain of BAT from Par-1a−/− and Par-1a+/+ mice; insets, interscapular fat pads. (F) Weight comparisons of BAT between WT, Par-1a−/−, and Par-1b−/− mice on CD (black) and after 16 weeks of a high-fat diet (red); weights were normalized to that of the WT on a chow diet. Statistical tests and indicated P values derived from original weights (provided below bars). (G) Representative coronal section microPET images of [18F]FDG uptake in Par-1a−/− (top) and Par-1b−/− mice (bottom) 1 h after [18F]FDG injection.
The dissection of the gonadal fat pads (Fig. 4B) representative of the WAT showed a disproportionate decrease in the fat mass of Par-1a null mice relative to that of WT mice on CD (Fig. 4C; raw weight values provided). The weight difference corresponds to ∼48% (Par-1a−/−) and ∼36% (Par-1b−/−) of the WT-WAT weight (Fig. 4C). Although we did not quantitatively assess abdominal fat, during dissection the dimensions of mesenteric root, omental, and retroperitoneal fat content were strikingly lower in Par-1a null mice, and this difference is not accounted for in the displayed values of gonadal WAT (Fig. 4C).
The dissection of the interscapular fat depot, representative of BAT, is shown in Fig. 4E. The weight difference corresponds to ∼52% (Par-1a−/−) and ∼38% (Par-1b−/−) of the WT-BAT weigh (Fig. 4F). For both adipose tissues, the difference significantly exceeded the overall ∼9% weight difference observed in Par-1a null mice. Adipocyte counts from six independent samples of WAT and BAT indicated no differences in adipocyte size from WT and Par-1a null mice (for WAT, WT mice showed 436 ± 100 adipocytes per mm2 and Par-1a−/− mice showed 419 ± 74; P = 0.137, t test; for BAT, WT mice showed 1,894 ± 349 and Par-1a−/− mice showed 1,385 ± 434; P = 0.382, t test). Thus, we conclude that the observed decrease in the adipose tissue of Par-1a null mice is due to decreases in total adipocyte cell number, not cell size, findings similar to those made for Par-1b null mice (43).
We also determined the weight of muscle samples (tibialis anterior, gastrocnemius, plantaris, diaphragm, and extensor digitorum longus) typically used for the assessment of myogenic phenotypes and fiber type composition (22, 84), which, under HFD conditions, showed significantly decreased weights (WT, P = 3.7e−6; Par-1−/−, P = 0.006; both t tests) (Fig. 4A, column e). Skeletal muscle plays an integral role in the coordination of fuel homeostasis (41) and is the most important site of insulin-stimulated glucose disposal (26). We therefore performed functional (motor, biodistribution, and microPET analysis), biochemical (glycogen quantification), and morphological (routine, PAS, fiber type analysis, and ultrastructure) screening of skeletal muscle but observed no differences between wild-type and Par-1a null mice (not shown). Thus, the reduced body weight of Par-1a mice on HFD likely is accounted for by a combination of reduced muscle mass (3, 92) and reduced adiposity, although the latter is not reflected in the gonadal WAT measurements (Fig. 4A, columns c and d).
Par-1a loss causes decreased WAT glucose uptake in the absence of BAT phenotype.
MicroPET imaging of WAT was performed (not shown); however, the reduced size of Par-1 null mice and high tracer concentrations in kidneys, bladder, and skeletal muscle precluded the meaningful quantification of uptake in retroperitoneal, gonadal, and calf fat deposits, respectively. We therefore applied invasive biodistribution analysis, and in contrast to Par-1b null mice, WAT of Par-1a null mice showed significantly decreased glucose uptake under normal dietary conditions (WT, 0.52 ± 0.08%ID/g [n = 9]; Par-1a−/−, 0.32 ± 0.05%ID/g [n = 8]; U [sum of signed ranks] = 14; P = 0.036, Mann-Whitney U test; P = 0.05, t test) (Fig. 4D). When challenged with HFD, Par-1a null mice showed a statistically significant increase in [18F]FDG uptake compared to that of Par-1a null mice on chow diet that eliminated the difference of glucose uptake from that of WT mice on HFD (Fig. 4D).
We next examined glucose uptake in BAT of CD-fed Par-1a null mice by microPET analyses of the interscapular BAT (Fig. 4G). We evaluated four different conditions at 1 h after [18F]FDG injection: the fed and fasted state using acute insulin challenge versus saline administration. As part of the HFD trial, we also examined BAT glucose uptake via biodistribution analysis in all genotypes. We found that the loss of Par-1a did not significantly alter BAT glucose uptake in any of these six tested conditions (not shown).
Although Par-1a null mice exhibited reduced fat mass on chow diet and delayed weight gain relative to that of WT mice on HFD, sampled gonadal adipose mass in Par-1a null mice was not significantly different from that of WT mice after 16 weeks of HFD conditions (P = 0.12 and 0.13, both t tests; for WAT/BAT, n = 8 Par-1a−/− mice and 17 WT mice) (Fig. 4C and F). In contrast, the BAT/WAT mass in Par-1b null mice remained reduced under HFD conditions compared to that of WT mice on the same diet (P = 0.002 and 0.04, t tests [WAT/BAT]; n = 6 for Par-1b−/− and 17 for WT) (Fig. 4C and F).
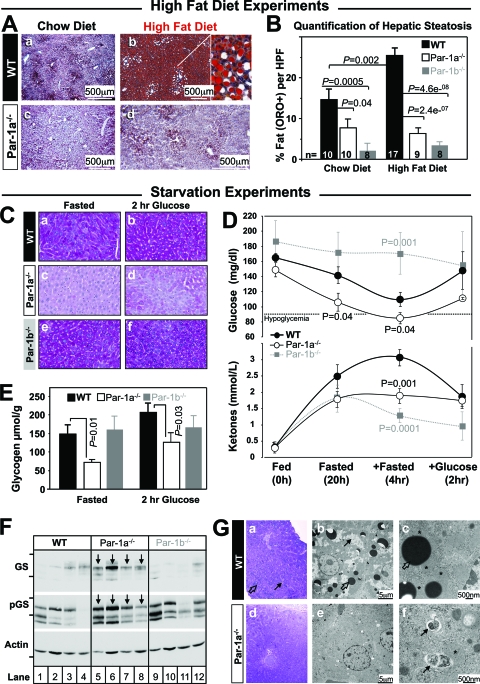
Par-1a−/− mice show resistance to hepatic steatosis.
The chronic exposure of mice to HFD causes liver injury via the accumulation of lipids, leading to fatty liver (steatosis) (13, 79). The absence of steatosis in Par-1a null mice was apparent from the gross examination of the liver, which was notably darker than that of the WT littermates (not shown), and histologic examination showed abundant panlobular, macrovesicular steatosis in WT (Fig. 5 A, image b, inset) but not Par-1a null livers (Fig. 5A, image d). Remarkably, with respect to hepatic steatosis, Par-1a null mice on HFD were no different than WT mice on CD (Fig. 5A, compare image d to image a). Based on previous findings (43), we quantified hepatic lipid content in fresh-frozen sections stained with oil red O and found 14.6 to 25% in WT versus 6.4 to 7.7% in Par-1a null mice when expressed as percent fat per high-power field (Fig. 5B). The quantitative difference in the WT on chow diet (Fig. 5B) corresponds to an absence of centrilobular (zone III) lipid accentuation in Par-1a null mice (Fig. 5A, compare image a to image c). In addition, biodistribution studies demonstrated identical [18F]FDG uptake in livers of Par-1a null and WT mice on CD and HFD (not shown). Similarly, the microPET examination of the liver using four different metabolic conditions (see Materials and Methods) did not show significant differences (P value range, 0.12 to 0.71; t tests). These findings indicate that Par-1a null mice show resistance to steatosis in the absence of altered hepatic glucose uptake under both normal and HFD conditions.
FIG. 5.
Liver findings in high-fat diet and starvation experiments. (A) Morphological comparison of oil red O-stained sections shows resistance to hepatic steatosis in Par-1a−/− mice. The inset shows abundant macrovesicular steatosis. (B) Semiautomated pixel quantification, expressed as the percentage of oil red O-positive structures (fat) per high-power field (HPF); genotypes, numbers of animals, and diet are indicated. (C) Periodic acid Schiff (PAS) staining of liver of 20-week-old WT, Par-1a−/−, and Par-1b−/− littermates following overnight fasting or 2 h after the administration of 2 mg/g glucose by i.p. injection. Images are representative of 96 fields from eight animals. (D) WT mice show a physiologic decrease in blood glucose in response to overnight starvation, with a nadir at the extended fasting time point (20 + 4 h [top]) and corresponding increased ketone production (bottom); note the restoration of blood glucose and ketones 2 h after intraperitoneal glucose injection (2 mg/g). Par-1a−/− mice show significantly lower blood glucose levels at both time points with associated hypoglycemia at 20 + 4 h. Fasting hypoglycemia is accompanied by a significant decrease in ketone levels (hypoketotic hypoglycemia). Par-1b−/− mice maintained glucose levels and exhibited expected lower ketone levels. (E) Glycogen content was measured in livers from 20-week-old Par-1a−/−, Par-1b−/−, and WT littermates following overnight fasting or 2 h after the i.p. injection of 2 mg/g glucose. Tissues were extracted and acid hydrolyzed, and glycosyl units were assayed using an amyloglucosidase/glucose oxidase method (see Materials and Methods). (F) Western blot for glycogen synthase (GS) and phosphorylated GS (pGS) in overnight fasted liver samples of four different animals per genotype. Weight markers: GS/pGS, 100 (top) and 75 kDa (bottom); actin, 37 kDa. Arrows indicate Par-1a−/−. (G) Ultrastructural characterization of liver tissue from WT (images a to c) and Par-1a−/− littermates (images d to f) after nutrient starvation for 24 h. (a) Toluidine blue-stained plastic section of starved WT liver at low magnification showing the accumulation of lipid droplets (open arrow) and glycogen (solid arrow). (b) Electron microscopic examination of WT hepatocytes at low-power magnification showing a large number of variable-sized lipid vacuoles (open arrow) and glycogen islands (solid arrow). (c) Detail of cytoplasmic organization with lipid droplets (open arrow) surrounded by numerous mitochondria (asterisks). (d) Toluidine blue-stained plastic section of starved Par-1a−/− liver at low magnification shows the absence of lipid droplets and glycogen. (e) Electron microscopic examination of Par-1a−/− hepatocytes at low-power magnification showing the absence of glycogen and lipid vacuoles. (f) Detailed EM view of cytoplasmic organization with numerous autophagosomes (arrows) consisting of isolated membranes surrounding cytosolic material (top, so-called myelin figures; bottom, ingested rough endoplasmic reticulum). Note the close proximity of autophagocytic vacuoles and mitochondria (asterisk).
Decreased glycogen deposition in Par-1a−/− livers leads to hypoketotic hypoglycemia.
The storage and utilization of glucose is one of the main contributors to the pathophysiologic changes in diabetes (21, 90). PAS staining was used to detect glycogen content (Fig. 5C). Fasted WT liver showed partially depleted liver glycogen stores (Fig. 5C, image a), whereas liver sections from fasted Par-1a null mice exhibited the complete depletion of hepatic glycogen content (PAS quantification [fasted]: WT, 23.6 ± 8.5; Par-1a−/−, 0.17 ± 0.13; P = 0.01, t test) (Fig. 5C, image c). This depletion was not observed in Par-1b null mice (26.68 ± 2.8; P = 0.73, t test) (Fig. 5C, image e). Two hours after glucose administration, glycogen levels were substantially elevated and visible and confluently PAS positive in livers of WT mice (Fig. 5C, image b). In contrast, Par-1a null mice showed the absence of the restoration of hepatic glycogen content with only spotty PAS-positive islands (Fig. 5C, image d). The quantitative enzymatic analysis of glycogen was consistent with the PAS staining such that fasted and glucose-stimulated liver glycogen was significantly decreased in Par-1a null animals (Fig. 5E) with no significant difference in Par-1b null mice. After mice were fed, glycogen levels were normal and no significant histological differences were observed between groups (not shown).
To test the physiologic significance of hepatic glycogen depletion in Par-1a null mice, we performed an extended fasting experiment and observed significantly decreased glucose levels in Par-1a null mice after overnight fasting (20 h) compared to that of WT littermates (Fig. 5D). After four additional hours without external energy sources, Par-1a null animal blood glucose levels further declined (P = 0.04 compared to results for the WT mice; t test), and the mice developed hypoketotic hypoglycemia (Fig. 5D). In contrast, Par-1b null mice showed an absence of physiologic decline of blood glucose levels with appropriately low ketone levels (Fig. 5D).
To test whether the tissue differences in glycogen levels relate to glycogen synthesis, we determined total and phosphoglycogen synthase (pGS) levels (Fig. 5F) and found increased levels of GS and pGS in the livers of Par-1a null mice. Since Par-1a null mice demonstrate resistance to steatosis as well as defective gluconeogenesis, we probed for UCP2 in fasted and nonfasted liver samples but found similar protein levels (not shown). Most recently, autophagy has been linked to lipid (91) as well as glycogen/glucose homeostasis (55, 56). Therefore, we monitored LC-3 as a marker for membrane formation and early stages of autophagy but observed no differences (not shown). The electron microscopic examination of livers from fasted WT mice demonstrated irregularly shaped glycogen islands (Fig. 5G, images a and b) and variably sized lipid vacuoles (5G, image b) in close proximity to mitochondria with normal morphology (Fig. 5G, image c). In striking contrast, livers from fasted Par-1a null mice did not have lipid vacuoles, were completely depleted of glycogen (Fig. 5G, images d and e), and had abundant autophagocytic vacuoles throughout the hepatocytes (Fig. 5G, images e and f). These autophagic vacuoles were normally configured and frequently associated with mitochondria (Fig. 5G, image f). Our ultrastructural findings with WT mice indicate a catabolic state, whereas the findings in Par-1a null liver are indicative of more extreme energy deprivation with the self digestion of organelles (Fig. 5G, image f).
Par-1b/Par-1a double mutants are not viable, and at least one allele is necessary for embryonic survival.
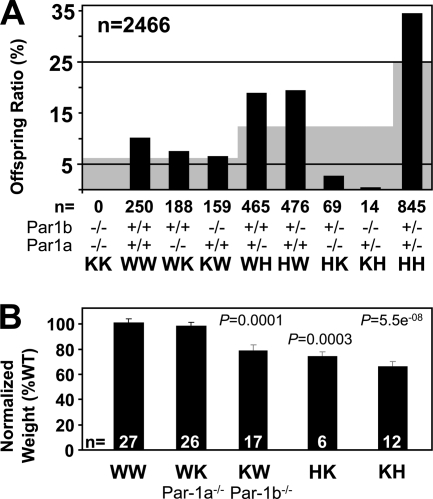
Cross-breeding experiments were performed to obtain mice disrupted for both Par-1a and Par-1b. After an initial experiment where 24 intercrosses were performed and only six of the nine possible genotypes were obtained, genotype frequencies in ongoing HH × HH crosses were monitored. Eventually two of the three missing genotypes were obtained (Fig. 6A) but at a substantial deviation from expected Mendelian ratio (probability of 2.176911e−198 using the chi-square test with 8 degrees of freedom) and with a complete absence of mice lacking both Par-1a and Par-1b (KK). In addition, there was a drastic reduction in the numbers of KH and HK mice (0.5 and 2.7%, respectively). Instead of the expected 12.5% (corresponding to an estimated 308 mice; gray background in Fig. 6A), we obtained a total of 14 KH and 69 HK mice (out of 2,466). Pregnant females that had been superovulated from HH intercrosses were sacrificed, and fetuses at 10.5 (n = 4) or 8.5 (n = 4) dpc were isolated. None of the obtained sets of embryos (total n = 31) included the KK genotype (expected frequency, ∼2 mice). All other genotypes were obtained (not shown). These results illustrate that Par-1b/Par-1a double mutants are not viable and that at least one allele of either Par-1a or Par-1b is necessary for viability and likely embryonic survival.
FIG. 6.
Par-1b−/−/Par-1a−/− double knockout mice are not viable, and breeding experiments support ranked redundancy between isoforms. (A) Survival analysis of the offspring from matings of double-heterozygous Par-1a/b mutant mice. The black bars plot the observed frequency of the indicated genotypes as a percentage of the total, whereas the gray background indicates the expected frequency based on Mendelian inheritance (n = 2,466). (B) Comparison of body weights for five of the obtained genotypes from panel A. Weight is normalized to that of WT littermates (HK, n = 6 mice at 15 to 20 weeks versus control WT [n = 6]; KH [embryonic day 17 postcoitus], n = 12 versus age-matched control WT [n = 8; from the same litter]).
When normalized to age-matched WT controls, Par-1b null mice are ∼20% reduced in body weight (43) and Par-1a null mice are ∼9% reduced in body weight (this study). The loss of one Par-1a allele in Par-1b null mice (KH) led to a more-pronounced reduction in body weight (30%) than that observed in Par-1a null mice containing a single allele of Par-1b (HK, 23%) (Fig. 6B). HK and KH mice were not healthy, and a high proportion of these animals died shortly after birth (not shown). The severity of phenotypes observed in each genotype followed the order KK > KH > HK > KW > WK > WW and suggests an intriguing gene dosage effect at the organism level.
DISCUSSION
This study describes metabolic phenotypes arising in mice disrupted for Par-1a and the consequences of intercrossing of Par-1a- with Par-1b-deficient mice. Par-1a null mice are hypermetabolic and show reduced body weight, decreased adiposity, resistance to hepatic steatosis, and hypofertility; these features are reminiscent of the Par-1b null phenotype (43). Metabolic phenotypes of Par-1a null mice not shared by Par-1b null mice include (i) hepatic glycogen depletion after starvation associated with hypoketotic hypoglycemia and increased autophagic vacuoles in the liver, and (ii) a modestly reduced glucose uptake by WAT when mice were fed a standard chow diet. Features observed in Par-1b null mice but not in Par-1a mice include (i) hyperphagy (the food intake of Par-1a null mice was equivalent to that of controls); (ii) reduced serum insulin levels under both fed and fasting conditions on a standard CD (serum insulin levels in Par-1a null mice were lower than those measured in WT littermates fed an HFD but not standard CD); (iii) increased insulin sensitivity on standard CD (Par-1a null mice were insulin sensitive on an HFD but not standard CD); (iv) resistance to HFD-induced weight gain (Par-1a null mice showed a reduced rate of weight gain relative to that of control littermates when fed an HFD); (v) enhanced glucose uptake in BAT (the loss of Par-1a did not affect glucose uptake in BAT under any condition); and (vi) resistance to hepatic steatosis accompanied by enhanced glucose uptake in liver (Par-1a null mice; resistance to hepatic steatosis was not accompanied by enhanced hepatic glucose uptake).
Strikingly, Par-1a null mice on HFD exhibited profound resistance to the development of glucose intolerance (Fig. 3D) as well as delayed weight gain on HFD (Fig. 2A). These findings most likely are related to peripheral insulin hypersensitivity, as illustrated by resistance to glucose intolerance and decreased insulin secretion during GTT (on HFD). Par-1a null livers maintained resistance to steatosis despite HFD challenge (Fig. 5A and B). However, the adiposity and WAT glucose uptake measured in Par-1a null mice fed an HFD were similar to those of WT littermates fed the same diet. Although this study does not provide conclusive evidence for a hepatocyte-autonomous role for either Par-1a or Par-1b in lipogenesis, the striking resistance to hepatic steatosis seen in both models suggests such a function. Upon short-term starvation, the livers of Par-1a null mice exhibited profound defects in both glycogen storage and glucose mobilization (gluconeogenesis), phenotypes not observed in Par-1b null mice. The depletion of both lipid and glycogen stores, the upregulation of glycogen synthase, and the activation of autophagy are observed in the livers of Par-1a null mice. The increase in total glycogen synthase might be a compensatory response in light of the decreased glycogen storage. However, we also observe an increase in the inhibited (phosphorylated) form of glycogen synthase. Thus, it is unclear whether the net glycogen synthase activity is altered in the livers of Par-1a−/− mice. Our ultrastructural findings of increased autophagy in the absence of glycogen or lipid vacuoles in the livers of Par-1a null mice suggests substantial energy deprivation with the self digestion of organelles. The absence of both hepatic glycogen mobilization and gluconeogenesis required to maintain normal blood glucose levels during starvation in turn leads to hypoglycemia and hypoketosis in Par-1a null mice.
The underlying molecular mechanisms driving the hypermetabolic phenotype, resistance to hepatic steatosis in Par-1a null mice, and the inability of these mice to maintain metabolic homeostasis during periods of starvation remain unclear. The regulation of lipogenesis, glucose metabolism, and autophagy are coordinated, at least in part, through the Par-1-related kinase AMPK. It is tempting to speculate that Par-1 regulates these pathways via a common mechanism. To test this, we monitored the activation state of the mTOR pathway by assessing levels of phospho- and total AKT, phospho- and total AMPK, phospho- and total p70S6K, phospho- and total pS6 ribosomal protein, and LC-3 in the livers of Par-1-deficient mice during starvation and upon refeeding. We did not observe any consistent alterations in the levels or activation state of these proteins. Curiously, although both AMPK and SIK kinases negatively regulate lipogenesis via SREBP1c, the phenotypes of Par-1 null mice suggest that Par-1 is a positive regulator for lipogenesis in adipocytes and hepatocytes. The molecular mechanism underlying the phenotypes of Par-1a null mice will require tissue-specific deletion and the identification of specific downstream _targets of Par-1a mediating these effects.
Another major finding of this study is that Par-1b compensates more effectively for the loss of Par-1a than the reverse, and that embryonic survival requires the presence of at least one allele of Par-1a/b. Moreover, the presence of only one Par-1a allele in the complete absence of Par-1b conveys the most severe phenotype (growth restriction), supporting the notion of ranked redundancy with the most severe phenotype observed in the very rare KH (Par-1b−/−/Par-1a+/−) mutant.
Acknowledgments
We thank Li He and Yihua Zhao for technical support during the early stages of this project, Mike White for performing blastocyst injections, Jinwu Sun for help with Southern blot analysis, Clay Semenkovich for helpful suggestions and comments, and Kelsey Tinkum for experimental and editorial assistance. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital for the use of the Embryonic Stem Cell Core and electroporation services; the Siteman Cancer Center Small Animal Imaging Core; L. Strong, J. Engelbach, J. Rutlin, and R. Laforest for assistance with microPET imaging; the Research Resource for Cancer Imaging for the production of [18F]FDG; the Washington University Diabetes Research Training Center (St. Louis, MO); the Mouse Metabolic Phenotyping Core at Yale University School of Medicine (New Haven, CT); and G. Cline for help with metabolic characterization. We thank A. Watson, V. Layton, K. Selle, D. Leahart, K. Keith, and R. Brown for expert histotechnical assistance, J. Rossi, J. Mills, and T. Stappenbeck for microscope use, W. Clermont, S. Yates, and M. Isaacs for slide scanning, and J. S. Lewis, Jr., and the research histology laboratory. We thank K. G. Green for help with electron microscopy; B. Alter, M. Montana, M. Morales, J. Golden, S. Vogt, G. Story, and T. Coleman for help with experiments; and H. Abel for help with statistical analysis. We thank C. F. Semenkovich, J. Pfeifer, E. Brunt, and S. Kulkarni at Washington University (St. Louis, MO) as well as B. Lennerz and J. Majzoub (Children's Hospital, Boston, MA) for thoughtful discussions.
This study was supported in part by P50 CA94056. The Siteman Cancer Center is supported in part by NCI Cancer Center support grant P30 CA91842. The Washington University Diabetes Research Training Center is supported in part by P60 DK020579. The Mouse Metabolic Phenotyping Core is supported in part by NIH/NIDDK U24 DK59635. This work was supported by an NIH Neuroscience Blueprint Interdisciplinary Center Core grant (P30 NS057105). H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 23 August 2010.
REFERENCES
- 1.Abramoff, M., P. Magelhaes, and S. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Alessi, D. R. 2001. Discovery of PDK1, one of the missing links in insulin signal transduction. Colworth Medal Lecture. Biochem. Soc. Trans. 29:1-14. [DOI] [PubMed] [Google Scholar]
- 3.Argilés, J. M., F. J. Lopez-Soriano, and S. Busquets. 2007. Mechanisms to explain wasting of muscle and fat in cancer cachexia. Curr. Opin. Support Palliat. Care 1:293-298. [DOI] [PubMed] [Google Scholar]
- 4.Arimura, N., and K. Kaibuchi. 2007. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 8:194-205. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann, M., H. Hennemann, P. X. Xing, I. Hoffmann, and T. Moroy. 2004. The oncogenic serine/threonine kinase Pim-1 phosphorylates and inhibits the activity of Cdc25C-associated kinase 1 (C-TAK1): a novel role for Pim-1 at the G2/M cell cycle checkpoint. J. Biol. Chem. 279:48319-48328. [DOI] [PubMed] [Google Scholar]
- 6.Banks, L., and P. O. Humbert. 2008. On the guardians of polarity and the disorientation of cancer. Oncogene 27:6876-6877. [DOI] [PubMed] [Google Scholar]
- 7.Benton, R., and D. St. Johnston. 2002. Cell polarity: posterior Par-1 prevents proteolysis. Curr. Biol. 12:R479-R481. [DOI] [PubMed] [Google Scholar]
- 8.Bernal-Mizrachi, C., S. Weng, B. Li, L. A. Nolte, C. Feng, T. Coleman, J. O. Holloszy, and C. F. Semenkovich. 2002. Respiratory uncoupling lowers blood pressure through a leptin-dependent mechanism in genetically obese mice. Arterioscler. Thromb. Vasc. Biol. 22:961-968. [DOI] [PubMed] [Google Scholar]
- 9.Bessone, S., F. Vidal, Y. Le Bouc, J. Epelbaum, M. T. Bluet-Pajot, and M. Darmon. 1999. EMK protein kinase-null mice: dwarfism and hypofertility associated with alterations in the somatotrope and prolactin pathways. Dev. Biol. 214:87-101. [DOI] [PubMed] [Google Scholar]
- 10.Betschinger, J., K. Mechtler, and J. A. Knoblich. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422:326-330. [DOI] [PubMed] [Google Scholar]
- 11.Biernat, J., Y. Z. Wu, T. Timm, Q. Zheng-Fischhofer, E. Mandelkow, L. Meijer, and E. M. Mandelkow. 2002. Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol. Biol. Cell 13:4013-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigott, H. M., J. L. Prior, D. R. Piwnica-Worms, and M. J. Welch. 2005. Imaging multidrug resistance P-glycoprotein transport function using microPET with technetium-94m-sestamibi. Mol. Imaging 4:30-39. [DOI] [PubMed] [Google Scholar]
- 13.Bradbury, M. W. 2006. Lipid metabolism and liver inflammation. I. Hepatic fatty acid uptake: possible role in steatosis. Am. J. Physiol. Gastrointest. Liver Physiol. 290:G194-G198. [DOI] [PubMed] [Google Scholar]
- 14.Brajenovic, M., G. Joberty, B. Kuster, T. Bouwmeester, and G. Drewes. 2004. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J. Biol. Chem. 279:12804-12811. [DOI] [PubMed] [Google Scholar]
- 15.Bronisz, A., S. M. Sharma, R. Hu, J. Godlewski, G. Tzivion, K. C. Mansky, and M. C. Ostrowski. 2006. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol. Biol. Cell 17:3897-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunt, E. M., C. G. Janney, A. M. Di Bisceglie, B. A. Neuschwander-Tetri, and B. R. Bacon. 1999. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am. J. Gastroenterol. 94:2467-2474. [DOI] [PubMed] [Google Scholar]
- 17.Bullock, S. L., and D. Ish-Horowicz. 2002. Cell polarity: Oskar seeks PARtner for a stable relationship. Nat. Cell Biol. 4:E117-E118. [DOI] [PubMed] [Google Scholar]
- 18.Chakravarthy, M. V., Z. Pan, Y. Zhu, K. Tordjman, J. G. Schneider, T. Coleman, J. Turk, and C. F. Semenkovich. 2005. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 1:309-322. [DOI] [PubMed] [Google Scholar]
- 19.Chen, Y. M., Q. J. Wang, H. S. Hu, P. C. Yu, J. Zhu, G. Drewes, H. Piwnica-Worms, and Z. G. Luo. 2006. Microtubule affinity-regulating kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc. Natl. Acad. Sci. U. S. A. 103:8534-8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirrito, J. R., R. Deane, A. M. Fagan, M. L. Spinner, M. Parsadanian, M. B. Finn, H. Jiang, J. L. Prior, A. Sagare, K. R. Bales, S. M. Paul, B. V. Zlokovic, D. Piwnica-Worms, and D. M. Holtzman. 2005. P-glycoprotein deficiency at the blood-brain barrier increases amyloid-beta deposition in an Alzheimer disease mouse model. J. Clin. Investig. 115:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cline, G. W., D. L. Rothman, I. Magnusson, L. D. Katz, and G. I. Shulman. 1994. 13C-nuclear magnetic resonance spectroscopy studies of hepatic glucose metabolism in normal subjects and subjects with insulin-dependent diabetes mellitus. J. Clin. Investig. 94:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corbett, M. A., C. S. Robinson, G. F. Dunglison, N. Yang, J. E. Joya, A. W. Stewart, C. Schnell, P. W. Gunning, K. N. North, and E. C. Hardeman. 2001. A mutation in alpha-tropomyosin(slow) affects muscle strength, maturation and hypertrophy in a mouse model for nemaline myopathy. Hum. Mol. Genet. 10:317-328. [DOI] [PubMed] [Google Scholar]
- 23.Cullis, D. N., B. Philip, J. D. Baleja, and L. A. Feig. 2002. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J. Biol. Chem. 277:49158-49166. [DOI] [PubMed] [Google Scholar]
- 24.D'Agostino, R. B. 1986. Tests for normal distribution, p. xviii. In R. B. D'Agostino and M. A. Stephens (ed.), Goodness-of-fit techniques. M. Dekker, New York, NY.
- 25.Dalal, S. N., C. M. Schweitzer, J. Gan, and J. A. DeCaprio. 1999. Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol. 19:4465-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeFronzo, R. A., E. Jacot, E. Jequier, E. Maeder, J. Wahren, and J. P. Felber. 1981. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes 30:1000-1007. [DOI] [PubMed] [Google Scholar]
- 27.de Leng, W. W., M. Jansen, R. Carvalho, M. Polak, A. R. Musler, A. N. Milne, J. J. Keller, F. H. Menko, F. W. de Rooij, C. A. Iacobuzio-Donahue, F. M. Giardiello, M. A. Weterman, and G. J. Offerhaus. 2007. Genetic defects underlying Peutz-Jeghers syndrome (PJS) and exclusion of the polarity-associated MARK/Par1 gene family as potential PJS candidates. Clin. Genet. 72:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dequiedt, F., M. Martin, J. Von Blume, D. Vertommen, E. Lecomte, N. Mari, M. F. Heinen, M. Bachmann, J. C. Twizere, M. C. Huang, M. H. Rider, H. Piwnica-Worms, T. Seufferlein, and R. Kettmann. 2006. New role for hPar-1 kinases EMK and C-TAK1 in regulating localization and activity of class IIa histone deacetylases. Mol. Cell. Biol. 26:7086-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Serio, C., S. Pellerito, M. Duarte, D. Massi, A. Naldini, G. Cirino, I. Prudovsky, M. Santucci, P. Geppetti, N. Marchionni, G. Masotti, and F. Tarantini. 2007. Protease-activated receptor 1-selective antagonist SCH79797 inhibits cell proliferation and induces apoptosis by a protease-activated receptor 1-independent mechanism. Basic Clin. Pharmacol. Toxicol. 101:63-69. [DOI] [PubMed] [Google Scholar]
- 30.Drewes, G. 2004. MARKing tau for tangles and toxicity. Trends Biochem. Sci. 29:548-555. [DOI] [PubMed] [Google Scholar]
- 31.Drewes, G., A. Ebneth, and E. M. Mandelkow. 1998. MAPs, MARKs and microtubule dynamics. Trends Biochem. Sci. 23:307-311. [DOI] [PubMed] [Google Scholar]
- 32.Drewes, G., A. Ebneth, U. Preuss, E. M. Mandelkow, and E. Mandelkow. 1997. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89:297-308. [DOI] [PubMed] [Google Scholar]
- 33.Drewes, G., B. Trinczek, S. Illenberger, J. Biernat, G. Schmitt-Ulms, H. E. Meyer, E. M. Mandelkow, and E. Mandelkow. 1995. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark). A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J. Biol. Chem. 270:7679-7688. [DOI] [PubMed] [Google Scholar]
- 34.Ducharme, N. A., C. M. Hales, L. A. Lapierre, A. J. Ham, A. Oztan, G. Apodaca, and J. R. Goldenring. 2006. MARK2/EMK1/Par-1Balpha phosphorylation of Rab11-family interacting protein 2 is necessary for the timely establishment of polarity in Madin-Darby canine kidney cells. Mol. Biol. Cell 17:3625-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu, A., A. C. Ng, C. Depatie, N. Wijesekara, Y. He, G. S. Wang, N. Bardeesy, F. W. Scott, R. M. Touyz, M. B. Wheeler, and R. A. Screaton. 2009. Loss of Lkb1 in adult beta cells increases beta cell mass and enhances glucose tolerance in mice. Cell Metab. 10:285-295. [DOI] [PubMed] [Google Scholar]
- 36.Geldner, N. 2009. Cell polarity in plants: a PARspective on PINs. Curr. Opin. Plant Biol. 12:42-48. [DOI] [PubMed] [Google Scholar]
- 37.Göransson, O., M. Deak, S. Wullschleger, N. A. Morrice, A. R. Prescott, and D. R. Alessi. 2006. Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation. J. Cell Sci. 119:4059-4070. [DOI] [PubMed] [Google Scholar]
- 38.Granot, Z., A. Swisa, J. Magenheim, M. Stolovich-Rain, W. Fujimoto, E. Manduchi, T. Miki, J. K. Lennerz, C. J. Stoeckert, Jr., O. Meyuhas, S. Seino, M. A. Permutt, H. Piwnica-Worms, N. Bardeesy, and Y. Dor. 2009. LKB1 regulates pancreatic beta cell size, polarity, and function. Cell Metab. 10:296-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gustot, T., A. Lemmers, C. Moreno, N. Nagy, E. Quertinmont, C. Nicaise, D. Franchimont, H. Louis, J. Deviere, and O. Le Moine. 2006. Differential liver sensitization to toll-like receptor pathways in mice with alcoholic fatty liver. Hepatology 43:989-1000. [DOI] [PubMed] [Google Scholar]
- 40.He, L., A. Sabet, S. Djedjos, R. Miller, X. Sun, M. A. Hussain, S. Radovick, and F. E. Wondisford. 2009. Metformin and insulin suppress hepatic gluconeogenesis through phosphorylation of CREB binding protein. Cell 137:635-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hegarty, B. D., N. Turner, G. J. Cooney, and E. W. Kraegen. 2009. Insulin resistance and fuel homeostasis: the role of AMP-activated protein kinase. Acta Physiol. (Oxford) 196:129-145. [DOI] [PubMed] [Google Scholar]
- 42.Hurov, J., and H. Piwnica-Worms. 2007. The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell Cycle 6:1966-1969. [DOI] [PubMed] [Google Scholar]
- 43.Hurov, J. B., M. Huang, L. S. White, J. Lennerz, C. S. Choi, Y. R. Cho, H. J. Kim, J. L. Prior, D. Piwnica-Worms, L. C. Cantley, J. K. Kim, G. I. Shulman, and H. Piwnica-Worms. 2007. Loss of the Par-1b/MARK2 polarity kinase leads to increased metabolic rate, decreased adiposity, and insulin hypersensitivity in vivo. Proc. Natl. Acad. Sci. U. S. A. 104:5680-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurov, J. B., T. S. Stappenbeck, C. M. Zmasek, L. S. White, S. H. Ranganath, J. H. Russell, A. C. Chan, K. M. Murphy, and H. Piwnica-Worms. 2001. Immune system dysfunction and autoimmune disease in mice lacking Emk (Par-1) protein kinase. Mol. Cell. Biol. 21:3206-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurov, J. B., J. L. Watkins, and H. Piwnica-Worms. 2004. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14:736-741. [DOI] [PubMed] [Google Scholar]
- 46.Hutchison, M., K. S. Berman, and M. H. Cobb. 1998. Isolation of TAO1, a protein kinase that activates MEKs in stress-activated protein kinase cascades. J. Biol. Chem. 273:28625-28632. [DOI] [PubMed] [Google Scholar]
- 47.Illenberger, S., G. Drewes, B. Trinczek, J. Biernat, H. E. Meyer, J. B. Olmsted, E. M. Mandelkow, and E. Mandelkow. 1996. Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. Phosphorylation sites and regulation of microtubule dynamics. J. Biol. Chem. 271:10834-10843. [DOI] [PubMed] [Google Scholar]
- 48.Jansen, M., W. W. de Leng, A. F. Baas, H. Myoshi, L. Mathus-Vliegen, M. M. Taketo, H. Clevers, F. M. Giardiello, and G. J. Offerhaus. 2006. Mucosal prolapse in the pathogenesis of Peutz-Jeghers polyposis. Gut 55:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jansson, D., A. C. Ng, A. Fu, C. Depatie, M. Al Azzabi, and R. A. Screaton. 2008. Glucose controls CREB activity in islet cells via regulated phosphorylation of TORC2. Proc. Natl. Acad. Sci. U. S. A. 105:10161-10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao, H. Y., A. Verdel, C. C. Tsai, C. Simon, H. Juguilon, and S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496-47507. [DOI] [PubMed] [Google Scholar]
- 51.Kato, T., S. Satoh, H. Okabe, O. Kitahara, K. Ono, C. Kihara, T. Tanaka, T. Tsunoda, Y. Yamaoka, Y. Nakamura, and Y. Furukawa. 2001. Isolation of a novel human gene, MARKL1, homologous to MARK3 and its involvement in hepatocellular carcinogenesis. Neoplasia. 3:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kemphues, K. J., J. R. Priess, D. G. Morton, and N. S. Cheng. 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52:311-320. [DOI] [PubMed] [Google Scholar]
- 53.Kesarwala, A. H., J. L. Prior, J. Sun, S. E. Harpstrite, V. Sharma, and D. Piwnica-Worms. 2006. Second-generation triple reporter for bioluminescence, micro-positron emission tomography, and fluorescence imaging. Mol. Imaging 5:465-474. [PubMed] [Google Scholar]
- 54.Kosuga, S., E. Tashiro, T. Kajioka, M. Ueki, Y. Shimizu, and M. Imoto. 2005. GSK-3beta directly phosphorylates and activates MARK2/PAR-1. J. Biol. Chem. 280:42715-42722. [DOI] [PubMed] [Google Scholar]
- 55.Kotoulas, O. B., S. A. Kalamidas, and D. J. Kondomerkos. 2004. Glycogen autophagy. Microsc. Res. Tech. 64:10-20. [DOI] [PubMed] [Google Scholar]
- 56.Kotoulas, O. B., S. A. Kalamidas, and D. J. Kondomerkos. 2006. Glycogen autophagy in glucose homeostasis. Pathol. Res. Pract. 202:631-638. [DOI] [PubMed] [Google Scholar]
- 57.Krahn, M. P., and A. Wodarz. 2009. Notch signaling: linking delta endocytosis and cell polarity. Dev. Cell 17:153-154. [DOI] [PubMed] [Google Scholar]
- 58.Krummel, M. F., and I. Macara. 2006. Maintenance and modulation of T cell polarity. Nat. Immunol. 7:1143-1149. [DOI] [PubMed] [Google Scholar]
- 59.Kusakabe, M., and E. Nishida. 2004. The polarity-inducing kinase Par-1 controls Xenopus gastrulation in cooperation with 14-3-3 and aPKC. EMBO J. 23:4190-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lennerz, J. K., V. Ruhle, E. P. Ceppa, W. L. Neuhuber, N. W. Bunnett, E. F. Grady, and K. Messlinger. 2008. Calcitonin receptor-like receptor (CLR), receptor activity-modifying protein 1 (RAMP1), and calcitonin gene-related peptide (CGRP) immunoreactivity in the rat trigeminovascular system: differences between peripheral and central CGRP receptor distribution. J. Comp. Neurol. 507:1277-1299. [DOI] [PubMed] [Google Scholar]
- 61.Lin, J., K. K. Hou, H. Piwnica-Worms, and A. S. Shaw. 2009. The polarity protein Par1b/EMK/MARK2 regulates T cell receptor-induced microtubule-organizing center polarization. J. Immunol. 183:1215-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindsey, K. 2004. Polarity in plants. Blackwell, Oxford, United Kingdom.
- 63.Lizcano, J. M., O. Goransson, R. Toth, M. Deak, N. A. Morrice, J. Boudeau, S. A. Hawley, L. Udd, T. P. Makela, D. G. Hardie, and D. R. Alessi. 2004. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lock, J. G., and J. L. Stow. 2005. Rab11 in recycling endosomes regulates the sorting and basolateral transport of E-cadherin. Mol. Biol. Cell 16:1744-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin, S. G., and D. St. Johnston. 2003. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature 421:379-384. [DOI] [PubMed] [Google Scholar]
- 66.Meyer, S. L. 1975. Data analysis for scientists and engineers. Wiley, New York, NY.
- 67.Mitchison, T., and M. Kirschner. 1984. Dynamic instability of microtubule growth. Nature 312:237-242. [DOI] [PubMed] [Google Scholar]
- 68.Müller, J., S. Ory, T. Copeland, H. Piwnica-Worms, and D. K. Morrison. 2001. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell 8:983-993. [DOI] [PubMed] [Google Scholar]
- 69.Müller, J., D. A. Ritt, T. D. Copeland, and D. K. Morrison. 2003. Functional analysis of C-TAK1 substrate binding and identification of PKP2 as a new C-TAK1 substrate. EMBO J. 22:4431-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy, J. M., D. M. Korzhnev, D. F. Ceccarelli, D. J. Briant, A. Zarrine-Afsar, F. Sicheri, L. E. Kay, and T. Pawson. 2007. Conformational instability of the MARK3 UBA domain compromises ubiquitin recognition and promotes interaction with the adjacent kinase domain. Proc. Natl. Acad. Sci. U. S. A. 104:14336-14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nance, J. 2005. PAR proteins and the establishment of cell polarity during C. elegans development. Bioessays 27:126-135. [DOI] [PubMed] [Google Scholar]
- 72.Ogg, S., B. Gabrielli, and H. Piwnica-Worms. 1994. Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J. Biol. Chem. 269:30461-30469. [PubMed] [Google Scholar]
- 73.Ossipova, O., S. Dhawan, S. Sokol, and J. B. Green. 2005. Distinct PAR-1 proteins function in different branches of Wnt signaling during vertebrate development. Dev. Cell 8:829-841. [DOI] [PubMed] [Google Scholar]
- 74.Ossipova, O., J. Ezan, and S. Y. Sokol. 2009. PAR-1 phosphorylates mind bomb to promote vertebrate neurogenesis. Dev. Cell 17:222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ossipova, O., J. Tabler, J. B. Green, and S. Y. Sokol. 2007. PAR1 specifies ciliated cells in vertebrate ectoderm downstream of aPKC. Development 134:4297-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parsa, I. 1988. Loss of a Mr 78,000 marker in chemically induced transplantable carcinomas and primary carcinoma of human pancreas. Cancer Res. 48:2265-2272. [PubMed] [Google Scholar]
- 77.Parsons, D. W., T. L. Wang, Y. Samuels, A. Bardelli, J. M. Cummins, L. DeLong, N. Silliman, J. Ptak, S. Szabo, J. K. Willson, S. Markowitz, K. W. Kinzler, B. Vogelstein, C. Lengauer, and V. E. Velculescu. 2005. Colorectal cancer: mutations in a signalling pathway. Nature 436:792. [DOI] [PubMed] [Google Scholar]
- 78.Peng, C. Y., P. R. Graves, S. Ogg, R. S. Thoma, M. J. Byrnes III, Z. Wu, M. T. Stephenson, and H. Piwnica-Worms. 1998. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 9:197-208. [PubMed] [Google Scholar]
- 79.Postic, C., and J. Girard. 2008. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: lessons from genetically engineered mice. J. Clin. Investig. 118:829-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rabinovich, S. G. 2000. Measurement errors and uncertainties: theory and practice, 2nd ed. AIP Press, New York, NY.
- 81.Rubio-Aliaga, I., D. Soewarto, S. Wagner, M. Klaften, H. Fuchs, S. Kalaydjiev, D. H. Busch, M. Klempt, B. Rathkolb, E. Wolf, K. Abe, S. Zeiser, G. K. Przemeck, J. Beckers, and M. H. de Angelis. 2007. A genetic screen for modifiers of the delta1-dependent notch signaling function in the mouse. Genetics 175:1451-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saadat, I., H. Higashi, C. Obuse, M. Umeda, N. Murata-Kamiya, Y. Saito, H. Lu, N. Ohnishi, T. Azuma, A. Suzuki, S. Ohno, and M. Hatakeyama. 2007. Helicobacter pylori CagA _targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature 447:330-333. [DOI] [PubMed] [Google Scholar]
- 83.Sammak, P. J., and G. G. Borisy. 1988. Direct observation of microtubule dynamics in living cells. Nature 332:724-726. [DOI] [PubMed] [Google Scholar]
- 84.Sanoudou, D., M. A. Corbett, M. Han, M. Ghoddusi, M. A. Nguyen, N. Vlahovich, E. C. Hardeman, and A. H. Beggs. 2006. Skeletal muscle repair in a mouse model of nemaline myopathy. Hum. Mol. Genet. 15:2603-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmidt, R. E., K. G. Green, L. L. Snipes, and D. Feng. 2009. Neuritic dystrophy and neuronopathy in Akita (Ins2(Akita)) diabetic mouse sympathetic ganglia. Exp. Neurol. 216:207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Segu, L., A. Pascaud, P. Costet, M. Darmon, and M. C. Buhot. 2008. Impairment of spatial learning and memory in ELKL motif kinase1 (EMK1/MARK2) knockout mice. Neurobiol. Aging 29:231-240. [DOI] [PubMed] [Google Scholar]
- 87.Sharma, V., G. D. Luker, and D. Piwnica-Worms. 2002. Molecular imaging of gene expression and protein function in vivo with PET and SPECT. J. Magn. Reson. Imaging 16:336-351. [DOI] [PubMed] [Google Scholar]
- 88.Sharma, V., J. L. Prior, M. G. Belinsky, G. D. Kruh, and D. Piwnica-Worms. 2005. Characterization of a 67Ga/68Ga radiopharmaceutical for SPECT and PET of MDR1 P-glycoprotein transport activity in vivo: validation in multidrug-resistant tumors and at the blood-brain barrier. J. Nucl. Med. 46:354-364. [PubMed] [Google Scholar]
- 89.Shin, K., V. C. Fogg, and B. Margolis. 2006. Tight junctions and cell polarity. Annu. Rev. Cell Dev. Biol. 22:207-235. [DOI] [PubMed] [Google Scholar]
- 90.Shulman, G. I., G. Cline, W. C. Schumann, V. Chandramouli, K. Kumaran, and B. R. Landau. 1990. Quantitative comparison of pathways of hepatic glycogen repletion in fed and fasted humans. Am. J. Physiol. 259:E335-E341. [DOI] [PubMed] [Google Scholar]
- 91.Singh, R., S. Kaushik, Y. Wang, Y. Xiang, I. Novak, M. Komatsu, K. Tanaka, A. M. Cuervo, and M. J. Czaja. 2009. Autophagy regulates lipid metabolism. Nature 458:1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sitnick, M., S. C. Bodine, and J. C. Rutledge. 2009. Chronic high fat feeding attenuates load-induced hypertrophy in mice. J. Physiol. 587:5753-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spicer, J., S. Rayter, N. Young, R. Elliott, A. Ashworth, and D. Smith. 2003. Regulation of the Wnt signalling component PAR1A by the Peutz-Jeghers syndrome kinase LKB1. Oncogene 22:4752-4756. [DOI] [PubMed] [Google Scholar]
- 94.Sun, T. Q., B. Lu, J. J. Feng, C. Reinhard, Y. N. Jan, W. J. Fantl, and L. T. Williams. 2001. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat. Cell Biol. 3:628-636. [DOI] [PubMed] [Google Scholar]
- 95.Suzuki, A., M. Hirata, K. Kamimura, R. Maniwa, T. Yamanaka, K. Mizuno, M. Kishikawa, H. Hirose, Y. Amano, N. Izumi, Y. Miwa, and S. Ohno. 2004. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14:1425-1435. [DOI] [PubMed] [Google Scholar]
- 96.Tian, A. G., and W. M. Deng. 2009. Par-1 and Tau regulate the anterior-posterior gradient of microtubules in Drosophila oocytes. Dev. Biol. 327:458-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Timm, T., K. Balusamy, X. Li, J. Biernat, E. Mandelkow, and E. M. Mandelkow. 2008. Glycogen synthase kinase (GSK) 3beta directly phosphorylates serine 212 in the regulatory loop and inhibits microtubule affinity-regulating kinase (MARK) 2. J. Biol. Chem. 283:18873-18882. [DOI] [PubMed] [Google Scholar]
- 98.Timm, T., X. Y. Li, J. Biernat, J. Jiao, E. Mandelkow, J. Vandekerckhove, and E. M. Mandelkow. 2003. MARKK, a Ste20-like kinase, activates the polarity-inducing kinase MARK/PAR-1. EMBO J. 22:5090-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uboha, N. V., M. Flajolet, A. C. Nairn, and M. R. Picciotto. 2007. A calcium- and calmodulin-dependent kinase Ialpha/microtubule affinity regulating kinase 2 signaling cascade mediates calcium-dependent neurite outgrowth. J. Neurosci. 27:4413-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, C., Z. L. You, Q. Xia, T. Xiong, Y. Xia, and D. Z. Yao. 2007. Upregulation of Mark3 and Rpgrip1 mRNA expression by jujuboside A in mouse hippocampus. Acta Pharmacol. Sin. 28:334-338. [DOI] [PubMed] [Google Scholar]
- 101.Watkins, J. L., K. T. Lewandowski, S. E. Meek, P. Storz, A. Toker, and H. Piwnica-Worms. 2008. Phosphorylation of the Par-1 polarity kinase by protein kinase D regulates 14-3-3 binding and membrane association. Proc. Natl. Acad. Sci. U. S. A. 105:18378-18383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Woods, D. F., and P. J. Bryant. 1991. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell 66:451-464. [DOI] [PubMed] [Google Scholar]
- 103.Zhang, S. H., R. Kobayashi, P. R. Graves, H. Piwnica-Worms, and N. K. Tonks. 1997. Serine phosphorylation-dependent association of the band 4.1-related protein-tyrosine phosphatase PTPH1 with 14-3-3beta protein. J. Biol. Chem. 272:27281-27287. [DOI] [PubMed] [Google Scholar]
- 104.Zhang, Y., H. Guo, H. Kwan, J. W. Wang, J. Kosek, and B. Lu. 2007. PAR-1 kinase phosphorylates Dlg and regulates its postsynaptic _targeting at the Drosophila neuromuscular junction. Neuron 53:201-215. [DOI] [PMC free article] [PubMed] [Google Scholar]