Abstract
Large-scale and in-depth characterization of the intestinal microbiota necessitates application of high-throughput 16S rRNA gene-based technologies, such as barcoded pyrosequencing and phylogenetic microarray analysis. In this study, the two techniques were compared and contrasted for analysis of the bacterial composition in three fecal and three small intestinal samples from human individuals. As PCR remains a crucial step in sample preparation for both techniques, different forward primers were used for amplification to assess their impact on microbial profiling results. An average of 7,944 pyrosequences, spanning the V1 and V2 region of 16S rRNA genes, was obtained per sample. Although primer choice in barcoded pyrosequencing did not affect species richness and diversity estimates, detection of Actinobacteria strongly depended on the selected primer. Microbial profiles obtained by pyrosequencing and phylogenetic microarray analysis (HITChip) correlated strongly for fecal and ileal lumen samples but were less concordant for ileostomy effluent. Quantitative PCR was employed to investigate the deviations in profiling between pyrosequencing and HITChip analysis. Since cloning and sequencing of random 16S rRNA genes from ileostomy effluent confirmed the presence of novel intestinal phylotypes detected by pyrosequencing, especially those belonging to the Veillonella group, the divergence between pyrosequencing and the HITChip is likely due to the relatively low number of available 16S rRNA gene sequences of small intestinal origin in the DNA databases that were used for HITChip probe design. Overall, this study demonstrated that equivalent biological conclusions are obtained by high-throughput profiling of microbial communities, independent of technology or primer choice.
The human gastrointestinal (GI) tract is inhabited by a microbiota that predominantly consists of bacteria and is dominated by the phyla Firmicutes, Bacteroidetes, and Actinobacteria (36). This community increases in numbers as well as diversity along the longitudinal axes of the GI tract and ultimately reaches populations as high as 1011 bacteria per gram of contents in the large intestine (6, 26, 46). The diversity and population dynamics of the lower GI tract microbiota have been well documented (26, 35, 51). In contrast, the microbiota of the upper GI tract has been poorly described, which is mainly due to sampling difficulties (5, 26). Recently, the human small intestinal microbiota was characterized using samples obtained from ileostomy subjects (5, 20), and samples from the small intestine of healthy individuals were obtained with an orally introduced catheter (E. G. Zoetendal et al., submitted for publication).
Much emphasis has been placed on understanding the dynamics and activities of the intestinal bacterial communities (5, 35, 45). The means by which this research has been conducted underwent a revolution from culture-based approaches to molecular technologies during the last few decades (see references 46 and 49 for reviews). Molecular technologies based on 16S rRNA and its encoding gene, such as fluorescent in situ hybridization (FISH) (19), quantitative PCR (qPCR) (38), denaturing gradient gel electrophoresis (DGGE) (4), and terminal-restriction fragment length polymorphism (T-RFLP) (13), as well as the classical 16S rRNA gene amplicon cloning and sequencing approach (12), are commonly used for compositional studies of the intestinal microbial ecosystem (47, 49). However, these approaches are laborious, especially when one aims for in-depth microbial community profiling (49). Phylogenetic microarrays (33, 35) and pyrosequencing technology (28) have become popular methods since they principally allow high-throughput and in-depth monitoring of microbial communities. While the former relies on 16S rRNA gene-_targeted oligonucleotide probes for detection of bacteria in environmental samples (35), the latter allows de novo community profiling by sequencing and subsequent identification of partial 16S rRNA gene amplicons (1).
Each of the above-mentioned approaches for characterization of microbial communities is limited in its correct assessment of microbial abundances due to the unspecified partial phylogenetic coverage by primers or probes that are used during the initial stages of sample preparation (3, 15). This notion is exemplified by the underestimation of Bifidobacterium abundances in fecal samples by the commonly used universal 27F primer (21). One constraint of phylogenetic arrays for microbial profiling is their being limited to detecting phylogenetic groups for which probes are represented on the array, although higher-taxonomic-level probes can still provide information for those groups (33), whereas analysis of data from pyrosequencing is challenging due to the vast number of obtained sequences that may contain sequence errors that hinder appropriate data interpretation (18).
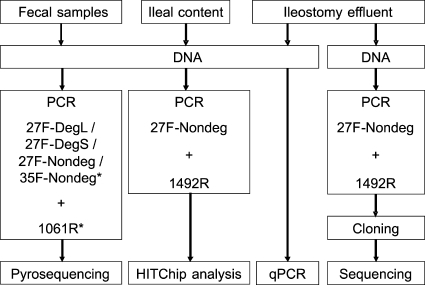
The aim of this research was to assess the accordance of barcoded pyrosequencing and a phylogenetic microarray, the human intestinal tract chip (HITChip) (35), for profiling of human fecal and small intestinal microbial communities. In addition, the use of different primer pairs in barcoded pyrosequencing was evaluated to answer whether their use in PCR-based approaches influences the outcome of microbial diversity estimates and profiling (Fig. 1).
FIG. 1.
Schematic representation of the experimental setup for characterization of the microbial composition in fecal samples, ileal content, and ileostomy effluent using molecular approaches. The asterisks indicate that forward primers and the reverse primer used for pyrosequencing were appended with adaptor A and adaptor B, respectively.
MATERIALS AND METHODS
Sample collection.
Fecal samples (F1 to F3) used in this study were collected at home from three healthy individuals (2 female and 1 male; aged 30 to 32 years), frozen in dry ice immediately, and transported to the laboratory, where they were kept at −80°C until further analysis.
Ileostomy effluent samples (S1 and S2) were previously collected (5) at least 3 h apart in the morning and afternoon, respectively, from a healthy 74-year-old male ileostomist as part of a previous project, results of which are reported elsewhere (5). The volunteer collected the ileostomy effluent samples by emptying the ileostomy effluent in freezer baskets as soon as the bulk of ileostomy effluent was collected in a clean empty ileostomy bag. Samples were stored on dry ice at approximately −80°C and were processed within 3 days after collection.
An ileum lumen sample (S3) was obtained from a 24-year-old healthy female individual by using an orally introduced catheter, which passed to the ileum (120 cm distal to the pylorus) by peristalsis. Sampling was done under gastroenterologist supervision, following flushing of the ileum with 10 ml physiological salt solution through a port of the catheter, after which the sample was frozen and stored at −80°C.
Bacterial reference strains and culture conditions.
Bifidobacterium longum (DSM 20219) was grown in ST medium as described in reference 41 with a substitution of proteose peptone for 1 g/liter Casitone (Becton Dickinson, Breda, Netherlands) and meat extract for 3 g/liter beef extract (Sigma, St. Louis, MO). Escherichia coli MC1061 was cultivated in Luria-Bertani (LB) broth at 37°C in a shaking incubator (135 rpm; Heidolph Instruments GmbH & Co., Schwabach, Germany). Streptococcus thermophilus (CNRZ 1066) was grown in M17 broth (Becton Dickinson) supplemented with 0.5% (wt/vol) glucose (Sigma) at 37°C. Veillonella atypica (DSM 20739) was grown in Veillonella medium described in the DSMZ catalogue (medium 136) under an N2 atmosphere.
DNA extraction.
Genomic DNA (gDNA) extractions from reference strains were performed using the FastDNA Spin kit for soil (MP Biomedicals, Solon, OH) with pelleted cells from 2 ml pure culture as starting material (data not shown).
Total DNA was extracted from 0.25 g fecal sample and 0.25 ml ileal content, using the repeated bead beating method described in reference 40, and from 0.2 g ileostomy effluent as previously described (50) by using the QIAamp DNA stool minikit (Qiagen GmbH, Hilden, Germany). A recent study by Salonen et al. (40) concluded that the difference in microbial compositions between DNA extraction methods is relatively small in relation to that between subjects.
DNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and adjusted to 10 to 20 ng/μl as template for subsequent 16S rRNA gene PCR amplification.
16S rRNA gene amplicon pyrosequencing.
Amplicons from the V1 to V6 region of 16S rRNA genes were generated by PCR using two degenerated (27F-DegL and 27F-DegS) and two nondegenerated (27F-Nondeg and 35F-Nondeg) primers in combination with a single reverse primer (1061R-Deg) (Table 1) for each fecal and small intestinal DNA extraction.
TABLE 1.
Adaptors and primers used in this study for 16S rRNA gene sequence PCR amplification for pyrosequencing, HITChip analysis, qPCR, and 16S rRNA gene cloning and sequencing
| _target bacteria or bacterium | Primera | Primer sequence (5′-3′)b | Applicationc and PCR annealing temp (°C)d | Source or reference |
|---|---|---|---|---|
| Adaptor A | CCATCTCATCCCTGCGTGTCTCCGACTCAG | P | Provided by GATC-Biotech | |
| Adaptor B | BioTEGg/CCTATCCCCTGTGTGCCTTGGCAGTCTCAG | P | Provided by GATC-Biotech | |
| Total bacteria | 27F-DegL | AGRGTTYGATYMTGGCTCAG | P (56) | 32 |
| 27F-DegS | GTTYGATYMTGGCTCAG | P (56) | This studye | |
| 27F-Nondeg | GTTTGATCCTGGCTCAG | P (56)/H (52)f/C (52) | 35 | |
| 35F-Nondeg | CCTGGCTCAGGATGAACG | P (56) | 21 | |
| 1061R-Deg | CRRCACGAGCTGACGAC | P (56) | 1 | |
| Uni-1492-rev | CGGCTACCTTGTTACGAC | H (52)/C (52) | 35 | |
| BACT1369F | CGGTGAATACGTTCYCGG | Q (56) | 44 | |
| PROK1492R | GGWTACCTTGTTACGACTT | Q (56) | 44 | |
| Bifidobacterium | g-Bifid-F | CTCCTGGAAACGGGTGG | Q (55) | 29 |
| g-Bifid-R | GGTGTTCTTCCCGATATCTACA | Q (55) | 29 | |
| Veillonella | Veil-F-Rinttilä | AYCAACCTGCCCTTCAGA | Q (57) | 38 |
| Veil-R-Rinttilä | CGTCCCGATTAACAGAGCTT | Q (57) | 38 | |
| Streptococcus | Strep-F-Rudney | AGATGGACCTGCGTTGT | Q (55) | 39 |
| Stherm 08 | GTGAACTTTCCACTCTCACAC | Q (55) | 16 | |
| Escherichia coli | E. coli-F-Huijsdens | CATGCCGCGTGTATGAAGAA | Q (57) | 23 |
| E. coli-R-Huijsdens | CGGGTAACGTCAATGAGCAAA | Q (57) | 23 |
Primer names may not correspond to original publication.
M = A or C; R = A or G; W = A or T; Y = C or T.
Abbreviations: P, pyrosequencing; H, HITChip analysis; Q, qPCR; C, 16S rRNA clone library construction and sequencing.
Annealing temperatures indicated in bold were determined as explained in Materials and Methods.
5′-end 3-nt-trimmed version of the 27F-DegL primer.
With a T7 promoter sequence appended on the 5′ end.
Biotin labeled.
To facilitate pyrosequencing using titanium chemistry, each forward primer was appended with the titanium sequencing adaptor A and an “NNNN” barcode sequence (Table 1) at the 5′ end, where NNNN is a sequence of four nucleotides that was unique for each sample and did not start with G or contain a triplicate of identical bases. The reverse primer carried the titanium adaptor B at the 5′ end.
PCRs were performed using a GS0001 thermocycler (Gene Technologies, Braintree, United Kingdom) in a total volume of 50 μl containing 1× PCR buffer, 1 μl PCR-grade nucleotide mix, 2.4 units of Faststart Taq DNA polymerase (Roche Diagnostics GmbH, Mannheim, Germany), 200 nM forward and reverse primers (Biolegio BV, Nijmegen, Netherlands), and 0.2 to 0.4 ng/μl of template DNA. The amplification program consisted of an initial denaturation step at 95°C for 5 min; 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 40 s, and elongation at 72°C for 70 s; and a final extension step at 72°C for 10 min. The size of the PCR products was confirmed by gel electrophoresis using 1 μl of the reaction mixture on a 1% (wt/vol) agarose gel containing 0.4 μg/ml ethidium bromide (Bio-Rad, Hercules, CA). Control PCRs were performed alongside each separate amplification without addition of template and consistently yielded no product. The optimal annealing temperature for primers (56°C) with attached adaptors and barcodes was determined by a 12-degree temperature gradient (49°C to 61°C) PCR using DNA from fecal sample F2 (data not shown).
PCR products were purified with the ZR-96 DNA Clean and Concentrator kit (Zymo Research, Orange, CA) followed by DNA yield quantification using a NanoDrop ND-1000 spectrophotometer.
Purified PCR products were mixed in equimolar amounts with a final DNA concentration of 100 ng/μl. The pooled amplicons were pyrosequenced using an FLX genome sequencer in combination with titanium chemistry (GATC-Biotech, Konstanz, Germany). Sequencing occurred on a picotiter plate, of which a quarter space was available for samples included in this study.
HITChip analysis.
Microbial community profiling was also performed using the HITChip (35), which is a phylogenetic microarray, produced by Agilent Technologies (Palo Alto, CA) in an 8 by 15K format (each chip has 8 arrays, each with 15,000 probes), with over 4,800 tiling oligonucleotides _targeting the V1 or the V6 region of the 16S rRNA gene from 1,132 microbial phylotypes present in the human gastrointestinal tract (35). (See the supplemental material for the hybridization and analysis procedure.)
Pyrosequence analysis and comparison with HITChip analysis.
Pyrosequences were sorted per barcode. To the best of our knowledge, no recommendations for quality filtering of reads generated by pyrosequencing using titanium chemistry have been published to date, and therefore, we applied previously reported recommendations for quality filtering of pyrosequences generated by the GS 20 platform (24). This filtering was performed using an in-house perl script that passed sequences with exact matches to the forward primer, no ambiguous bases (N), and read lengths not longer or shorter than 1 standard deviation (SD) from the average sequence length (>87 to 157 and <314 to 359 nucleotides; see Table S2 in the supplemental material for the actual upper and lower read length limits per sample). Additionally, primer sequences were removed from the pyrosequencing reads, and the remaining sequences were analyzed. The numbers of operational taxonomic units (OTUs), rarefaction curves, and total species richness estimations (abundance-based coverage estimators [ACE] and Chao1) (22) for the quality filtered sequences per sample were calculated using ESPRIT (43) with default settings (without removing low-quality reads) at an 0.02 distance level.
Taxonomic classification of sequencing reads employed a locally installed version of the Ribosomal Database Project (RDP) classifier (48), which by default produces classifications into the new higher-order taxonomy as proposed in Bergey's Manual of Systematic Bacteriology (17). The corresponding assignments differ from those that are produced by HITChip analysis, which has a standard output of the relative contributions of 1,132 microbial phylotypes at level 1 (phylum-like with Firmicutes divided into classes or clusters), level 2 (genus-like), and/or level 3 (phylotypes based on >98% sequence identity) (35) to the overall microbial community per sample in the phylogeny as proposed by Collins et al. (11). The 16S rRNA gene sequences of these phylotypes are present in a nonredundant ARB (27) database that was used for HITChip probe design, the human unique OTU database (35, 36). The sequences with corresponding assignments present in this database were exported and used to train the RDP classifier. This yielded a classifier that (in combination with a trial multiclassifier provided by the RDP staff) could classify pyrosequencing reads originating from different samples on a large scale with the same assignments as those produced by HITChip analysis. The confidence threshold used for classification of the pyrosequences was kept at 80%. Moreover, the multiclassifier summarized the assignments per taxon, which facilitated calculation of relative contributions and subsequent construction of microbial profiles for comparison with those that were generated by HITChip analysis.
Hierarchical cluster analysis of the microbial profiles was done in R version 2.9.2 by computing a distance matrix that was based on Pearson product-moment correlation coefficients (r) between pairs of profiles with level 2 community data. Visualization of hierarchical clustering was done by using the distance matrix in the hclust function in R with Ward's minimum variance method as agglomeration method. The Shannon diversity index was calculated in R using the diversity function with the level 2 community data from each sample. Screening of sequences for exact matches with the HITChip probes or primers was performed using in-house perl scripts.
Quantification of bacterial community members by qPCR.
All qPCRs were performed in 96-well PCR plates (Bio-Rad) sealed with Microseal B film (Bio-Rad) using a MyIQ Icycler with MyIQ software version 1.0.410 (Bio-Rad). Each reaction was carried out in a total volume of 25 μl using IQ SYBR green Supermix (Bio-Rad) according to the manufacturer's instructions with 200 nM forward and reverse primer in combination with 5 μl template DNA.
From a literature survey, group-specific primers were chosen (Table 1) that were deemed optimal in their phylogenetic specificity and coverage (based on results of the probe match tool offered in the Ribosomal Database Project [http://rdp.cme.msu.edu/] [10]) as well as the minimal tendency to form secondary structures, including hairpin loops, heterodimers, and homodimers (assessed using the IDTDNA Oligoanalyzer 3.1 [Integrated DNA Technologies]) that may interfere with PCR efficiency (25). The optimal annealing temperature for each primer pair was determined by an 8-degree temperature (53°C to 64°C) gradient PCR using gDNA from _target bacterial reference strains as template (data not shown).
The amplification program for most qPCR assays consisted of an initial denaturation step at 95°C for 5 min; 40 cycles of denaturation at 95°C for 15 s, annealing at the optimal temperature for 30 s (with data acquisition), and elongation at 72°C for 30 s; and a final extension step at 72°C for 10 min. The elongation time for the Streptococcus qPCR assay was set at 20 s, whereas the denaturation and the elongation times for the Bifidobacterium qPCR assay were set at 30 s and 40 s, respectively. This was done, for practical reasons, to reduce the time to complete the Streptococcus qPCR assay and to provide sufficient time for denaturation and elongation of the relatively large amplicon (550 bp) produced during the Bifidobacterium qPCR assay. Melting curve analysis was carried out by incrementally increasing the temperature from 55°C to 95°C at 30 s per 0.5°C with continuous fluorescence collection.
For each qPCR assay, a standard curve comprising 8 serial 10-fold dilutions of full-length 16S rRNA gene PCR products was generated from _target gDNA preparations of the respective reference strains. For the total bacterial qPCR assay, a standard curve was generated using E. coli MC1061 gDNA. The standard curves of each qPCR assay were used to determine the relative contributions of _target bacterial groups to the total bacterial community in sample DNA preparations.
16S rRNA gene library construction and analysis.
The 27F-Nondeg and Uni-1492-rev primers (Table 1) were used for PCR amplification of 16S rRNA gene sequences from the undiluted extracted DNA of samples S1 and S2. Each reaction was performed in quadruplicate in a total volume of 50 μl containing 1× PCR buffer (Promega), 200 nM (each) primer (Biolegio), 200 μM (each) deoxyribonucleotide triphosphate (Promega), 1.25 U GoTaq DNA polymerase (Promega) and 1 μl of the extracted DNA. The amplification program was performed on a T1 thermocycler (Biometra, Göttingen, Germany) and consisted of an initial denaturation step at 94°C for 2 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 52°C for 40 s, and elongation at 72°C for 90 s; and a final extension step at 72°C for 5 min. PCR products were verified by gel electrophoresis using 5 μl of the reaction mixture on a 1.0% (wt/vol) agarose gel containing 0.4 μg/ml ethidium bromide (Bio-Rad). Quadruplicate PCR products from the same sample were pooled and subsequently purified with a High Pure cleanup microkit (Roche) using 10 μl elution buffer (Roche) for elution. The purified PCR products were diluted 10 times, of which 1 μl was used for ligation into the pGEM-T Easy vector (Promega) overnight at 4°C according to the manufacturer's instructions. XL1-Blue competent cells (75 μl; Stratagene, La Jolla, CA) were transformed with 2 μl ligation mixture according to the manufacturer's instructions and subsequently plated on LB agar containing ampicillin (100 μg/ml; Sigma), isopropyl-β-d-thiogalactopyranoside (IPTG; 0.16 mM; Carl Roth GmbH, Karlsruhe, Germany), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 100 μg/ml; Invitrogen, Carlsbad, CA) for blue-white color screening. White colonies were randomly selected and separately cultured overnight at 37°C in LB medium containing ampicillin. Subsequently, 2 sets of 96 clones from each clone library were randomly selected and the cloned inserts were sequenced from both ends using the T7 and SP6 priming sites (GATC-Biotech, Konstanz, Germany). The obtained sequences per clone were assembled using Clone Manager 9 Professional Edition (Scientific & Educational Software, Cary, NC), yielding nearly full-length 16S rRNA sequences, which were analyzed with DNA Baser v2.71.0 (Heracle Software, Lilienthal, Germany) to trim vector sequences. Subsequently, sequences were tested for chimeras using Mallard (2) according to the instructions of the authors with the default settings to identify chimeric sequences. Putatively anomalous 16S rRNA sequences identified by Mallard were further analyzed according to the anomaly confirmation protocol suggested by the authors, and unambiguously anomalous sequences were excluded from further microbiome interpretations. Nonchimeric sequences were taxonomically classified using the in-house customized RDP classifier described above. Sequences for which no classification could be obtained above the 80% confidence threshold were classified using the locally installed version of the RDP classifier version 2.2 (48) with a default confidence threshold of 80%.
The nonchimeric 16S rRNA gene sequences from each clone library were aligned using the SILVA Webaligner (34) and subsequently imported into ARB. Each clone library was manually screened (using a neighbor joining distance matrix, employing no correction, generated in ARB) for sequences showing <98% identity to 16S rRNA sequences represented in the human unique OTU database that was used for HITChip probe design.
Nucleotide sequence accession numbers.
The cloned 16S rRNA gene sequences from ileostomy effluent were deposited in the GenBank database and are available under accession numbers HQ176022 to HQ176318.
RESULTS
Analysis of pyrosequencing reads from 16S rRNA gene amplicons.
Pyrosequencing of the 16S rRNA gene PCR amplicons from fecal samples (F1 to F3), ileostomy effluent (S1 and S2), and ileal lumen content (S3) yielded in total 190,652 sequences with 7,944 ± 2,201 sequences per sample. Quality filtering passed approximately 50% of the pyrosequencing reads, with an average length of 224 nucleotides (nt) (Table 2; see Table S1 in the supplemental material for a comprehensive overview of the characteristics of pyrosequencing reads before and after quality filtering). Detailed analysis revealed that the majority (74.80% ± 4.74%) of the sequences that failed to pass quality filtering were due to sizes that were outside the sequence length thresholds (see Table S2).
TABLE 2.
Characteristics of sequence analysis before and after quality filtering
| Sample and primer | Characteristic before quality filtering |
Characteristic after quality filtering |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of sequences | Sequence length (nt) |
No. of sequences | Avg sequence length (nt) | % remaining quality filtered sequences | OTUa | Chao1a |
ACEa | Shannon diversity index | ||||
| Value | 95% CIa,b |
|||||||||||
| Avg | SD | Upper limit | Lower limit | |||||||||
| F1 | ||||||||||||
| 27F-DegL | 5,051 | 248.77 | 105.78 | 2,562 | 236.07 | 50.72 | 712 | 1,029 | 1,141 | 946 | 1,049 | 3.139 |
| 27F-DegS | 7,554 | 253.98 | 105.43 | 3,767 | 244.11 | 49.87 | 993 | 1,420 | 1,553 | 1,318 | 1,341 | 3.168 |
| 27F-Nondeg | 5,465 | 232.13 | 114.40 | 2,652 | 227.05 | 48.53 | 775 | 1,078 | 1,181 | 1,002 | 1,118 | 2.981 |
| 35F-Nondeg | 7,909 | 220.36 | 118.15 | 3,747 | 219.15 | 47.38 | 1,060 | 1,572 | 1,717 | 1,460 | 1,554 | 3.019 |
| F2 | ||||||||||||
| 27F-DegL | 10,425 | 247.38 | 98.65 | 5,431 | 233.67 | 52.10 | 1,199 | 1,591 | 1,704 | 1,503 | 1,587 | 3.269 |
| 27F-DegS | 11,591 | 258.18 | 100.93 | 5,803 | 245.61 | 50.06 | 1,217 | 1,618 | 1,735 | 1,527 | 1,597 | 3.275 |
| 27F-Nondeg | 9,524 | 230.07 | 110.93 | 4,704 | 225.01 | 49.39 | 1,139 | 1,572 | 1,697 | 1,476 | 1,565 | 3.231 |
| 35F-Nondeg | 7,932 | 228.53 | 110.98 | 4,071 | 223.96 | 51.32 | 1,070 | 1,569 | 1,715 | 1,457 | 1,524 | 3.190 |
| F3 | ||||||||||||
| 27F-DegL | 4,782 | 230.79 | 100.25 | 2,595 | 218.93 | 54.27 | 703 | 1,103 | 1,245 | 997 | 1,058 | 2.845 |
| 27F-DegS | 5,349 | 227.53 | 105.71 | 2,784 | 220.06 | 52.05 | 709 | 1,036 | 1,156 | 949 | 1,019 | 2.828 |
| 27F-Nondeg | 7,430 | 204.91 | 110.82 | 3,606 | 207.95 | 48.53 | 981 | 1,424 | 1,553 | 1,324 | 1,487 | 2.832 |
| 35F-Nondeg | 4,584 | 201.40 | 113.57 | 2,191 | 202.30 | 47.80 | 649 | 985 | 1,111 | 894 | 950 | 2.774 |
| S1 | ||||||||||||
| 27F-DegL | 10,393 | 237.59 | 96.99 | 5,016 | 217.23 | 48.26 | 687 | 910 | 1,008 | 842 | 864 | 2.105 |
| 27F-DegS | 10,320 | 243.42 | 96.71 | 5,136 | 227.11 | 49.77 | 738 | 978 | 1,076 | 908 | 960 | 2.084 |
| 27F-Nondeg | 9,593 | 217.34 | 100.03 | 4,660 | 203.31 | 48.58 | 664 | 838 | 914 | 785 | 844 | 1.914 |
| 35F-Nondeg | 11,378 | 234.07 | 108.65 | 5,822 | 227.00 | 51.17 | 842 | 1,067 | 1,154 | 1,004 | 1,056 | 2.190 |
| S2 | ||||||||||||
| 27F-DegL | 7,925 | 246.04 | 97.97 | 3,993 | 227.30 | 50.38 | 722 | 964 | 1,061 | 894 | 928 | 2.329 |
| 27F-DegS | 10,053 | 241.03 | 101.26 | 4,930 | 226.91 | 49.04 | 778 | 973 | 1,051 | 917 | 980 | 2.385 |
| 27F-Nondeg | 7,628 | 228.75 | 106.49 | 3,752 | 216.33 | 49.19 | 669 | 852 | 929 | 797 | 862 | 2.160 |
| 35F-Nondeg | 10,294 | 227.87 | 113.33 | 4,509 | 216.00 | 43.80 | 758 | 965 | 1,047 | 907 | 983 | 2.285 |
| S3 | ||||||||||||
| 27F-DegL | 5,924 | 247.74 | 100.82 | 3,127 | 234.86 | 52.79 | 792 | 1,079 | 1,179 | 1,005 | 1,092 | 2.899 |
| 27F-DegS | 6,501 | 240.44 | 105.29 | 3,456 | 235.31 | 53.16 | 901 | 1,301 | 1,428 | 1,204 | 1,294 | 3.021 |
| 27F-Nondeg | 6,759 | 218.29 | 114.20 | 3,217 | 216.92 | 47.60 | 917 | 1,374 | 1,514 | 1,267 | 1,355 | 3.159 |
| 35F-Nondeg | 6,288 | 218.97 | 114.26 | 3,072 | 213.99 | 48.85 | 859 | 1,196 | 1,305 | 1,114 | 1,215 | 3.163 |
Calculated at an 0.02 distance level.
95% CI, 95% confidence interval.
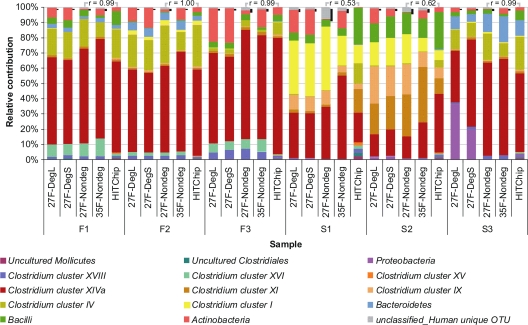
Microbial profiles, based on the level 1 (phylum-like [Fig. 2]) and level 2 (genus-like [see Fig. S1 in the supplemental material]) taxonomic assignments of the pyrosequences that passed quality filtering, were constructed for all samples. As anticipated, most pyrosequences were assigned to the Firmicutes (85.6%), Actinobacteria (7.6%), and Bacteroidetes (2.9%; predominantly encountered in fecal samples F1 and F2 as well as in the ileal lumen content sample S3). Notably, only 1.5% of all pyrosequences could not be classified using the confidence threshold of 80% and are represented as Unclassified_Human unique OTU (Fig. 2). Furthermore, microbial profiles obtained using all pyrosequences (without quality filtering) revealed essentially the same profiles as those obtained with the quality-filtered sequences, albeit with a raised abundance of the Unclassified_Human unique OTU (see Fig. S2), indicating that the quality filtering step does not drastically influence the reconstruction of the microbial community but predominantly eliminates noise.
FIG. 2.
Relative contributions of detected bacterial phyla with pyrosequencing using four different forward primers and HITChip analysis for community data at level 1. Pearson product-moment correlation coefficients (r) between pairs of profiles are shown above the bars. The phylum Firmicutes was subdivided into Bacilli, Clostridium clusters, uncultured Mollicutes, and uncultured Clostridiales. Pyrosequences that could not be classified above the confidence threshold of 80% are grouped to Unclassified_Human unique OTU, which is indicated in the microbial profiles with shadowing (black bars). Phylogenetic groups that contribute at least 1% to one of the profiles are indicated in the color key.
Hierarchical clustering of the microbial profiles revealed separate grouping of fecal samples and ileum-lumen content from ileostomy effluent samples (see Fig. S3A in the supplemental material). The divergence between these clusters was most apparent for phylogenetic groups belonging to the Firmicutes, with fecal samples and ileum-lumen content being abundant in Clostridium clusters IV (12.3% ± 7.3%), XIVa (58.6% ± 8.6%), XVI (4.4% ± 3.8%), and XVIII (3.0% ± 1.8%), while Bacilli (9.2% ± 4.5%) and Clostridium clusters I (24.0% ± 11.5%), IX (13.3% ± 7.1%), XI (14.0% ± 14.1%), and XIVa (27.2% ± 13.4%) were predominant in ileostomy effluent. Moreover, species richness, as reflected by Chao1 and ACE supported by the rarefaction curves, as well as the Shannon diversity index, was higher in fecal samples and ileum-lumen content than in ileostomy effluent (Table 2; see also Fig. S4).
The effects of different forward primers on microbial profiling by barcoded pyrosequencing.
To determine the effects of different primers on microbial profiling, 16S rRNA gene PCR amplicons were generated for each intestinal sample using four different forward primers (27F-DegL, 27F-DegS, 27F-Nondeg, and 35F-Nondeg). Microbial profiles constructed on the basis of pyrosequences per sample were highly correlated for the different primers (average r of 0.88 ± 0.14 at level 2 community data [data not shown]). Furthermore, hierarchical clustering (see Fig. S3A in the supplemental material) of the microbial profiles revealed distinct clusters of microbial profiles for each of the samples using the four forward primers, except for sample S3 as discussed below, indicating that the effect of primers on microbial profiling is smaller than the sample-specific effect and supporting a high level of technical reproducibility of the pyrosequencing method. Analogously, comparison of rarefaction curves (see Fig. S4) and species richness estimators Chao1 and ACE, as well as Shannon diversity indices (Table 2), did not reveal a particular primer giving consistently the highest or lowest value for any of these ecological metrics. Nonetheless, qualitative comparison demonstrated that the microbial profiles deduced from pyrosequencing using amplicons generated with the 27F-DegL, 27F-DegS, and 35F-Nondeg primers were notably more abundant in Actinobacteria than were those using the 27F-Nondeg primer (Fig. 2), confirming previous reports on the underestimation of the Actinobacteria using the 27F-Nondeg primer (21).
Because the PCR annealing temperatures used for amplicon generation differed between pyrosequencing and HITChip analysis (56°C versus 52°C, respectively), both annealing temperatures were employed for HITChip analysis of sample F3 to investigate the effects of different annealing temperatures on microbial profiling. Results demonstrated highly similar microbial profiles (r ≥ 0.99; data not shown), and therefore, comparison of microbiota profiling by pyrosequencing and that by HITChip analysis was not biased by different PCR annealing temperatures.
Comparison of barcoded pyrosequencing with HITChip analysis.
The concordance between microbial profiling by pyrosequencing and that by phylogenetic microarray analysis was evaluated. Although the principles for classification and abundance estimations of microbial community members differ between these technologies, hierarchical clusterings of the microbial profiles from the two methods matched (see Fig. S3 in the supplemental material).
Microbial profiles as a result of HITChip analysis were compared to those from pyrosequencing using the 27F-Nondeg primer, as this forward primer is used for amplicon generation in the HITChip analytic procedure. The resulting comparison of the community data at level 1 (phylum-like) showed a high correlation for the fecal samples (F1 to F3; r = 0.99 to 1.00) and ileum-lumen content (S3; r = 0.99), while the correlation was lower for ileostomy effluent samples (S1 and S2; r = 0.53 to 0.62) (Fig. 2). Correlations for the community data at level 2 (genus-like) were significantly lower but remained highest for the fecal samples (r = 0.63 to 0.78) and ileum-lumen content (r = 0.71) and lowest for the ileostomy effluent samples (r = 0.31 to 0.49) (see Fig. S1 in the supplemental material). This difference between ileostomy effluent and other intestinal samples is also demonstrated in the community data scatter plots at levels 1 and 2 (see Fig. S5). Remarkably, numerous phylogenetic groups at level 2 showed significant abundances with HITChip analysis but were absent from the pyrosequence data set (see Fig. S5 inset), suggesting that HITChip analysis enables detection of low-abundance bacterial groups by its dynamic range being broader than that of pyrosequencing.
To verify if suboptimal HITChip probe matches could potentially explain higher abundances per cluster with grouping of fecal samples and ileal lumen content (cluster I) apart from ileostomy effluent samples (cluster II) of some phylogenetic _targets in the pyrosequence data relative to HITChip analysis (see Fig. S6 in the supplemental material), pyrosequences were screened for exact matches with the HITChip probes designed for detection of these phylogenetic groups (see Fig. S7). In general, the two clusters showed approximately the same fractions (∼90%) of pyrosequences that had a perfect match with at least one of the HITChip probes. For cluster I, Eubacterium rectale et rel. and Ruminococcus obeum et rel. contained the most pyrosequences that lack a HITChip probe perfect match, while Veillonella showed the highest number of sequences (19.3%) without a perfect match for the HITChip probes for cluster II.
Phylogenetic groups for which the abundance estimates were higher in the HITChip analyses than in pyrosequencing included Streptococcus spp. (Bacilli), for which a deviation in relative abundance estimations between the two profiling technologies was as high as ∼7% (see Fig. S6 in the supplemental material). The abundances of this phylogenetic group and others were further investigated by means of qPCR as well as 16S rRNA gene cloning and sequencing (see below).
Quantification of bacterial groups by qPCR.
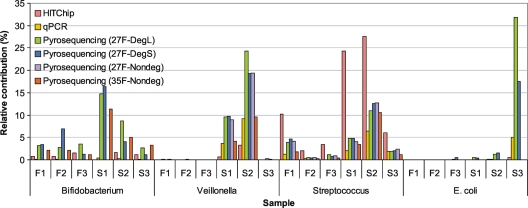
To evaluate the performances of pyrosequencing and HITChip analysis in estimating relative abundances of microbial community members, the results of the two techniques were compared with those obtained by means of group-specific qPCR, focusing on Bifidobacterium, Veillonella, Streptococcus, and E. coli (Fig. 3).
FIG. 3.
Comparison of the relative contributions as determined by means of HITChip, qPCR assays, and pyrosequencing for 4 phylogenetic groups in fecal (F) and small intestinal (S) samples. Relative contributions as assessed by qPCR assays were not determined for sample F3.
The estimated community proportion of Streptococcus spp. was consistently highest with HITChip analysis relative to qPCR assays and pyrosequencing, while Bifidobacterium abundance levels were expectedly low when determined with the HITChip as a result of using the 27F-Nondeg primer for the initial sample preparation. Surprisingly, Bifidobacterium abundances assessed with the qPCR assay were relatively low as well, whereas estimations by pyrosequencing using the 27F-DegL, 27F-DegS, and 35F-Nondeg primers showed relative abundances varying from 1.10% to 6.98% for samples F1, F2, F3, and S3 and from 4.04% to 16.37% for samples S1 and S2. For Veillonella and E. coli, relative abundances were highest by pyrosequencing analysis, followed by intermediate values obtained from qPCR and lowest values assessed by HITChip analysis. Sample S3 was the only sample for which an E. coli abundance above 1.5% was detected, and it showed relative contributions as high as 31.8% and 17.4% in microbial profiles from pyrosequencing using the 27F-DegL and 27F-DegS primers, respectively. Although the proportion of E. coli bacteria as assessed by the HITChip was considerably lower, with an abundance of 0.44%, the E. coli-specific qPCR assay revealed a contribution of 5% and confirms the abundant presence of E. coli in sample S3. Additionally, this observation suggests that microbial profiling using primers 27F-DegL and 27F-DegS results in a more accurate representation of the microbial composition in intestinal samples that contain higher proportions of E. coli.
Screening ileostomy effluent for novel bacterial phylotypes.
Based on the prominent deviations between pyrosequencing and HITChip analysis for ileostomy effluent, these small intestinal samples are a potential source for a range of bacteria with 16S rRNA sequences that are absent from the human unique OTU database to date. To support this notion, 16S rRNA clone libraries for the two ileostomy effluent samples were constructed and analyzed (Fig. 4).
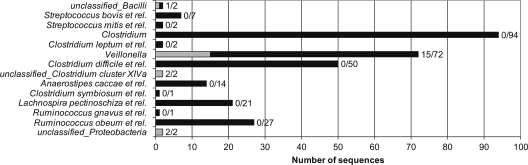
FIG. 4.
Total numbers of cloned sequences detected per phylogenetic group (black) and numbers of sequences showing <98% identity to the 16S rRNA sequences represented in the human unique OTU database (gray) for ileostomy samples S1 and S2. The ratio of number of sequences showing <98% identity to the 16S rRNA sequences represented in the human unique OTU database to the total number of cloned sequences is provided for each level 2 group. Sequences that could not be classified above the confidence threshold of 80% are grouped to Unclassified_ at the specific rank per taxon.
A total number of 139 and 158 cloned nonanomalous 16S rRNA gene sequences were obtained for the S1 and S2 16S rRNA clone libraries, respectively. Out of the total of 297 cloned sequences, 6 could not be classified at level 2 assignments above the 80% confidence threshold using the RDP classifier trained with the human unique OTU database. Consequently, these sequences were grouped to Unclassified_ with the specific level 1 assignment (Fig. 4). To determine if the resolution of the identifications could be improved, sequences were reclassified using the standard RDP classifier (see Materials and Methods), which is based on a set of 16S rRNA gene sequences more exhaustive than that of the human unique OTU database. This showed that both Unclassified_Bacilli sequences were assigned to the order Lactobacillales, though deviating in their genus-level classifications as Granulicatella for one and Streptococcus for the other. The sequence classified as the latter also showed <98% identity to 16S rRNA gene sequences represented in the human unique OTU database. Sequences of the Unclassified_Clostridium cluster XIVa group could be classified no further than the family Lachnospiraceae, whereas the Unclassified_Proteobacteria were assigned to the genus Variovorax belonging to Betaproteobacteria.
Further analysis of the libraries revealed that 20 sequences, predominantly belonging to Veillonella, showed <98% identity to 16S rRNA gene sequences represented in the human unique OTU database and were therefore considered to be phylotypes not previously reported as being associated with the human intestine (Fig. 4). The finding of a relatively large proportion of these phylotypes among the Veillonella 16S rRNA gene sequences supports the suggestion that the HITChip probes display relatively poor sequence matches with these sequences (see above). Detailed analysis indeed showed that 5 out of 7 HITChip probes specific for Veillonella had more than 2 mismatches with sequences classified as Veillonella, whereas the remaining two probes at most had 1 mismatch (see Fig. S8 in the supplemental material).
The same screening strategy was applied to determine if the cloned sequences classified as Veillonella had exact matches with the forward and reverse primers used for the Veillonella qPCR assay. Out of the 72 cloned sequences classified as Veillonella, only one sequence did not show a perfect match with the reverse primer. In contrast, 16 sequences had a single mismatch with the forward primer (see Fig. S8 in the supplemental material). Interestingly, 13 of these sequences were also identified as novel intestinal phylotypes (see above). This shows that the forward primer for the Veillonella qPCR assay is not in agreement with the 16S rRNA gene sequences of Veillonella and could also explain the lower Veillonella abundances as determined by qPCR in comparison with those estimated by pyrosequencing (Fig. 3). Taken together, these results suggest that the human small intestine is inhabited by novel Veillonella phylotypes that previously have not been reported to inhabit this niche.
DISCUSSION
In this study, the performances of two culture-independent techniques, barcoded pyrosequencing and phylogenetic microarray analysis using the HITChip, were compared and contrasted for profiling of human fecal and small intestinal microbial communities. The two techniques generated similar microbial composition profiles for fecal and terminal ileum-lumen samples, whereas more distinct profiles were obtained for ileostomy effluent samples. Ileostomy effluent, in comparison to fecal samples, contained less rich and diverse microbial communities, which were abundant in Streptococcus spp., Veillonella spp., and members of several Clostridium clusters. These findings are consistent with results published by Booijink et al. (5) as well as with the recent study by Zoetendal et al. that concluded that the phylogenetic composition in ileostomy effluent is different from that of the ileum and resembles the microbiota in the proximal small intestine, i.e., the jejunum and proximal ileum (Zoetendal et al., submitted).
The high comparability of the pyrosequencing- and HITChip-derived microbial profiles obtained for fecal samples is in agreement with previously published results (8). Thereby, this study confirms that the two profiling technologies facilitate robust microbial profiling and generate essentially equivalent biological conclusions regarding compositions of microbial communities. Nonetheless, abundance estimates for several phylogenetic groups deviated between pyrosequencing and HITChip analysis. Determining the exact cause for the technical divergence is not trivial, but possible reasons for this are (i) probe-based versus sequence-based quantification, (ii) failure to detect species that were not represented in the reference sequences used for probe design and cognate overestimation of relative abundances of the detected phylogenetic groups, (iii) sequencing errors, (iv) incorrect taxonomic classification of sequences, and/or (v) a difference in dynamic ranges between technologies. The last was also apparent from comparison of HITChip analyses with very deep pyrosequencing, resulting in a level of depth that is comparable to close to 200,000 reads per sample (8). Sequencing depth could be improved by employing the Illumina sequencing platform, with which microbial community diversity is analyzed with increased depth relative to pyrosequencing (7, 9). However, to date, this approach is still challenging due to the limited phylogenetic resolution obtained from such short sequence reads and the increasing sequence error rates for reads extended beyond 60 bp.
A challenging yet essential part of pyrosequencing analysis is quality control of the acquired data set. Here, a strict quality filtering procedure that eliminated approximately half of the pyrosequences, most of which were deemed either too short or too long, was employed. The proportion of sequences excluded from further analysis due to quality filtering was higher than those in other studies applying similar exclusion criteria. Those studies, however, employed the older GS 20 (1, 24) or 454 GS FLX (8) sequencing platform, while in this study the titanium method was applied.
HITChip probe design is based on a 16S rRNA gene database that predominantly contains sequences with a fecal or colonic origin. Since the two profiling techniques correlated strongly for fecal samples and the HITChip offers a broader dynamic range of detection, HITChip analysis is preferred for profiling of the lower GI tract microbiota. However, HITChip coverage of the small intestinal microbiota appeared to be more incomplete (5), and therefore, pyrosequencing would be the method of choice for de novo profiling of these microbial communities. Furthermore, this finding exemplifies the intrinsic constraint of microarray approaches that are limited to detection of phylogenetic groups for which sequences were included during array design. This may to a large extent explain the lower correlations between pyrosequencing and HITChip analysis that were obtained for the ileostomy effluent samples and suggests that ileostomy effluent harbors novel intestinal bacteria that have not been detected in feces or other large intestinal samples. To identify these phylotypes, pyrosequences were screened per phylogenetic group for perfect matches with the respective HITChip probes. All groups had multiple sequences without a probe match, which may indicate the presence of novel intestinal phylotypes, suggesting that HITChip probe design may be improved by the addition of probes to detect this expanding community. The latter would require a flexible array design strategy as suggested by Rajilić-Stojanović et al. (35). However, despite quality filtering to improve overall pyrosequence data set reliability, this technology still suffers from a relatively high sequence error rate (∼0.5% for the GS 20 and GS FLX platform [31]), which erroneously may contribute to the number of sequences that mismatch with the HITChip probes. Analogously, the pyrosequencing technology was reported to overestimate microbial diversity as a consequence of these sequencing errors (37).
Results from screening cloned 16S rRNA gene sequences from the ileostomy effluent samples showed that 7% of the cloned sequences represented novel intestinal phylotypes, which appears to be lower than might have been anticipated on the basis of previous studies (5, 20) The novel phylotypes encountered here predominantly belonged to the Veillonella group, which is in agreement with the relatively large fraction of pyrosequences corresponding to this group that lacked a perfect-match HITChip probe. Other novel phylotypes from the clone libraries were identified as Variovorax bacteria, which have been cultured from soil (42) and have been detected in the rabbit cecum (30). However, to the best of our knowledge, Variovorax spp. have not been identified as inhabitants of the human GI tract to date. Therefore, the human small intestine contains a range of species that were not previously associated with this niche, and elucidating the role of these microorganisms, especially of the abundant Veillonella, in their environment is a task for the future.
Quantification of bacterial groups by means of qPCR was chosen as a benchmark technology to investigate the deviations in profiling between pyrosequencing and HITChip analysis. Discrepancies were observed between relative abundances as determined by qPCR and the two profiling techniques. This can at least in part be attributed to the difference in abundance calculations, which are based on a separate total bacterial assessment for qPCR, whereas for the HITChip and pyrosequencing relative abundances are calculated based on probe signals or number of pyrosequences per phylogenetic group as part of the total, respectively. Streptococcus abundance levels were consistently highest with HITChip analysis relative to qPCR assays and pyrosequencing. This observation corroborates the results previously published by Rajilić-Stojanović and colleagues (35), who reported a significantly higher relative abundance estimate of Streptococcus spp. in fecal samples based on HITChip analysis than based on group-specific FISH analysis.
PCR amplification was performed using four different forward primers for each intestinal sample to assess the impact of PCR primer choice on microbial profiling by means of pyrosequencing. With the exception of sample S3, microbiota compositions per sample were highly similar for the different primers. This underpins the degree of reproducibility of microbial profiling by means of pyrosequencing and suggests that correlations between technical replicates can be expected to be even higher. Moreover, primer choice did not profoundly affect species richness and diversity estimates, which is in agreement with a recent study that showed consistent species evenness estimates when using different primer pairs _targeting the same region of the 16S rRNA gene (14). Qualitative analysis of the microbial profiles, however, clearly revealed a lower abundance of Actinobacteria found by the 27F-Nondeg primer, which confirms the observations by Hayashi et al. (21) showing that the 27F-Nondeg primer is incomplete in its coverage of Bifidobacterium spp.
In conclusion, this paper demonstrates that different primers and high-throughput 16S rRNA profiling technologies like barcoded pyrosequencing and HITChip analysis provide overall similar results. However, this similarity is dependent on the origin of the samples, which relates to the sequences used during array design and may thus be influenced by updated microarray design to accommodate novel sequences. Nonetheless, based on the results described here, it is our recommendation to use either the 27F-DegL or the 27F-DegS primer, since both these multiple-degenerate primers appear to provide a more complete assessment of Actinobacteria and E. coli abundances.
Supplementary Material
Acknowledgments
We appreciate the help of Carien Booijink and Freddy Troost in providing ileostomy effluent and ileum-lumen samples and DNA isolated therefrom. We thank Sebastian Tims and Hans Heilig for providing fecal sample DNA and assistance with the HITChip analyses, Muriel Derrien and Odette Pérez Gutiérrez for their help in phylogenetic analysis of cloned sequences, and Hauke Smidt and Mirjana Rajilić-Stojanović for critical reading of the manuscript. We thank Christopher Bauser, Andrea Bolte, and Manuela Hinz of GATC-Biotech (Konstanz, Germany) for assistance in the setup of the pyrosequencing experiments and Benli Chai from RDP (East Lansing, MI) for his assistance in customizing the RDP classifier.
This project was supported by the Netherlands Bioinformatics Centre (NBIC).
Footnotes
Published ahead of print on 21 January 2011.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersson, A. F., et al. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Amor, K., et al. 2005. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl. Environ. Microbiol. 71:4679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booijink, C. C., et al. 2010. High temporal and inter-individual variation detected in the human ileal microbiota. Environ. Microbiol. 12:3213-3227. [DOI] [PubMed] [Google Scholar]
- 6.Booijink, C. C., E. G. Zoetendal, M. Kleerebezem, and W. M. de Vos. 2007. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol. 2:285-295. [DOI] [PubMed] [Google Scholar]
- 7.Caporaso, J. G., et al. 3 June 2010, posting date. Microbes and Health Sackler Colloquium: global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. doi:10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed]
- 8.Claesson, M. J., et al. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claesson, M. J., et al. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, J. R., et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, M. D., et al. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 12.Eckburg, P. B., et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egert, M., et al. 2007. Identification of glucose-fermenting bacteria present in an in vitro model of the human intestine by RNA-stable isotope probing. FEMS Microbiol. Ecol. 60:126-135. [DOI] [PubMed] [Google Scholar]
- 14.Engelbrektson, A., et al. 2010. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 4:642-647. [DOI] [PubMed] [Google Scholar]
- 15.Frank, J. A., et al. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74:2461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furet, J. P., P. Quenee, and P. Tailliez. 2004. Molecular quantification of lactic acid bacteria in fermented milk products using real-time quantitative PCR. Int. J. Food Microbiol. 97:197-207. [DOI] [PubMed] [Google Scholar]
- 17.Garrity, G. M., J. A. Bell, and T. G. Lilburn. 2004. Taxonomic outline of the prokaryotes, p. 401. Bergey's manual of systematic bacteriology, 2nd ed., release 5.0. Springer-Verlag, New York, NY.
- 18.Hamady, M., and R. Knight. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 19:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen, H. J., G. C. Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman, A. L., et al. 2009. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc. Natl. Acad. Sci. U. S. A. 106:17187-17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi, H., M. Sakamoto, and Y. Benno. 2004. Evaluation of three different forward primers by terminal restriction fragment length polymorphism analysis for determination of fecal bifidobacterium spp. in healthy subjects. Microbiol. Immunol. 48:1-6. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huijsdens, X. W., et al. 2002. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40:4423-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huse, S. M., J. A. Huber, H. G. Morrison, M. L. Sogin, and D. M. Welch. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubista, M., et al. 2006. The real-time polymerase chain reaction. Mol. Aspects Med. 27:95-125. [DOI] [PubMed] [Google Scholar]
- 26.Leser, T. D., and L. Molbak. 2009. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ. Microbiol. 11:2194-2206. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig, W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margulies, M., et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuki, T., et al. 2002. Development of 16S rRNA-gene-_targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteils, V., L. Cauquil, S. Combes, J. J. Godon, and T. Gidenne. 2008. Potential core species and satellite species in the bacterial community within the rabbit caecum. FEMS Microbiol. Ecol. 66:620-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu, B., L. Fu, S. Sun, and W. Li. 2010. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paliy, O., H. Kenche, F. Abernathy, and S. Michail. 2009. High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray. Appl. Environ. Microbiol. 75:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer, C., et al. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 34:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pruesse, E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajilic-Stojanovic, M., et al. 2009. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 11:1736-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajilic-Stojanovic, M., H. Smidt, and W. M. de Vos. 2007. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9:2125-2136. [DOI] [PubMed] [Google Scholar]
- 37.Reeder, J., and R. Knight. 2009. The ‘rare biosphere’: a reality check. Nat. Methods 6:636-637. [DOI] [PubMed] [Google Scholar]
- 38.Rinttila, T., A. Kassinen, E. Malinen, L. Krogius, and A. Palva. 2004. Development of an extensive set of 16S rDNA-_targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166-1177. [DOI] [PubMed] [Google Scholar]
- 39.Rudney, J. D., Y. Pan, and R. Chen. 2003. Streptococcal diversity in oral biofilms with respect to salivary function. Arch. Oral Biol. 48:475-493. [DOI] [PubMed] [Google Scholar]
- 40.Salonen, A., et al. 2010. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 81:127-134. [DOI] [PubMed] [Google Scholar]
- 41.Schwiertz, A., G. Le Blay, and M. Blaut. 2000. Quantification of different Eubacterium spp. in human fecal samples with species-specific 16S rRNA-_targeted oligonucleotide probes. Appl. Environ. Microbiol. 66:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorensen, S. R., et al. 2005. Elucidating the key member of a linuron-mineralizing bacterial community by PCR and reverse transcription-PCR denaturing gradient gel electrophoresis 16S rRNA gene fingerprinting and cultivation. Appl. Environ. Microbiol. 71:4144-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, Y., et al. 2009. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 37:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turnbaugh, P. J., et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van den Bogert, B., M. M. Leimena, W. M. de Vos, E. G. Zoetendal, and M. Kleerebezem. Functional intestinal metagenomics. In F. J. de Bruin (ed.), Handbook of molecular microbial ecology II: metagenomics in different habitats, in press. Wiley, New York, NY.
- 47.Vaughan, E. E., et al. 2000. A molecular view of the intestinal ecosystem. Curr. Issues Intest. Microbiol. 1:1-12. [PubMed] [Google Scholar]
- 48.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoetendal, E. G., C. T. Collier, S. Koike, R. I. Mackie, and H. R. Gaskins. 2004. Molecular ecological analysis of the gastrointestinal microbiota: a review. J. Nutr. 134:465-472. [DOI] [PubMed] [Google Scholar]
- 50.Zoetendal, E. G., et al. 2006. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 1:870-873. [DOI] [PubMed] [Google Scholar]
- 51.Zoetendal, E. G., M. Rajilic-Stojanovic, and W. M. de Vos. 2008. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57:1605-1615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.