Abstract
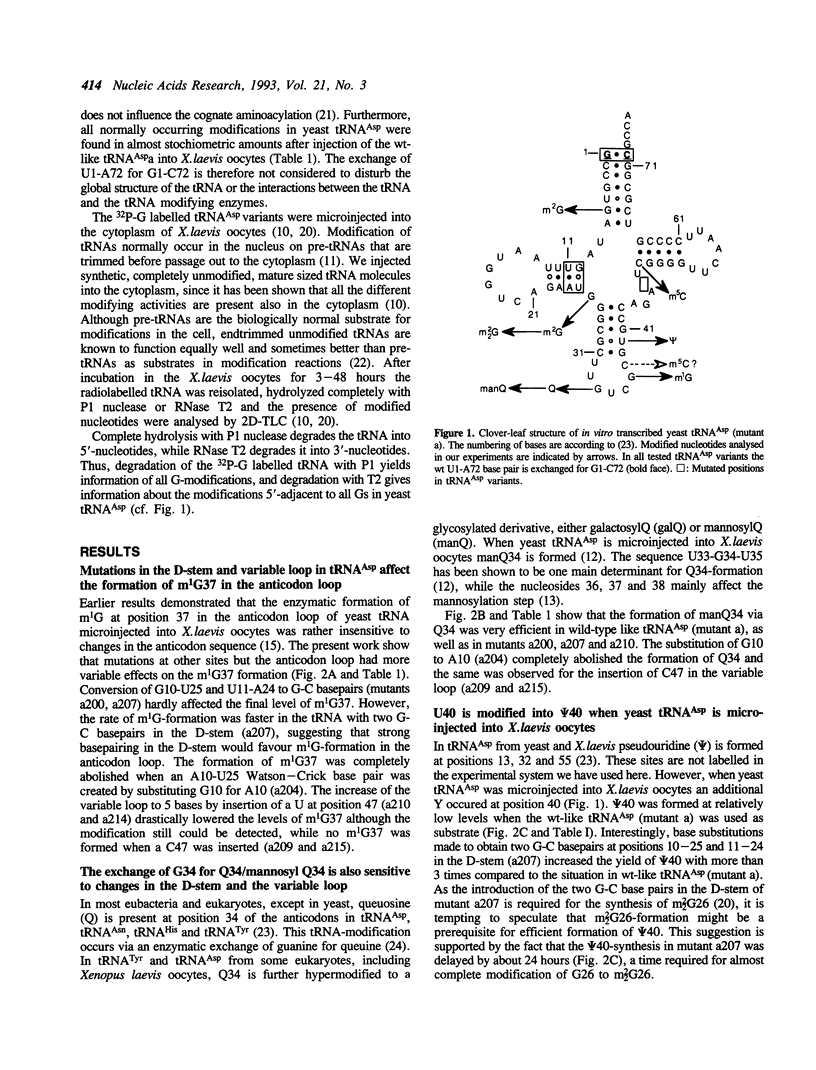
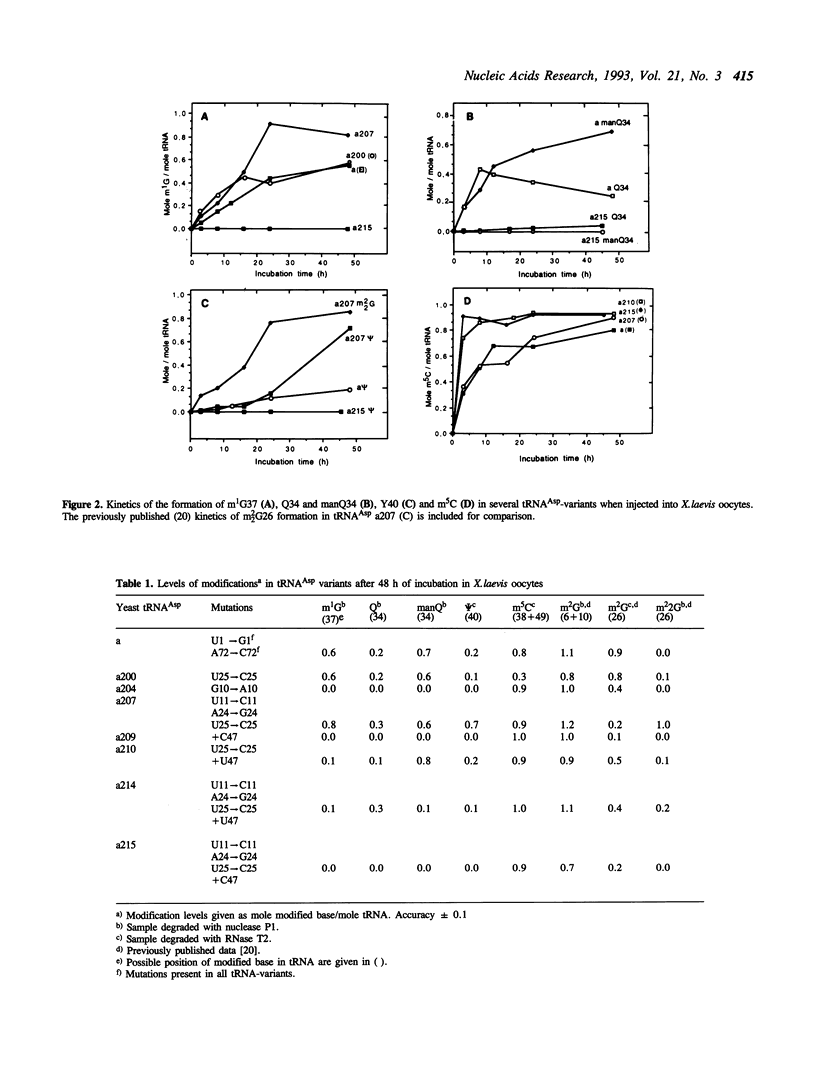
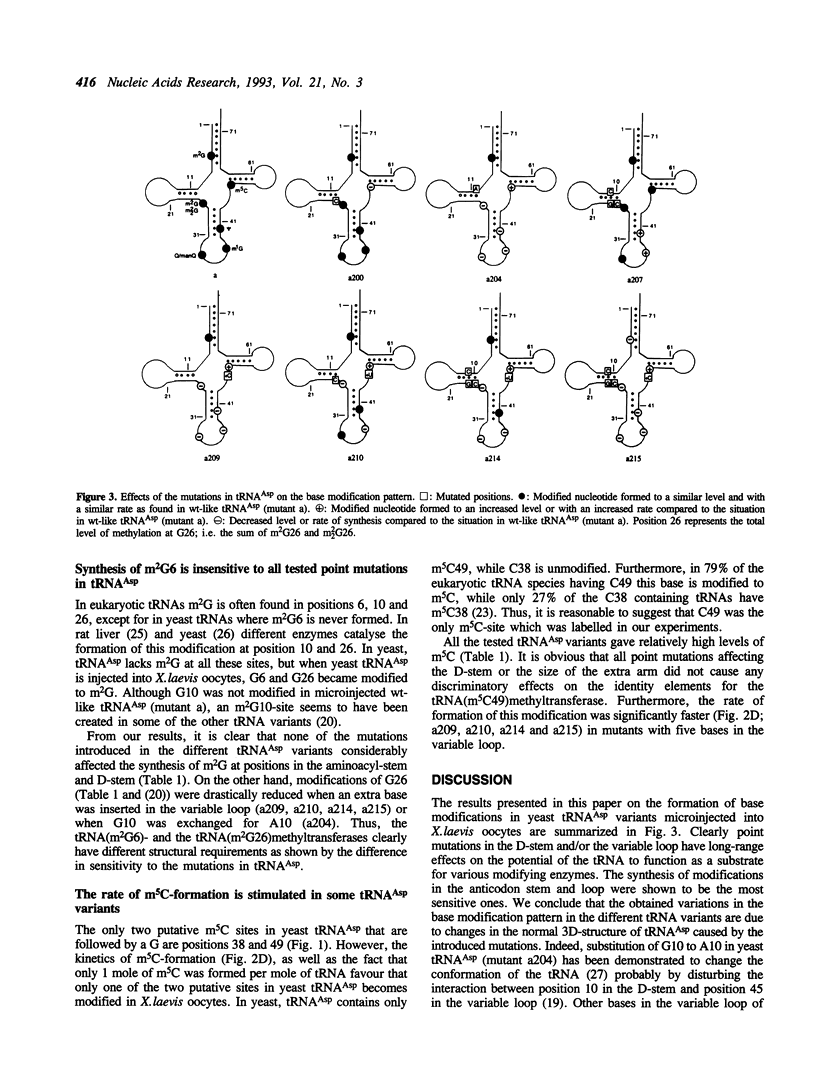
The base-modification pattern has been studied in several synthetic variants of yeast tRNA(Asp) injected into Xenopus laevis oocytes. Certain point mutations in the D-stem and the variable loop of the tRNA led to considerably decreased levels of m1G37, psi 40 and Q34/manQ34 in the anticodon stem or loop and an increased rate of synthesis for m5C49 in the T-stem. The formation of m2G6 in the aminoacyl-stem was not affected in any of the tRNA-variants. Thus, mutations in one part of the tRNA-molecule can have long-range effects on the interactions between another part of the tRNA and the tRNA modifying enzymes.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Björk G. R., Ericson J. U., Gustafsson C. E., Hagervall T. G., Jönsson Y. H., Wikström P. M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- Carbon P., Haumont E., Fournier M., de Henau S., Grosjean H. Site-directed in vitro replacement of nucleosides in the anticodon loop of tRNA: application to the study of structural requirements for queuine insertase activity. EMBO J. 1983;2(7):1093–1097. doi: 10.1002/j.1460-2075.1983.tb01551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. R., Nierlich D. P. The methylation of transfer RNA in Escherichia coli. Biochim Biophys Acta. 1974 Nov 20;374(1):23–37. doi: 10.1016/0005-2787(74)90196-8. [DOI] [PubMed] [Google Scholar]
- Droogmans L., Grosjean H. Enzymatic conversion of guanosine 3' adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: dependence on the anticodon sequence. EMBO J. 1987 Feb;6(2):477–483. doi: 10.1002/j.1460-2075.1987.tb04778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droogmans L., Haumont E., de Henau S., Grosjean H. Enzymatic 2'-O-methylation of the wobble nucleoside of eukaryotic tRNAPhe: specificity depends on structural elements outside the anticodon loop. EMBO J. 1986 May;5(5):1105–1109. doi: 10.1002/j.1460-2075.1986.tb04329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds C. G., Crain P. F., Gupta R., Hashizume T., Hocart C. H., Kowalak J. A., Pomerantz S. C., Stetter K. O., McCloskey J. A. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J Bacteriol. 1991 May;173(10):3138–3148. doi: 10.1128/jb.173.10.3138-3148.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., De Henau S., Doi T., Yamane A., Ohtsuka E., Ikehara M., Beauchemin N., Nicoghosian K., Cedergren R. The in vivo stability, maturation and aminoacylation of anticodon-substituted Escherichia coli initiator methionine tRNAs. Eur J Biochem. 1987 Jul 15;166(2):325–332. doi: 10.1111/j.1432-1033.1987.tb13518.x. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Droogmans L., Giégé R., Uhlenbeck O. C. Guanosine modifications in runoff transcripts of synthetic transfer RNA-Phe genes microinjected into Xenopus oocytes. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):267–273. doi: 10.1016/0167-4781(90)90179-6. [DOI] [PubMed] [Google Scholar]
- Gu X. R., Santi D. V. The T-arm of tRNA is a substrate for tRNA (m5U54)-methyltransferase. Biochemistry. 1991 Mar 26;30(12):2999–3002. doi: 10.1021/bi00226a003. [DOI] [PubMed] [Google Scholar]
- Haumont E., Droogmans L., Grosjean H. Enzymatic formation of queuosine and of glycosyl queuosine in yeast tRNAs microinjected into Xenopus laevis oocytes. The effect of the anticodon loop sequence. Eur J Biochem. 1987 Oct 1;168(1):219–225. doi: 10.1111/j.1432-1033.1987.tb13408.x. [DOI] [PubMed] [Google Scholar]
- Haumont E., Fournier M., de Henau S., Grosjean H. Enzymatic conversion of adenosine to inosine in the wobble position of yeast tRNAAsp: the dependence on the anticodon sequence. Nucleic Acids Res. 1984 Mar 26;12(6):2705–2715. doi: 10.1093/nar/12.6.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. M., Andraos-Selim C., Roberts I., Wahab S. Z. Structural requirements for tRNA methylation. Action of Escherichia coli tRNA(guanosine-1)methyltransferase on tRNA(1Leu) structural variants. J Biol Chem. 1992 Jul 5;267(19):13440–13445. [PubMed] [Google Scholar]
- Hori H., Saneyoshi M., Kumagai I., Miura K., Watanabe K. Effects of modification of 4-thiouridine in E. coli tRNA(fMet) on its methyl acceptor activity by thermostable Gm-methylases. J Biochem. 1989 Nov;106(5):798–802. doi: 10.1093/oxfordjournals.jbchem.a122933. [DOI] [PubMed] [Google Scholar]
- Kraus J., Staehelin M. N2-guanine specific transfer RNA methyltransferase II from rat liver. Nucleic Acids Res. 1974 Nov;1(11):1479–1496. doi: 10.1093/nar/1.11.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., De Robertis E. M., Cortese R. Order and intracellular location of the events involved in the maturation of a spliced tRNA. Nature. 1980 Mar 13;284(5752):143–148. doi: 10.1038/284143a0. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Structure, biosynthesis, and function of queuosine in transfer RNA. Prog Nucleic Acid Res Mol Biol. 1983;28:49–73. doi: 10.1016/s0079-6603(08)60082-3. [DOI] [PubMed] [Google Scholar]
- Perret V., Florentz C., Puglisi J. D., Giegé R. Effect of conformational features on the aminoacylation of tRNAs and consequences on the permutation of tRNA specificities. J Mol Biol. 1992 Jul 20;226(2):323–333. doi: 10.1016/0022-2836(92)90950-o. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Grosjean H., Ebel J. P., Florentz C., Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990 Apr 19;344(6268):787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Dumas P., Ebel J. P., Giegé R. Comparison of the tertiary structure of yeast tRNA(Asp) and tRNA(Phe) in solution. Chemical modification study of the bases. J Mol Biol. 1987 May 5;195(1):193–204. doi: 10.1016/0022-2836(87)90336-6. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Sindhuphak T., Hellman U., Svensson I. Site specificities of three transfer RNA methyltransferases from yeast. Biochim Biophys Acta. 1985 Jan 29;824(1):66–73. doi: 10.1016/0167-4781(85)90030-2. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Dank N., Nock S., Schön A. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2127–2171. doi: 10.1093/nar/19.suppl.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweykowska-Kulinska Z., Beier H. Sequence and structure requirements for the biosynthesis of pseudouridine (psi 35) in plant pre-tRNA(Tyr). EMBO J. 1992 May;11(5):1907–1912. doi: 10.1002/j.1460-2075.1992.tb05243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wildenauer D., Gross H. J., Riesner D. Enzymatic methylations: III. Cadaverine-induced conformational changes of E. coli tRNA fMet as evidenced by the availability of a specific adenosine and a specific cytidine residue for methylation. Nucleic Acids Res. 1974 Sep;1(9):1165–1182. doi: 10.1093/nar/1.9.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]


