Abstract
Wildlife may facilitate the spread of antibiotic resistance (AR) between human-dominated habitats and the surrounding environment. Here, we use functional metagenomics to survey the diversity and genomic context of AR genes in gulls. Using this approach, we found a variety of AR genes not previously detected in gulls and wildlife, including class A and C β-lactamases as well as six tetracycline resistance gene types. An analysis of the flanking sequences indicates that most of these genes are present in Enterobacteriaceae and various Gram-positive bacteria. In addition to finding known gene types, we detected 31 previously undescribed AR genes. These undescribed genes include one most similar to an uncharacterized gene in Verrucomicrobium and another to a putative DNA repair protein in Lactobacillus. Overall, the study more than doubled the number of clinically relevant AR gene types known to be carried by gulls or by wildlife in general. Together with the propensity of gulls to visit human-dominated habitats, this high diversity of AR gene types suggests that gulls could facilitate the spread of AR.
Keywords: herring gulls, antibiotic resistance, Verrucomicrobia, Larus argentatus, metagenomics
Introduction
The vast majority of research on emerging antibiotic resistance (AR) has focused on AR in clinical settings. However, there is growing recognition that resistance acquired outside of hospital settings (community-acquired infections) is a serious and growing threat (Pitout et al., 2005). Antibiotics are likely important for bacterial interactions in natural communities as resistance genes originated in environmental bacteria long before the antibiotic era (Aminov and Mackie, 2007; Martinez, 2008; Davies and Davies, 2010). The collection of all resistance genes and their precursors in pathogenic and non-pathogenic microorganisms has been termed the antibiotic “resistome” (Wright, 2007).
Bacteria can readily exchange DNA via lateral transfer of mobile genetic elements (e.g., plasmids or transposons). Thus, AR genes from non-pathogenic environmental microorganisms have the potential to be transferred to pathogens infecting humans, especially when use of antibiotics imposes heavy selective pressure. Indeed, as a result of widespread antibiotic use in medicine and agriculture, multidrug resistant bacteria and resistance genes have been detected in many environments (e.g., Sayah et al., 2005; Seyfried et al., 2010). Antibiotics and AR bacteria originating from human-made AR “reservoirs” may be released into nearby groundwater, streams, rivers, and estuaries (Li et al., 2009; Martinez, 2009), potentially leading to emergence of novel mechanisms of resistance in pathogenic organisms. However, discovery of new AR mechanisms is limited because most studies focus on culturable bacteria and known genes detected via PCR. This approach has resulted in a heavily biased and incomplete understanding of the broader environmental resistome.
Wildlife may facilitate exchange of AR bacteria and genes between human-made and natural environments. AR bacteria and, more recently, AR genes have been detected in a variety of wild birds and mammals (e.g., Mallon et al., 2002; Literak et al., 2009; Marrow et al., 2009). Many species of wildlife visit human-made areas such as farms and sewage lagoons where they may acquire AR (Osterblad et al., 2001; Blanco et al., 2009). Further, some studies have discovered high abundances of AR bacteria in wildlife with little or no apparent exposure to anthropogenic sources (Williams et al., 2011).
Gulls may be a particularly important carrier of AR because of their close association with humans. During the past several decades, gull populations have increased worldwide due in large part to burgeoning human populations along coasts and the associated increase in availability of human-derived foods (Duhem et al., 2008; Schwemmer et al., 2008). Gulls carry a diverse array of potentially pathogenic bacteria including Salmonella spp. (Skov et al., 2008), Campylobacter spp. (Waldenstrom et al., 2007), Listeria spp. (Fenlon, 1985), and vero cytotoxin-producing Escherichia coli O157 (Wallace et al., 1997), which they likely acquire by feeding and loafing at sewage lagoons, garbage dumps, and farms (Benskin et al., 2009). Moreover, several recent studies have detected clinically relevant AR bacteria (Cizek et al., 2007; Rose et al., 2009) and AR genes (e.g., Dolejska et al., 2007; Bonnedahl et al., 2010) in gull feces.
Here, we use functional metagenomics to characterize the diversity of the gull resistome. Previous studies of AR genes in gulls and other wildlife have relied on initial cultivation of bacteria or PCR of specific, known gene variants. However, studies using functional metagenomics have revealed a diverse array of previously undescribed AR genes in soils (Rondon et al., 2000) and humans (Diaz-Torres et al., 2006; Sommer et al., 2009). We assay: (1) the most abundant AR classes and gene types within these classes that confer resistance to four antibiotics; (2) information about the genes’ genomic context (i.e., the genomic neighborhood) as revealed by the flanking sequences; and (3) microdiversity within the abundant gene types. Specifically, we screened the libraries for resistance against ampicillin, amoxicillin, penicillin, and tetracycline. These four antibiotics were chosen due their historical importance, differences in resistance mechanism, and range of _target organisms. Penicillin, ampicillin, and amoxicillin are β-lactam antibiotics. In the past 20 years, bacterial strains that produced extended-spectrum β-lactamases (ESBLs) – enzymes conferring broad resistance to β-lactam antibiotics – have emerged posing a major threat to public health (Pitout and Laupland, 2008). Tetracyclines have been used for more than 40 years in clinical and agricultural settings and are broad-spectrum agents, exhibiting activity against a wide range of organisms (Chopra and Roberts, 2001). Resistance to tetracyclines is also widespread in the environment (e.g., Nascimento et al., 2003; Bryan et al., 2004).
Materials and Methods
Sampling
Fecal samples were collected from four Herring Gulls in a breeding colony at the Shoals Marine Laboratory on Appledore Island, ME (42° 58′ N, 70° 36′ W) located approximately 10 km off the coast of mainland United States. Approximately 650 pairs of Herring Gulls nest on the island (Ellis, unpublished data). We collected fresh samples by observing individual birds until defecation occurred and immediately collecting samples from the ground using a sterile syringe. For comparison, one soil and two gull samples were also collected from the mainland (Rochester, NH, USA). Samples were immediately placed on ice. DNA extraction was performed with the FastDNA Spin for Soil kit (MP Biomedical, CA, USA) using the protocol of the manufacturer with the following changes. Cells were lysed using the MP Biomedicals FastPrep24 (setting 5.0 twice for 45 s and once for 20 s, cooling on ice in between). The precipitation step was carried out twice with 250 μl protein precipitation solution (PPS) from the FastDNA Spin for Soil kit (MP Biomedical, CA, USA). We used 1.5 ml binding matrix and inverted for 4 min. We discarded 1 ml of the supernatant. The final pellet was resuspended in 75 μl of the provided water.
Cloning and screening
Three micrograms of genomic DNA from gull feces was sheared to a _target size of 3 kb with a Covaris S2 instrument (Covaris Inc., MA, USA). The 3-kb fragments were end-repaired with the DNA Terminator End Repair kit (Lucigen, WI, USA). The DNA was loaded on a 1% low melt agarose gel and fragments between 1 and 3 kb were extracted with Zymoclean Gel Recovery kit (Zymo Research, CA, USA). DNA was eluted in 10 μl TE elution buffer and ligated into pSMART–HCkan vector (accession number AF532107) using the Clonesmart blunt end cloning kit (Lucigen, WI, USA). Ligation was carried out overnight at room temperature. E. coli 10G electrocompetent cells (Lucigen, WI, USA) were transformed by electroporation using a BTX electroporator and 1.0 mm gap cuvettes. Electroporation conditions were 25 μF, 200 Ω, and 1800 V. The transformation was incubated for 1 h at 37°C and, after adding 30 μg/ml kanamycin, the transformation mixture was incubated another 2 h at 37°C. We plated 0.1 μl of the transformation mixture on kanamycin LB plates for titer counts and 100–200 μl on ampicillin (50 μg/ml), amoxicillin (20 μg/ml), penicillin (50 μg/ml), and tetracycline (8 μg/ml) containing plates. Screened clones were picked and grown up over night without shaking in LB + 10% glycerol. Sequencing was done using the SL1 forward primer (5′-CAGTCCAGTTACGCTGGAGTC-3′) and SR2 reverse primer (5′-GGTCAGGTATGATTTAAATGGTCAGT-3′; as suggested by Lucigen) and sequenced at Beckman Coulter Genomics.
Minimal inhibitory concentration
Antibiotic resistance clones were tested in duplicate for growth in the following concentrations of ampicillin: 60, 100, 200, 350, 500, 800, and 1000 μg/ml. Five microliters of live culture was inoculated into 96-well plates containing 225 μl LB Broth plus kanamycin (30 μg/ml) and the above concentrations of ampicillin. We also included a “no-cell” control for contamination and background OD. The cultures were incubated at 37°C and growth was measured at OD600 every 15 min for 24 h.
Tn5 transposon “knockout” of AR genes
To identify the AR gene in selected inserts, we randomly inserted a Tn5 transposon in the clones using the EZ-TN5 <TET-1> Insertion Kit (Epicentre, CA, USA). We used a molar ratio Tn5 to AR plasmid of one to minimize multiple insertion events. One microliter of a transposon reaction was electroporated (BioRad BTX) into 50 μl of E. Cloni 10G Electrocompetent Cells (Lucigen). Cells were plated on LB Agar containing kanamycin (30 μg/ml) and tetracycline (10 μg/ml). Colonies were next screened for Tn5 “knockouts” based on lack of ampicillin resistance in LB Broth plus kanamycin (30 μg/ml), tetracycline (10 μg/ml), and ampicillin (60 μg/ml). A replicate control 96-plate was also inoculated with the same colony in LB Broth plus kanamycin (30 μg/ml) and tetracycline (10 μg/ml). Both plates were grown at 37°C (without shaking) overnight. Sequencing was done using the TN5 <TET-1> forward and reverse primers (provided in Epicentre kit).
Sequence analysis
We trimmed the paired-end sequences using Phrap and analyzed the inserts using blast against both GenBank nr and the Antibiotic Resistance Genes Database (ARDB; Liu and Pop, 2009). For β-lactamases, the ARDB classification scheme generally follows other systems for organizing resistance genes (Bush and Jacoby, 2010). However, here we rely entirely on a sequence-based comparison. Using ARDB, genes are divided into the four major β-lactamase classes (A–D), then into specific ARDB types, and finally into sequence variants within each type. We detected some sequences that were identical to β-lactamases used in common cloning vectors; because we cannot determine whether these sequences are contaminants we removed them from the analysis.
Sequences from each ARDB type were assembled into contigs using Geneious (Biomatters). For selected inserts, we aligned and translated the sequences and built phylogenetic trees with Phylip (100 bootstraps) using neighbor-joining (protein distance matrix with Jones–Taylor–Thornton correction) and maximum likelihood methods (Jones–Taylor–Thornton correction and constant rate of change among sites; Felsenstein, 2006). The phylogenetic trees were visualized with the Interactive Tree of Life (iTOL, http://itol.embl.de/index.shtml; Letunic and Bork, 2007). Sequences from this study have been submitted to GenBank under accession numbers JM426721–JM427844.
Results
AR gene diversity
To _target AR in gulls, we sampled feces from four gulls on Appledore Island, Maine, and two gulls in Rochester, NH, USA. For comparison, we also sampled soil in Rochester without any gulls nearby (Table 1). We extracted and sheared the DNA, then cloned 3 kbp DNA fragments into E. coli. The number of resulting clones ranged between 1 × 105 and 4.4 × 106 per library. Based on these numbers, we estimate that up to 5% of the bacteria in gull feces are resistant to antibiotics (Table 1). We found ampicillin, amoxicillin, and penicillin resistance genes in all gulls assayed, whereas we detected tetracycline AR genes in only two out of six gulls. Although it is difficult to compare across antibiotics due to differences in potency and concentration, resistance to ampicillin was found in higher occurrence than amoxicillin and penicillin.
Table 1.
Metagenomic sample overview, including a list of sample IDs and description, titer of each clone library, and the number of positive (resistant) clones to each antibiotic.
| Sample id | Location | Environment | Antibiotic | Total titer | + Clones | % AR cells |
|---|---|---|---|---|---|---|
| 10-126 | Appledore | Gull feces | Ampicillin | 3.7 × 106 | 72 | 3.9 |
| 10-126 | Appledore | Gull feces | Amoxicillin | – | 58 | 3.1 |
| 10-126 | Appledore | Gull feces | Penicillin | – | 58 | 3.1 |
| 10-126 | Appledore | Gull feces | Tetracycline | – | 4 | 0.2 |
| 10-131 | Appledore | Gull feces | Ampicillin | 9.4 × 105 | 8 | 1.7 |
| 10-131 | Appledore | Gull feces | Tetracycline | – | 0 | 0 |
| 10-145 | Appledore | Gull feces | Ampicillin | 4.0 × 106 | 20 | 1.0 |
| 10-145 | Appledore | Gull feces | Amoxicillin | – | 15 | 0.8 |
| 10-145 | Appledore | Gull feces | Penicillin | – | 20 | 1.0 |
| 10-145 | Appledore | Gull feces | Tetracycline | – | 0 | 0 |
| 10-186 | Appledore | Gull feces | Ampicillin | 5.25 × 105 | 5 | 1.9 |
| 10-186 | Appledore | Gull feces | Amoxicillin | – | 3 | 1.1 |
| 10-186 | Appledore | Gull feces | Penicillin | – | 2 | 0.8 |
| 10-186 | Appledore | Gull feces | Tetracycline | – | 0 | 0 |
| 10-240 | Rochester | Gull feces | Ampicillin | 9.2 × 105 | 19 | 4.1 |
| 10-240 | Rochester | Gull feces | Amoxicillin | – | 20 | 4.3 |
| 10-240 | Rochester | Gull feces | Penicillin | – | 23 | 5.0 |
| 10-240 | Rochester | Gull feces | Tetracycline | – | 0 | 0 |
| 10-241 | Rochester | Gull feces | Ampicillin | 1.7 × 105 | N/A | N/A |
| 10-241 | Rochester | Gull feces | Amoxicillin | – | N/A | N/A |
| 10-241 | Rochester | Gull feces | Penicillin | – | N/A | N/A |
| 10-241 | Rochester | Gull feces | Tetracycline | – | 4 | 4.7 |
| 10-245 | Rochester | Soil outside gull area | Ampicillin | 4.4 × 106 | 18 | 0.8 |
| 10-245 | Rochester | Soil outside gull area | Amoxicillin | – | 9 | 0.4 |
| 10-245 | Rochester | Soil outside gull area | Penicillin | – | 12 | 0.5 |
| 10-245 | Rochester | Soil outside gull area | Tetracycline | – | 8 | 0.4 |
Percent AR cells are calculated based on an average insert size of 2 kb and an average gull bacteria genome size of 4 Mbp.
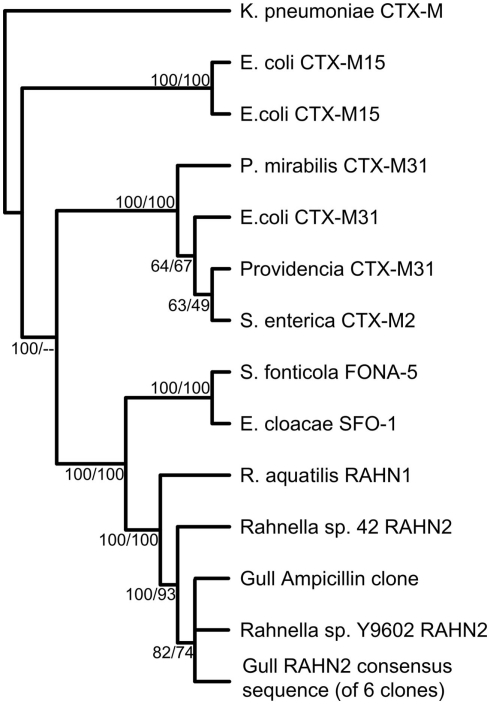
Gulls carried an extensive genetic diversity of both known and previously undescribed AR genes. To characterize this diversity in the metagenomic libraries, we paired-end sequenced the resistant clones and first classified the genes using ARDB (Liu and Pop, 2009; Tables 2 and 3). Overall, we found that similar genes conferred resistance to ampicillin, amoxicillin, and penicillin, including nine different class A and C β-lactamase ARDB categories. The most common category was bl1_ec, which was 100% similar to a β-lactamase in E. coli strain 1.1.43. Another common category detected was the extended-spectrum class A β-lactamase (ARDB category: bl2b_tem1). However, the bl2b_tem1 types were not identical to known variants in the database and on average shared only 92% sequence similarity. Combined, these two ARDB categories constituted more than 50% of the AR clones (Table 2). The last common ARDB category was the class A CTX-M type. However, these genes only shared 72% amino acid sequence similarity to CTX-M genes in E. coli (Table 3). A broader search in GenBank and a phylogenetic analysis revealed that the gene was 100% similar to the β-lactamase variant RAHN-2 in the Gram-negative enteric bacterium Rahnella aquatilis Y9602 (Ruimy et al., 2010) and therefore, was likely not a CTX-M variant (Figure 1).
Table 2.
The abundance of known AR genes (ARDB classification) from seven metagenomic libraries sorted by antibiotics used for selection.
| Gene | Description | Organism | Genomic neighborhood | Prev. detected |
Total | Amp |
AX |
Pen |
Tet |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gulls1 | Wildlife1 | Gull N = 6 | Soil N = 1 | Gull N = 6 | Soil N = 1 | Gull N = 6 | Soil N = 1 | Gull N = 6 | Soil N = 1 | |||||
| acrA | RND efflux pump | Shigella | N/A | No | No | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Bl1_ampC | Class C β-lactamase | Enterobacter | Transcriptional regulator | No | No | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Bl1_cmy2 | Class C β-lactamase | Enterobacteriaceae | On insertion element | No | Yes | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Bl1_ec | Class C β-lactamase | E. coli | Chromosomal | No | No | 69 | 20 | 9 | 13 | 5 | 17 | 5 | 0 | 0 |
| Bl1_sm | Class C β-lactamase | Providencia rettgeri | xre toxin system | No | No | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Bl2a_1 | Class A β-lactamase | Bacillus+ | Hypo. protein | No | No | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Bl2a_iii | Class A β-lactamase | Carnobacterium+ | Transcriptional regulator | No | No | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| Bl2b_tem1 | Class A β-lactamase | Many different | Yes | Yes | 14 | 6 | 1 | 1 | 0 | 5 | 1 | 0 | 0 | |
| Bl2be_ctxm | Class A β-lactamase | Rahnella | Chemotaxis protein | Yes2 | Yes2 | 6 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Bl2be_oxy1 | Class A β-lactamase | Klebsiella | Inner membrane protein | No | No | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| macB | RND efflux pump | E. coli, Gram+ | Transcriptional regulator | No | No | 3 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 |
| pbp1a | Penicillin resistance | Enterococcus+ | N/A | No | No | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| tetC | Tet. efflux pump | Clostridium+ | ATPase | No | Yes | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| tetJ | Tet. efflux pump | Proteus | Phage lysis protein | No | No | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| tetL | Tet. efflux pump | Enterococcus+ | On insertion element or plasmid | Yes | Yes | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| tetM | Ribosomal protection | Gram+ | Elongation factor G | Yes | Yes | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| tetO | Ribosomal protection | Roseburia+ | GTP binding protein | No | Yes | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| tetW | Ribosomal protection | Gram+ | On insertion element | No | Yes | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Unknown | No | No | 32 | 14 | 0 | 8 | 0 | 3 | 1 | 4 | 2 | |||
N is the number of libraries screened.
1These two columns indicate whether the AR gene type has previously been detected in either gulls or wildlife in general. This is based on the followed studies: Costa et al. (2006, 2008), Dolejska et al. (2007), Bonnedahl et al. (2009, 2010), Drobni et al. (2009), Figueiredo et al. (2009), Jeters et al. (2009), Literak et al. (2009), Hernandez et al. (2010), Radhouani et al. (2010), Rybarikova et al. (2010), Silva et al. (2010), Simoes et al. (2010), Ahmad et al. (2011), Alroy and Ellis (2011).
2The Rahnella variant has not been observed in gulls or wildlife before.
Table 3.
Amino acid similarity of AR genes and flanking regions to sequences in ARDB and GenBank databases.
| ARDB |
GenBank (AR gene) |
GenBank (genome neighborhood) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Sim (%) | Organism | Best hit | Accession no. | Sim (%) | Organism | Best hit | Accession no. | Sim (%) |
| acrA | 93 | Shigella flexneri | acrA | ADA72786 | 98 | N/A | |||
| Bl1_ampC | 85 | Enterobacter | bla(ampC) | BAA32077 | 100 | Enterobacter | Trans. regulator entR | ZP_05970670 | 90 |
| Bl1_cmy2 | 92 | Citrobacter | bla(CMY-45) | ACV32310 | 95 | Salmonella | sugE | YP_209396 | 59 |
| Bl1_ec | 100 | E. coli 1.1.43 | bla(ampC) | YP_002806354 | 100 | E. coli | blc, frdB, frdC, frdD, sugE, and hyp. protein | See text | 90–100 |
| Bl1_sm | 50 | Providencia | bla | ZP_06126521 | 85 | Providencia | xre toxin system | ZP_06126519 | 97 |
| Bl2a_1 | 100 | Bacillus | bla | ZP_04276394 | 100 | B. anthracis | Hyp. protein BA_3499 | NP_845779 | 100 |
| Bl2a_iii | 49 | Carnobacterium | bla | AAV65950 | 80 | Carnobacterium | Trans. regulator | ZP_02183939 | 61 |
| Bl2b_tem1 | 92 | ||||||||
| Bl2be_ctxm | 72 | Rahnella | bla(RAHN-2) | AAK83220 | 100 | Rahnella | Chemotaxis protein | AAK83219 | 94 |
| Bl2be_oxy1 | 85 | Klebsiella | bla | CAQ76676 | 85 | Klebsiella | Inner membrane protein | YP_001335817 | |
| macB | 37 | ABC transporter | YP_817928 or ZP_071822747 | 95 | Trans. regulator | ZP_04782719 or ZP_07165374 | 70–90 | ||
| pbp1a | 45 | Enterococcus | Glycosyl transferase | EFM68814 | 89 | N/A | N/A | N/A | N/A |
| tetC | 100 | Plasmid | IncN | NP_511233 | 100 | Clostridium | ATPase | ZP_02633149 | 98 |
| tetJ | 54 | Proteus | tetJ | ZP_03841720 | 100 | Proteus | phage lysis protein | YP_002152118 | 100 |
| tetL | 96 | Enterococcus | Tn916 | YP_133685 | 100 | Enterococcus | Hyp. protein | ZP_05563488 | 100 |
| tetM | 100 | Streptococcus | tetM | ADM91220 | 95 | Enterococcus | Elongation factor G | EFM68952 | 95 |
| tetO | 100 | Streptococcus | tetO | ACT76131 | 96 | Roseburia | GTP binding protein | CBL13258 | 93 |
| tetW | 92 | Lawsonia | tetW | YP_594556 | 96 | Lactobacillus | Transposase | YP_003601920 | 100 |
| unk1 | N/A | Lactobacillus | Hydrolase | EFG55459 | 48 | Enterococcus | PilT domain protein | ZP_03985770 | 84 |
| unk2 | N/A | Burkholderia | Flagellar asso. protein | ZP_02357438 | 53 | Nakamurella | UDP-glucosyltransferase | YP_003204087 | 71 |
| unk3 | N/A | Methylobacterium | Hypothetical protein | YP_001691243 | 79 | Rhizobium | Hypothetical protein | YP_471752 | 60 |
| unk4 | N/A | Enterococcus | Dihydroorotate dehydrogenase | ZP_06697492 | 69 | Mycoplasma | Dihydroorotate dehydrogenase | NP_758176 | 68 |
| unk5 | N/A | Pseudomonas phage EL | Hypothetical protein | YP_418084 | 52 | Pseudomonas phage EL | Hypothetical protein | YP_418084 | 52 |
| unk6 | N/A | E. coli | Methionyl-tRNA synthetase | ZP_07099092 | 93 | E. coli | Methionyl-tRNA synthetase | ZP_07099092 | 100 |
| unk7 | N/A | Enterococcus | Hypothetical protein | YP_398703 | 79 | Enterococcus | Hypothetical protein | YP_398703 | 79 |
| unk8 | N/A | Enterococcus | Hydrolase | EFM78292 | 56 | Enterococcus | Hydrolase | EFM78292 | 56 |
| unk9 | N/A | Citrobacter | ATP synthase subunit | YP_003367440 | 99 | Citrobacter | ATP synthase subunit | YP_003367440 | 99 |
| unk10 | N/A | E. coli | Hpt domain protein | ZP_07146767 | 97 | E. coli | Hpt domain protein | ZP_07146767 | 100 |
| unk11 | N/A | Enterococcus | NOL1/NOP2/sun family protein | ZP_05646561 | 85 | Enterococcus | NOL1/NOP2/sun family protein | ZP_05646561 | 83 |
| unk12 | N/A | Bacillus | Uracil-DNA glycosylase | YP_001377098 | 74 | Burkholderia | Short-chain dehydrogenase | YP_002909251 | 95 |
| unk13 | N/A | E. coli | Multidrug transporter | ZP_06938716 | 93 | E. coli | Multidrug transporter | YP_002385928 | 91 |
| unk14 | N/A | Streptococcus | Integrase | ZP_04061436 | 91 | Enterobacter | AraC family transcriptional regulator | YP_001175810 | 82 |
| unk15 | N/A | Verrucomicrobium | Hypothetical protein | ZP_02929363 | 84 | Gemmata | Protease | ZP_02733734 | 45 |
| unk16 | N/A | Arcobacter | Auxin efflux protein | YP_001489985 | 92 | N/A | N/A | N/A | N/A |
| unk17 | N/A | P. putida | Alcohol dehydrogenase | YP_001668264 | pantetheine-phosphate adenylyltransferase | ZP_07057662 | |||
| unk18 | N/A | Photobacterium | Hypothetical gene | ZP_06155446 | 93 | Pseudomonas | TrkH transporter | YP_791383 | 80 |
| unk19 | N/A | E. coli | Hypothetical gene | CAA74339 | 70 | E. coli | IS1 transposase B | NP_414562 | 91 |
| unk20 | N/A | Bacillus | β-Lactamase precursor | ZP_06873125 | 47 | Bacillus | Hypothetical protein | ZP_03104389 | 60 |
| unk21 | N/A | Proteus | Chondroitin ABC lyase precursor | ZP_03840872 | 93 | Proteus | Chondroitin ABC lyase precursor | YP_002151847 | 87 |
| unk22 | N/A | Bacteroides | Tyrosine type site-specific recombinase | ZP_06094951 | 98 | Bacteroides | Tyrosine type site-specific recombinase | YP_100741 | 88 |
| unk23 | N/A | Proteus | Colicin V production protein | YP_002151514 | 85 | Proteus | Hypothetical protein | YP_002151515 | 82 |
| unk24 | N/A | Providencia | TPR repeat protein | ZP_05971549 | 62 | Proteus | Hypothetical protein | ZP_03802426 | 85 |
| unk25 | N/A | Proteus | Iron ABC transporter | YP_002152660 | 93 | Proteus | Iron ABC transporter | YP_002152659 | 94 |
| unk26 | N/A | Lactobacillus | Transposase | ZP_05866387 | 94 | Faecalibacterium | Hypothetical protein | ZP_02091680 | 96 |
| unk27 | N/A | Pantoea | Phosphoglycerate mutase | ZP_07378382 | 94 | Yersinia | Hypothetical protein | ZP_04630452 | 88 |
| unk28 | N/A | Streptomyces | Hypothetical protein | NP_822969 | 74 | N/A | N/A | N/A | N/A |
| unk29 | N/A | Bacillus | Histidine kinase | YP_175245 | 100 | N/A | N/A | N/A | N/A |
| unk30 | N/A | Clostridium | D-3-phosphoglycerate transferase | ZP_02639608 | 100 | Clostridium | D-3-phosphoglycerate transferase | ZP_02639608 | 100 |
| unk31 | N/A | Lactobacillus | DNA repair gene | ZP_05752538 | 92 | Lactobacillus | Hypothetical protein | ZP_03963367 | 68 |
Figure 1.
Phylogenetic tree of the bl2be_ctxm type class A β-lactamase gene from metagenomic clones. The phylogenetic tree is a majority rule consensus tree based on protein similarity using neighbor-joining. Bootstrap values (total 100) are calculated with neighbor-joining and maximum likelihood methods. Klebsiella pneumoniae CTX-M is the outgroup.
Beyond the class A and class C β-lactamases, we also found ampicillin- and penicillin- resistant clones encoding for efflux pumps (acrA and macB type) and a membrane carboxypeptidase (ARDB category: pbp1a). Finally, we observed a high proportion of clones (38%) that had no clear match in ARDB.
The tetracycline resistant genes detected belonged to several ARDB categories including tetC, tetJ, and tetL efflux pumps and tetM, tetO, and tetW ribosomal modification proteins. These genes were generally very similar to known genes (Table 3) and likely do not constitute new variants.
For comparison, we also analyzed the genetic diversity of AR in a nearby soil sample. Keeping in mind that we only analyzed one soil sample, we found a lower frequency of AR genes in the soil sample compared to the gulls. With the exception of tetW, all ARDB categories observed in soil were also detected in gulls. Furthermore, we did not detect the RAHN-2/CTX-M category in soil. Thus, it appears that many AR genes in gulls are also found in soil (and vice-versa), with perhaps some exceptions.
Genomic neighborhood
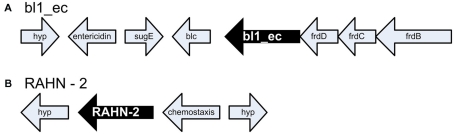
Antibiotic resistance genes are known to move around between hosts via lateral gene transfer, thus the AR gene sequence itself is not a good indicator of the organism with the gene. Therefore, we analyzed the flanking sequences around the AR genes (Figure 2; Tables 2 and 3). Data regarding the genomic neighborhood can give information about the host organism and/or whether the gene was located on a mobile genetic element. Our attempts to assemble clones belonging to the same ARDB category yielded good contigs, indicating that the detected AR genes shared flanking sequences. An example of that was the common class C β-lactamase belonging to the ARDB category bl1_ec. All clones associated with this category were located on a genomic fragment syntenous with E. coli strain K-12 substr. DH10B and several other E. coli strains (Figure 2A). Similar to DH10B, the gene was flanked by fumarate reductase subunits frdB, frdC, and frdD, an outer membrane lipoprotein (Blc), a quaternary ammonium compound-resistance protein (sugE), and the toxin entericidin (Durfee et al., 2008). Overall, this suggests that the most common class C β-lactamase found in the gulls was located on a genomic fragment similar to that found in E. coli. Similarly, the β-lactamase variant RAHN-2 also assembled into one contig which suggested a fragment syntenous with the genome from R. aquatilis Y9602 (Figure 2B). This contig including a chemotaxis gene and a hypothetical protein. This supports the hypothesis that this variant is indeed located in a Rahnella genome.
Figure 2.
Genome neighborhood of antibiotic resistance genes. (A) Genomic configuration of bl1_ec. frdBCD encode fumarate reductase subunits, Blc encodes an outer membrane lipoprotein, sugE encodes a quaternary ammonium compound-resistance protein and hyp. encodes a hypothetical protein (B) bl2be_ctxm. Hyp. encodes two hypothetical proteins adjacent to the β-lactamase RAHN-2 in Rahnella.
In contrast to the bl1_ec and RAHN-2 variants, clones matching the TEM class A β-lactamases (bl2b_tem1) were found in a range of genetic neighborhoods. The genes flanking bl2b_tem1 sequences were in some cases most closely related to Gram-positive bacteria (e.g., Lactobacillus, Enterococcus, and Clostridium) and in other cases, to Gram-negative bacteria (e.g., E. coli, Shigella, and Salmonella).
There were two major types of tetracycline resistance genes in our samples – efflux pumps (tetC, tetJ, and tetL) and ribosomal modification proteins (tetM, tetO, and tetW; Table 2). With the exception of tetJ, these were all associated with Gram-positive bacteria including Bacillus, Enterococcus, and Clostridium. Several of them were located on what appeared to be mobile elements including plasmids and transposons.
AR gene microdiversity
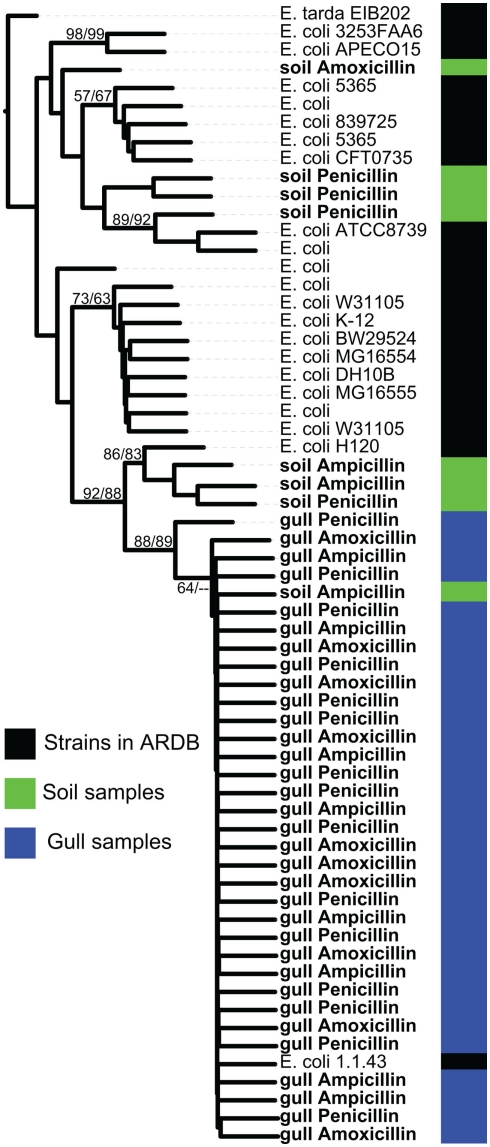
In addition to detecting multiple classes of AR genes, we found several DNA polymorphisms in the bl1_ec type. To further explore this result, we aligned the sequences matching this ARDB type and built a phylogenetic tree (Figure 3). All gull bl1_ec sequences grouped with E. coli 1.1.43 with high bootstrap support. In contrast, sequences retrieved from the soil samples formed three separate clades, with only one soil clone clustering with the gull variants. Instead, the one clade of soil AR sequences were 100% similar to E. coli H120, whereas the other genes branched away from known E. coli genes. Overall, this indicates that the bl1_ec diversity is related to its environmental origin. We also analyzed the sequence variation among clones from the RAHN-2 β-lactamase genes but only observed one variant.
Figure 3.
Phylogenetic tree of the bl1_ec type class C β-lactamase gene from metagenomic clones and sequenced representative strains from ARDB. The sequenced clones are from samples from gulls or soil, whereas we did not detect any bl1_ec type clones in samples from wastewater. Metagenomic clones were isolated on ampicillin, amoxicillin, or penicillin as listed for each entry. The phylogenetic tree is a majority rule consensus tree based on protein similarity using neighbor-joining. Bootstrap values (total 100) are calculated with neighbor-joining and maximum likelihood methods. Edwardsiella tarda sp. EIB202 is the outgroup.
Characterization of undescribed AR genes
In addition to the known AR gene categories in ARDB, we detected 31 gene types not previously described (Table 3). The novel AR genes were associated with a wide range of phylogenetic lineages including Pseudomonas, Bacteroides, Gram-positives, Verrucomicrobia, and different genera affiliated with Enterobacteriaceae. To further characterize these, we first measured the minimum inhibitory concentration (MIC) for ampicillin. All ampicillin/penicillin/amoxicillin resistant clones had an MIC above 1000 μg/ml ampicillin and thus clearly conferred resistance to this antibiotic.
Two of these clones assembled to a 2800-bp DNA fragment (unk15 in Table 3). One end of the fragment shared 84% amino acid similarity to the hypothetical protein VspiD_21980 in Verrucomicrobium spinosum from the phylum Verrucomicrobia. The other part of the fragment was 45% similar to a C-terminal protease in Gemmata obscuriglobus from the phylum Planctomycetes.
We next identified the genomic region in the clone that encoded for AR by randomly inserting a Tn5 transposon followed by testing for ampicillin resistance and DNA sequencing of the clones. The Tn5 “knockout” revealed that insertions in both genes interrupted the resistance whereas several insertions between the two genes had no effect. This represents a completely novel AR gene system that confers resistance in E. coli, but at present, we do not know the specific mechanism. To the best of our knowledge, this is the first study to identify an AR gene in either the phylum Verrucomicrobia or Planctomycetes.
A second previously uncharacterized AR gene (unk1) shared 85% amino acid similarity to a protein in the Gram-positive Carnobacterium sp. AT7 that was annotated as a putative DNA repair protein. A Tn5 insertion in this gene interrupted the resistance. Next to this putative AR gene was a gene 85% similar to pilT in another Gram-positive lineage, Enterococcus faecalis. Another previous unrecognized AR gene (unk31) was 92% similar to a DNA repair gene of the ImpB/MucB/SamB family in Lactobacillus coryniformis subsp. torquens KCTC 3535. Multiple Tn5 insertions in this gene also interrupted the resistance. None of these DNA repair genes have previously been shown to confer AR, so the exact mechanism of resistance is unclear.
Discussion
Using a functional metagenomics approach, we detected an extensive diversity of AR genes in gulls, including both well-known and undescribed AR genes. Less than half of the gene types detected have been reported previously in gulls (Table 2). We also detected several completely unknown AR genes including one associated with the lesser-known phyla Verrucomicrobia and Planctomycetes. These results highlight the strength of the functional metagenomic approach: the ability to reveal the most abundant genes conferring resistance to a particular antibiotic, without requiring prior knowledge of the gene classes of interest. In contrast, most studies of antimicrobial resistance have used PCR to characterize specific AR genes, often known from clinically relevant bacterial isolates (such as, E. coli, Salmonella, Enterococcus spp. – see references in Table 2). Our results suggest that these previous studies, while useful, may impart a heavily biased view of the AR genes carried by wild animals.
The functional metagenomic approach also has several advantages over a straight sequencing-based metagenomic analysis of a community. By including a functional screening, we can identify previously unknown AR genes that we would not have been able to identify by sequence homology searches (e.g., Sommer et al., 2009). Further, most metagenomic analyses only _target the most abundant lineages, whereas the functional metagenomic approach allows for screening even rare members in the community.
Of course, there are limitations to the method used here. The approach is unlikely to detect mechanisms that require large (>3 kb) gene cassettes or that confer resistance by modifying regulation of genes in another genomic region (Viveiros et al., 2007). Further, it will only find AR genes that confer resistance in E. coli. Nonetheless, 42% of the gene types we detected originated from Gram-positive hosts (e.g., Enterococcus and Clostridium) as well as lesser-studied phyla like Verrucomicrobia and Bacteroidetes. This indicates that this bias may not be as dramatic as would be expected. Further, the functional assay yields the additional useful information that the genes are expressed in E. coli and therefore have the potential to be transferred and functional in human pathogens.
Wild animals are probably natural reservoirs of AR bacteria and genes, carrying them as part of their commensal flora (Wright, 2010). Alternatively, animals may acquire AR genes through exposure to human activities, including agriculture and animal husbandry (Osterblad et al., 2001). Without a much broader understanding of natural AR gene diversity, we cannot determine the source of AR genes in gulls. However, the gulls sampled in this study carried a variety of AR genes that are commonly found in bacteria isolated from humans and domestic animals. This result, similar to that of other recent studies of gulls and other wildlife, suggests that AR genes arising from the use of antibiotics in humans and animals may have spread to wildlife. The genes may then be further disseminated by wild animals, particular those that migrate long distances such as birds (Sjolund et al., 2008; Drobni et al., 2009).
Studies of gull ecology conducted at Appledore Island, Maine (where most of our samples were taken) indicate that gulls not only carry AR genes, but also have the potential to be a vector of the genes between human-dominated habitats and the environment. Appledore-banded gulls have been found at a variety of human habitats across New England and as far away as Florida (Ellis, unpublished data). They have been observed loafing at landfills and wastewater treatment plants and share strains of E. coli in common with wastewater from these sites (Nelson et al., 2008). Garbage brought back from the mainland is also a large part of the diet of gulls on Appledore Island (Ellis et al., 2005). Finally, the birds are frequently observed at public beaches in coastal New Hampshire and Maine during the summer, where they come into close contact with humans. Thus, the combination of the traveling patterns of gulls and the extensive diversity of AR genes that they carry suggest that gulls may be important vectors for spreading AR genes in areas where humans live.
The most common gene detected in our study was bl1_ec, a class C β-lactamase. The microdiversity within this gene type provides further evidence that gulls could move AR genes from human habitats to the environment. The gull variants were highly similar (some sharing 100% aa identity) to that of E. coli 1.1.43, an isolate from a human gastrointestinal tract. In contrast, while the soil sample contained the gull variant, it also contained sequence diversity that did not match any known clinical isolates. Hence, gull AR gene diversity does not appear to simply reflect the diversity found in soils (and therefore, perhaps, the broader environment), although further work is needed to confirm this hypothesis.
Class A β-lactamases were also common in our samples. In a recent study of AR in Alaskan soil, class B β-lactamases were most common (Allen et al., 2009). We did not detect any class B types from our samples, however, indicating that the relative abundance of β-lactamase classes may vary by environment or location.
Only two of the ARDB β-lactamase gene types that we identified have been reported previously in gulls (Table 2). Notably, one of these genes was classified as a CTX-M-like ESBL. ESBL-producing organisms are a major public health concern in clinics and communities (Pitout and Laupland, 2008). However, the variant was also 100% similar to that previously found in R. aquatilis (Enterobacteriaceae), and the flanking sequences also suggest a Rahnella host. Rahnella has been implicated in several instances of infectious disease in humans, causing bacteremia (Chang et al., 1999; Tash, 2005). R. aquatilis has also been found in the guts of insects (e.g., Morales-Jimenez et al., 2009) and a wild fish (Skrodenyte-Arbaciauskiene et al., 2008); however, as far as we know, this is the first report of the genus in a wild bird.
Although we generally detected few tetracycline resistance genes in our samples, we identified five known tetracycline resistance gene types in gulls and a sixth type in soil. tetL and tetM have been reported before in a few studies of gulls and other wildlife (Table 1). Notably we did not detect tetA and tetB, which have been widely reported in other wildlife (e.g., Silva et al., 2010; Alroy and Ellis, 2011). The gene tetW has only been reported in one study of wild animals (Jeters et al., 2009). tetC has also only been reported in one other study of wildlife; the gene was found in small rodents trapped at swine farms, suggesting that the presence of the gene may have resulted from agricultural use of antibiotics. Interestingly, several other studies were unable to detect tetC in wildlife using PCR (e.g., Rybarikova et al., 2010; Ahmad et al., 2011). One other study of wild animals detected tetO in houseflies and German cockroaches at swine farms (Ahmad et al., 2011). Thus, gulls from Appledore Island may have acquired some tet genes from agricultural sources.
In addition to these well-known AR genes, both gulls and soil also harbor a variety of previously unknown AR genes. Given that the genes confer resistance in E. coli, they clearly have the potential to be functional in human pathogens. Some of these genes may have originally derived from human bacteria and have yet to be detected in clinical settings. Indeed, Sommer et al. (2009) detected a variety of novel AR genes in healthy humans using the same method. Alternatively, the genes may be intrinsic to the birds’ commensal flora or more broadly, to the environment. In this case, the mixing of human and gull fecal bacteria could result in novel combinations and mechanisms of AR in human beings.
In conclusion, by using a functional metagenomics approach, this study more than doubled the number of clinically relevant AR gene types known to be carried by gulls or by wildlife in general. Together with the propensity of gulls to visit human-dominated habitats, this high diversity of AR gene types suggests that gulls can facilitate the spread of AR. Further, the large number of novel genes that we identified suggests that gulls also have the potential to introduce new mechanisms of AR back to the human microflora.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the staff of the Shoals Marine Laboratory for sampling assistance, George Jacoby for helpful comments on the manuscript, the University of California, Irvine (to Jennifer B. H. Martiny), and Tufts Cummings School of Veterinary Medicine (to Julie C. Ellis) for supporting the work.
References
- Ahmad A., Ghosh A., Schal C., Zurek L. (2011). Insects in confined swine operations carry a large antibiotic resistant and potentially virulent enterococcal community. BMC Microbiol. 11, 23. 10.1186/1471-2180-11-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H. K., Moe L. A., Rodbumrer J., Gaarder A., Handelsman J. (2009). Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J 3, 243–251 10.1038/ismej.2008.86 [DOI] [PubMed] [Google Scholar]
- Alroy K., Ellis J. C. (2011). Pilot study of antimicrobial resistant Escherichia coli in herring gulls (Larus argentatus) and wastewater in the Northeast, United States. J. Zoo Wildl. Med. 42, 160–163 10.1638/2010-0130.1 [DOI] [PubMed] [Google Scholar]
- Aminov R. I., Mackie R. I. (2007). Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271, 147–161 10.1111/j.1574-6968.2007.00757.x [DOI] [PubMed] [Google Scholar]
- Benskin C. M. H., Wilson K., Jones K., Hartley I. R. (2009). Bacterial pathogens in wild birds: a review of the frequency and effects of infection. Biol. Rev. 84, 349–373 10.1111/j.1469-185X.2008.00076.x [DOI] [PubMed] [Google Scholar]
- Blanco G., Lemus J. A., Grande J. (2009). Microbial pollution in wildlife: linking agricultural manuring and bacterial antibiotic resistance in red-billed choughs. Environ. Res. 109, 405–412 10.1016/j.envres.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Bonnedahl J., Drobni M., Gauthier-Clerc M., Hernandez J., Granholm S., Kayser Y., Melhus A., Kahlmeter G., Waldenstrom J., Johansson A., Olsen B. (2009). Dissemination of Escherichia coli with CTX-M Type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE 4, e5958. 10.1371/journal.pone.0005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnedahl J., Drobni P., Johansson A., Hernandez J., Melhus A., Stedt J., Olsen B., Drobni M. (2010). Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65, 1939–1944 10.1093/jac/dkq222 [DOI] [PubMed] [Google Scholar]
- Bryan A., Shapir N., Sadowsky M. J. (2004). Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains, isolated from diverse human and animal sources. Appl. Environ. Microbiol. 70, 2503–2507 10.1128/AEM.70.4.2503-2507.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Jacoby G. A. (2010). Updated Functional Classification of beta-lactamases. Antimicrob. Agents Chemother. 54, 969–976 10.1128/AAC.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. L., Jeong J., Shin J. H., Lee E. Y., Son H. C. (1999). Rahnella aquatilis sepsis in an immunocompetent adult. J. Clin. Microbiol. 37, 4161–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., Roberts M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260 10.1128/MMBR.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizek A., Dolejska M., Karpiskova R., Dedicova D., Literak I. (2007). Wild black-headed gulls (Larus ridibundus) as an environmental reservoir of Salmonella strains resistant to antimicrobial drugs. Eur. J. Wildl. Res. 53, 55–60 10.1007/s10344-006-0054-2 [DOI] [Google Scholar]
- Costa D., Poeta P., Saenz Y., Vinue L., Rojo-Bezares B., Jouini A., Zarazaga M., Rodrigues J., Torres C. (2006). Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 58, 1311–1312 10.1093/jac/dkl411 [DOI] [PubMed] [Google Scholar]
- Costa P. M., Vaz-Pires P., Bernardo F. (2008). Antimicrobial resistance in Escherichia coli isolated in wastewater and sludge from poultry slaughterhouse wastewater plants. J. Environ. Health 70, 40–45 [PubMed] [Google Scholar]
- Davies J., Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Torres M. L., Villedieu A., Hunt N., Mcnab R., Spratt D. A., Allan E., Mullany P., Wilson M. (2006). Determining the antibiotic resistance potential of the indigenous oral microbiota of humans using a metagenomic approach. FEMS Microbiol. Lett. 258, 257–262 10.1111/j.1574-6968.2006.00221.x [DOI] [PubMed] [Google Scholar]
- Dolejska M., Cizek A., Literak I. (2007). High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 103, 11–19 10.1111/j.1365-2672.2006.03241.x [DOI] [PubMed] [Google Scholar]
- Drobni M., Bonnedahl J., Hernandez J., Haemig P., Olsen B. (2009). Vancomycin-resistant enterococci, point barrow, Alaska, USA. Emerging Infect. Dis. 15, 838–839 10.3201/eid1505.081219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhem C., Roche P., Vidal E., Tatoni T. (2008). Effects of anthropogenic food resources on yellow-legged gull colony size on Mediterranean islands. Popul. Ecol. 50, 91–100 10.1007/s10144-007-0059-z [DOI] [Google Scholar]
- Durfee T., Nelson R., Baldwin S., Plunkett G., III, Burland V., Mau B., Petrosino J. F., Qin X., Muzny D. M., Ayele M., Gibbs R. A., Csorgo B., Posfai G., Weinstock G. M., Blattner F. R. (2008). The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 190, 2597–2606 10.1128/JB.01092-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. C., Chen W., O’Keefe B., Shulman M. J., Witman J. D. (2005). Predation by gulls on crabs in rocky intertidal and shallow subtidal zones of the Gulf of Maine. J. Exp. Mar. Biol. Ecol. 324, 31–43 10.1016/j.jembe.2005.04.001 [DOI] [Google Scholar]
- Felsenstein J. (2006). PHYLIP (Phylogeny Inference Package), 3.65 Edn. Seattle: Department of Genome Sciences, University of Washington [Google Scholar]
- Fenlon D. R. (1985). Wild birds and silage as reservoirs of Listeria in the agricultural environment. J. Appl. Bacteriol. 59, 537–543 10.1111/j.1365-2672.1985.tb03357.x [DOI] [PubMed] [Google Scholar]
- Figueiredo N., Radhouani H., Gonçalves A., Rodrigues J., Carvalho C., Igrejas G., Poeta P. (2009). Genetic characterization of vancomycin-resistant enterococci isolates from wild rabbits. J. Basic Microbiol. 49, 491–494 10.1002/jobm.200800387 [DOI] [PubMed] [Google Scholar]
- Hernandez J., Bonnedahl J., Eliasson I., Wallensten A., Comstedt P., Johansson A., Granholm S., Melhus A., Olsen B., Drobni M. (2010). Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring bla(CTX-M-15), found in glaucous-winged gull at remote Commander Islands, Russia. Environ. Microbiol. Rep. 2, 329–332 10.1111/j.1758-2229.2010.00142.x [DOI] [PubMed] [Google Scholar]
- Jeters R. T., Rivera A. J., Boucek L. M., Stumpf R. M., Leigh S. R., Salyers A. A. (2009). Antibiotic resistance genes in the vaginal microbiota of primates not normally exposed to antibiotics. Microb. Drug Resist. 15, 309–315 10.1089/mdr.2009.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2007). Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- Li D., Yang M., Hu J. Y., Zhang J., Liu R. Y., Gu X., Zhang Y., Wang Z. Y. (2009). Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environ. Microbiol. 11, 1506–1517 10.1111/j.1462-2920.2008.01810.x [DOI] [PubMed] [Google Scholar]
- Literak I., Dolejska M., Radimersky T., Klimes J., Friedman M., Aarestrup F. M., Hasman H., Cizek A. (2009). Antimicrobial-resistant faecal Escherichia coli in wild mammals in central Europe: multiresistant Escherichia coli producing extended-spectrum beta-lactamases in wild boars. J. Appl. Microbiol. 108, 1702–1711 10.1111/j.1365-2672.2009.04572.x [DOI] [PubMed] [Google Scholar]
- Liu B., Pop M. (2009). ARDB – antibiotic resistance genes database. Nucleic Acids Res. 37, D443–D447 10.1093/nar/gkp644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon D. J., Corkill J. E., Hazel S. M., Wilson J. S., French N. P., Bennett M., Hart C. A. (2002). Excretion of vancomycin-resistant enterococci by wild mammals. Emerging Infect. Dis. 8, 636–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrow J., Whittington J. K., Mitchell M., Hoyer L. L., Maddox C. (2009). Prevalence and antibiotic-resistance characteristics of Enterococcus spp. Isolated from free-living and captive raptors in Central Illinois. J. Wildl. Dis. 45, 302–313 [DOI] [PubMed] [Google Scholar]
- Martinez J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367 10.1126/science.1159483 [DOI] [PubMed] [Google Scholar]
- Martinez J. L. (2009). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902 10.1016/j.envpol.2009.05.051 [DOI] [PubMed] [Google Scholar]
- Morales-Jimenez J., Zuniga G., Villa-Tanaca L., Hernandez-Rodriguez C. (2009). Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microb. Ecol. 58, 879–891 10.1007/s00248-009-9548-2 [DOI] [PubMed] [Google Scholar]
- Nascimento A. M. A., Cursino L., Goncalves-Dornelas H., Reis A., Chartone-Souza E., Marini M. A. (2003). Antibiotic-resistant Gram-negative bacteria in birds from the Brazilian Atlantic forest. Condor 105, 358–361 10.1650/0010-5422(2003)105[0358:AGBIBF]2.0.CO;2 [DOI] [Google Scholar]
- Nelson M., Jones S. H., Edwards C., Ellis J. C. (2008). Characterization of Escherichia coli populations from gulls, landfill trash, and wastewater using ribotyping. Dis. Aquat. Org. 81, 53–63 10.3354/dao01937 [DOI] [PubMed] [Google Scholar]
- Osterblad M., Norrdahl K., Korpimaki E., Huovinen P. (2001). Antibiotic resistance. How wild are wild mammals? Nature 409, 37–38 10.1038/35051173 [DOI] [PubMed] [Google Scholar]
- Pitout J. D. D., Laupland K. B. (2008). Extended-spectrum beta-lactamase-producing enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- Pitout J. D. D., Nordmann P., Laupland K. B., Poirel L. (2005). Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56, 52–59 10.1093/jac/dki166 [DOI] [PubMed] [Google Scholar]
- Radhouani H., Poeta P., Pinto L., Miranda J., Coelho C., Carvalho C., Rodrigues J., Lopez M., Torres C., Vitorino R., Domingues P., Igrejas G. (2010). Proteomic characterization of vanA-containing Enterococcus recovered from Seagulls at the Berlengas Natural Reserve, W Portugal. Proteome Sci. 8, 48. 10.1186/1477-5956-8-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon M. R., August P. R., Bettermann A. D., Brady S. F., Grossman T. H., Liles M. R., Loiacono K. A., Lynch B. A., Macneil I. A., Minor C., Tiong C. L., Gilman M., Osburne M. S., Clardy J., Handelsman J., Goodman R. M. (2000). Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66, 2541–2547 10.1128/AEM.66.6.2541-2547.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. M., Gast R. J., Bogomolni A., Ellis J. C., Lentell B. J., Touhey K., Moore M. (2009). Occurrence and patterns of antibiotic resistance in vertebrates off the Northeastern United States coast. FEMS Microbiol. Ecol. 67, 421–431 10.1111/j.1574-6941.2009.00648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruimy R., Meziane-Cherif D., Momcilovic S., Arlet G., Andremont A., Courvalin P. (2010). RAHN-2, a chromosomal extended-spectrum class A beta-lactamase from Rahnella aquatilis. J. Antimicrob. Chemother. 65, 1619–1623 10.1093/jac/dkq178 [DOI] [PubMed] [Google Scholar]
- Rybarikova J., Dolejska M., Materna D., Literak I., Cizek A. (2010). Phenotypic and genotypic characteristics of antimicrobial resistant Escherichia coli isolated from symbovine flies, cattle and sympatric insectivorous house martins from a farm in the Czech Republic (2006-2007). Res. Vet. Sci. 89, 179–183 10.1016/j.rvsc.2010.02.016 [DOI] [PubMed] [Google Scholar]
- Sayah R. S., Kaneene J. B., Johnson Y., Miller R. (2005). Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71, 1394–1404 10.1128/AEM.71.3.1394-1404.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmer P., Garthe S., Mundry R. (2008). Area utilization of gulls in a coastal farmland landscape: habitat mosaic supports niche segregation of opportunistic species. Landsc. Ecol. 23, 355–367 10.1007/s10980-008-9194-y [DOI] [Google Scholar]
- Seyfried E. E., Newton R. J., Rubert K. F., Pedersen J. A., Mcmahon K. D. (2010). Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb. Ecol. 59, 799–807 10.1007/s00248-009-9624-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N., Igrejas G., Figueiredo N., Goncalves A., Radhouani H., Rodrigues J., Poeta P. (2010). Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 408, 4871–4876 10.1016/j.scitotenv.2010.07.023 [DOI] [PubMed] [Google Scholar]
- Simoes R. R., Poirel L., Da Costa P. M., Nordmann P. (2010). Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerging Infect. Dis. 16, 110–112 10.3201/eid1601.090896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund M., Bonnedahl J., Hernandez J., Bengtsson S., Cederbrant G., Pinhassi J., Kahlmeter G., Olsen B. (2008). Dissemination of multidrug-resistant bacteria into the Arctic. Emerging Infect. Dis. 14, 70–72 10.3201/eid1412.080494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skov M. N., Madsen J. J., Rahbek C., Lodal J., Jespersen J. B., Jorgensen J. C., Dietz H. H., Chriel M., Baggesen D. L. (2008). Transmission of Salmonella between wildlife and meat-production animals in Denmark. J. Appl. Microbiol. 105, 1558–1568 10.1111/j.1365-2672.2008.03914.x [DOI] [PubMed] [Google Scholar]
- Skrodenyte-Arbaciauskiene V., Sruoga A., Butkauskas D., Skrupskelis K. (2008). Phylogenetic analysis of intestinal bacteria of freshwater salmon Salmo salar and sea trout Salmo trutta trutta and diet. Fish. Sci. 74, 1307–1314 10.1111/j.1444-2906.2008.01656.x [DOI] [Google Scholar]
- Sommer M. O. A., Dantas G., Church G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131 10.1126/science.1176950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tash K. (2005). Rahnella aquatilis bacteremia from a suspected urinary source. J. Clin. Microbiol. 43, 2526–2528 10.1128/JCM.43.5.2526-2528.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros M., Dupont M., Rodrigues L., Couto I., Davin-Regli A., Martins M., Pages J.-M., Amaral L. (2007). Antibiotic stress, genetic response and altered permeability of E. coli. PLoS ONE 2, e365. 10.1371/journal.pone.0000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenstrom J., On S. L. W., Ottvall R., Hasselquist D., Olsen B. (2007). Species diversity of campylobacteria in a wild bird community in Sweden. J. Appl. Microbiol. 102, 424–432 10.1111/j.1365-2672.2006.03090.x [DOI] [PubMed] [Google Scholar]
- Wallace J. S., Cheasty T., Jones K. (1997). Isolation of vero cytotoxin-producing Escherichia coli O157 from wild birds. J. Appl. Microbiol. 82, 399–404 10.1046/j.1365-2672.1997.00378.x [DOI] [PubMed] [Google Scholar]
- Williams N. J., Sherlock C., Jones T. R., Clough H. E., Telfer S. E., Begon M., French N., Hart C. A., Bennett M. (2011). The prevalence of antimicrobial-resistant Escherichia coli in sympatric wild rodents varies by season and host. J. Appl. Microbiol. 110, 962–970 [DOI] [PubMed] [Google Scholar]
- Wright G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186 10.1038/nrmicro1614 [DOI] [PubMed] [Google Scholar]
- Wright G. D. (2010). Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13, 589–594 10.1016/j.mib.2010.08.005 [DOI] [PubMed] [Google Scholar]