Abstract
Genome sequencing studies indicate that all humans carry many genetic variants predicted to cause loss of function (LoF) of protein-coding genes, suggesting unexpected redundancy in the human genome. Here we apply stringent filters to 2,951 putative LoF variants obtained from 185 human genomes to determine their true prevalence and properties. We estimate that human genomes typically contain ~100 genuine LoF variants with ~20 genes completely inactivated. We identify rare and likely deleterious LoF alleles, including 26 known and 21 predicted severe disease-causing variants, as well as common LoF variants in non-essential genes. We describe functional and evolutionary differences between LoF-tolerant and recessive disease genes, and a method for using these differences to prioritize candidate genes found in clinical sequencing studies.
Introduction
Genetic variants predicted to severely disrupt protein-coding genes, collectively known as loss-of-function (LoF) variants, are of considerable scientific and clinical interest. Traditionally such variants have been regarded as rare and having a high probability of being deleterious, on the basis of their well-established causal roles in severe Mendelian diseases such as cystic fibrosis and Duchenne muscular dystrophy. However, recent studies examining the complete genomes of apparently healthy subjects have suggested that such individuals carry at least 200 (1, 2) and perhaps as many as 800 (3) predicted LoF variants. These numbers imply a previously unappreciated robustness of the human genome to gene-disrupting mutations, and have important implications for the clinical interpretation of human genome sequencing data.
Comparison of reported LoF variants between published genomes is complicated by differences in sequencing technology, variant-calling algorithms and gene annotation sets between studies (4, 5), and by the expectation that LoF variants will be highly enriched for false positives. The basis for this predicted enrichment is that strong negative natural selection is expected to act against the majority of variants inactivating protein-coding genes, thereby reducing the amount of true variation at these sites relative to the genome average, while sequencing error is expected to be approximately uniformly distributed; as a result, highly functionally constrained sites should show lower levels of observed polymorphism and substantially higher false positive rates (4). To date, no large-scale attempt has been made to validate the LoF variants reported in published human genome sequences.
LoF variants found in healthy individuals will fall into several overlapping categories: severe recessive disease alleles in the heterozygous state; alleles that are less deleterious but nonetheless have an impact on phenotype and disease risk; benign LoF variation in redundant genes; genuine variants that do not seriously disrupt gene function; and, finally, a wide variety of sequencing and annotation artifacts. Distinguishing between these categories will be crucial for the complete functional interpretation of human genome sequences.
Obtaining and filtering candidate LoF variants
We identified 2,951 candidate LoF variants using whole-genome sequencing data from 185 individuals analyzed as part of the pilot phase of the 1000 Genomes Project (2), as well as detailed analysis of high-coverage whole-genome sequencing data from a single anonymous European individual (6). The individuals represented 3 population groups: Yoruba individuals from Ibadan, Nigeria (YRI), 60 individuals of Northern and Western European origin from Utah (CEU) and 30 Chinese individuals from Beijing and 30 Japanese individuals from Tokyo that were analyzed jointly (CHB+JPT).
We adopted a definition for LoF variants expected to correlate with complete loss of function of the affected transcripts: stop codon-introducing (nonsense) or splice site-disrupting single nucleotide variants (SNVs), insertion/deletion (indel) variants predicted to disrupt a transcript’s reading frame, or larger deletions removing either the first exon or more than 50% of the protein-coding sequence of the affected transcript. We further sub-divided these variants into “full” LoF variants predicted to affect all known protein-coding transcripts of the affected gene, and “partial” variants affecting only a fraction of known coding transcripts. All annotation was performed against the Gencode v3b annotation (7) using the algorithm VAT (8).
We then subjected our candidate list to a series of stringent informatic and experimental validation steps (9). Informatic filtering was based on local sequence context (such as the presence of highly repetitive sequence), gene annotation (such as variants affecting non-canonical splice sites, or located close to the end of the affected open reading frame), analysis of the effects of nearby variants (such as neighboring SNVs altering the predicted functional effect of the candidate LoF variant), and measures of sequence read mapping and quality (Fig. S1). Where possible, thresholds for filtering were derived from the experimental validation experiments below.
We validated all candidate LoF SNVs and indels that were not excluded by other filters and for which we could design assays (n = 1,877) with experimental genotyping using three Illumina genotyping arrays and 819 custom Sequenom assays run, where possible, on all 185 samples from the low- and high-coverage 1000 Genomes pilot projects. Large deletions had previously been subjected to extensive validation (10), while those identified in NA12878 were assessed by comparison with independent 454 sequencing and array-based data from the same individual, as well as _targeted capillary sequencing of variants in highly repetitive regions. Finally, 786 variants were re-examined by complete manual reannotation of the 689 affected gene models by experienced curators, using the HAVANA annotation pipeline (7), to identify annotation errors and flag variants unlikely to profoundly affect gene function. All 589 candidate LoF variants identified in NA12878 were subjected to independent genotype validation and complete gene model reannotation.
As expected, the proportion of likely sequencing and annotation errors in the initial candidate set was high, with overlapping sets of 25.0%, 26.8% and 11.1% examined LoF variants being excluded as representing likely sequencing/mapping errors, annotation/reference sequence errors, and variants unlikely to cause genuine LoF, respectively. Candidate LoF variants removed by filtering tended to be more common than high-confidence variants (Fig. 1A). False positive rates due to sequencing errors (Fig. 1B) were higher for LoF variants than for missense and synonymous variants in the CHB+JPT and YRI populations (P < 10−8 for all comparisons) and significantly higher than for missense variants in CEU (P < 0.05). Because most variants in a given genome are common, the comparatively high rate of annotation errors among high-frequency LoF variants meant that filtering resulted a large reduction in LoF variants per individual (Table 1).
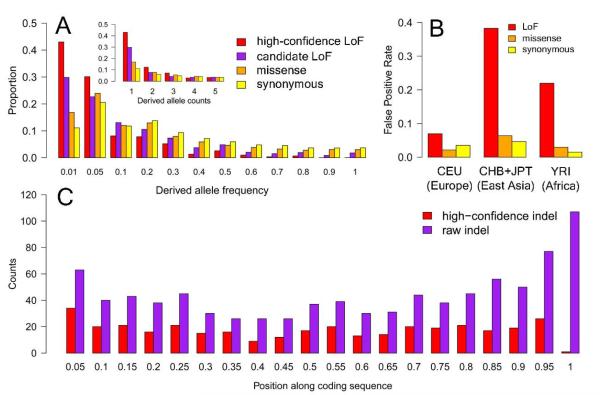
Figure 1.
A. Derived allele frequency distribution in the CEU population for raw and high-confidence LoF variants, compared to missense and synonymous coding variants. Inset, distribution of the proportion of SNVs in each class at low allele counts (1-5). B. False positive rates (based on independent array genotyping) for LoF variants filtered for annotation artifacts and frequency-matched missense and synonymous SNVs. C. Distribution of frameshift indels along the coding region of affected genes, before and after filtering (a similar pattern is also seen for nonsense SNVs; data not shown).
Table 1. Numbers of LoF variants before and after filtering.
Total numbers of candidate LoF variants and average number of LoF sites per individual (homozygous sites in brackets) are shown for each LoF class. For large deletions, numbers represent total number of genes predicted to be inactivated.
| before filtering | after filtering | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| variant type |
total | 1000G low-coverage average per individual |
NA12878 | total | 1000G low-coverage average per individual |
NA12878 | ||||
| CEU | CHB+JPT | YRI | CEU | CHB+JPT | YRI | |||||
| stop | 1111 | 85.7 (21.8) |
113.4 (26.7) |
109.1 (23.7) |
115 (25) | 565 | 26.2 (5.2) |
27.4 (6.9) |
37.2 (6.3) |
23 (2) |
|
| ||||||||||
| splice | 658 | 80.5 (29.5) |
98.1 (35.6) |
89.0 (30.4) |
95 (32) | 267 | 11.2 (1.9) |
13.2 (2.5) |
13.7 (1.9) |
12 (1) |
|
| ||||||||||
| frameshift indel |
1040 | 217.8 (112.1) |
225.5 (121.7) |
247.2 (118.7) |
348 (159) | 337 | 38.2 (9.2) |
36.2 (9.0) |
44.0 (8.0) |
38 (11) |
|
| ||||||||||
| large deletion |
142 | 32.4 (12.2) |
31.2 (11.8) |
31.4 (9.7) |
31 (5) | 116 | 28.3 (6.2) |
26.7 (5.9) |
26.6 (5.5) |
24 (4) |
|
| ||||||||||
| total | 2951 | 416.4 (175.6) |
468.2 (195.8) |
476.7 (316.0) |
654 (286) | 1285 | 103.9 (22.5) |
103.5 (24.3) |
121.5 (21.7) |
97 (18) |
We identified several sources of false positive LoF annotation that will require careful consideration in clinical sequencing projects. For instance, the predicted functional effect of a nonsense or frameshift variant can be altered by other nearby variants on the same chromosome (Table S1; Fig. S2), and predicted splice-disrupting SNVs and indels can be rescued by nearby alternative splice sites (Fig. S3). Both nonsense SNVs and frameshift indels are enriched towards the 3′ end of the affected gene, consistent with a greater tolerance to truncation close to the end of the coding sequence (Fig. 1C); putative LoF variants identified in the last 5% of the coding region were thus systematically removed from our high-confidence set, with the single exception of a known LoF indel in the NOD2 gene. There is also a discernible peak close to the 5′ end of genes, suggesting that some disrupted transcripts are rescued by transcriptional reinitiation at an alternative start codon (Fig. 1C).
Importantly, 415 (32.3%) of our high-confidence LoF variants are partial LoF variants, affecting only a subset of the known transcripts from the affected gene, meaning that functional protein may still be produced. We chose not to discard such cases, as it is currently impossible to assess the relative functional importance of different transcripts for most genes, and partial LoF mutations have previously been shown to be causal in Mendelian diseases (11).
In total, 43.5% (1,285/2,951) of our candidate LoF variants survived filtering. The resulting catalogue of high-confidence LoF variants is not complete: the 1000 Genomes pilot projects had low power to detect extremely rare variants (2), and we will not have detected certain classes of LoF variants, such as large gene-disrupting duplications, non-coding variants that disrupt gene expression or splicing regulation, or coding variants that destroy protein function without overtly disrupting an open reading frame (such as missense SNVs or in-frame indels). Several known LoF variant-containing genes such as ACTN3 (12) and CASP12 (13) were labeled as “polymorphic pseudogenes”, meaning that the reference genome contains non-functional allele of the gene, whereas in other haplotypes the gene is functional (14); it is likely that we missed LoF variants in other uncharacterized genes from this class.
Nonetheless, this catalogue represents the largest available set of high-confidence human variants predicted to disrupt protein-coding genes. We note that the majority of the LoF variants identified here are novel: 70% of the high-confidence LoF SNVs and indels were not present in dbSNP prior to the 1000 Genomes pilot project.
The true number of LoF variants in an individual genome
Using the systematically curated list of variants from NA12878, we estimate that this anonymous individual with European ancestry carries 97 LoF variants, with 18 present in a homozygous state (Tables 1, S2). These numbers, while still indicating an unexpected tolerance for gene inactivation in humans and being considerably higher than those based on genotyping known nonsense SNVs alone (15), are substantially lower than most previously published estimates based on whole-genome sequencing (e.g. (2, 3, 16), and provide a benchmark for further studies of individual variation in functional gene content. This analysis also provides a robust estimate of different variant classes on gene inactivation: for instance, we find that 39% of genes inactivated in the NA12878 genome are the result of frame-shifting indels, a potentially serious concern given that indels are typically under-called using short-read sequencing approaches (2). Over a quarter (28.7%) of the LoF SNVs and indels in NA12878 affect only a subset of the known transcripts from the affected genes, emphasizing the need to consider alternative splicing in the annotation of functional effects.
Properties of LoF variants and affected genes
LoF SNVs are strikingly enriched for low-frequency alleles compared to synonymous and missense SNVs (Fig. 1A), suggesting that many LoF variants are deleterious to human health and hence are prevented from increasing in frequency by purifying natural selection. Interestingly, the number of high-confidence LoF variants per individual is 25% higher in the YRI (Nigerian) sample than in the three non-African populations (P = 5.0 × 10−21; Table 1), suggesting a higher level of variation in functional gene content in African individuals consistent with their greater overall genetic diversity. However, we caution that larger samples with more homogeneous sequencing quality across populations will be required to confirm this finding and assess its likely functional impact.
We compared the properties of genes carrying at least one high-confidence LoF variant with those of other protein-coding genes. Genes containing high-confidence LoF alleles are relatively less evolutionarily conserved, showing a higher ratio of protein-altering to silent substitutions in coding regions between human and macaque (P = 2.8 × 10−52) and less evolutionary conservation in their promoter regions (GERP score; P = 3.7 × 10−16). On average, they have more closely related gene family members (paralogs) than other genes (P = 0.0058) and show greater sequence identity to paralogs (P = 0.0068), suggesting that in many cases their function may be partially redundant, and also increasing the possibility that LoF variants may be gained or lost through the process of gene conversion (17) as has recently been reported for disease mutations (18). They also have lower connectivity in both protein-protein interaction (P = 6.8 × 10−6) and gene interaction (P = 4.2 × 10−19) networks, suggesting that LoF-containing genes are generally less central to key cellular pathways, although there are caveats to this interpretation (9). LoF-containing genes are strongly enriched for functional categories related to olfactory reception, and depleted for genes implicated in protein-binding, transcriptional regulation and anatomical development (Table S8).
We estimated the probability that heterozygous inactivation of a given gene will be deleterious (a state known as haploinsufficiency) using a combination of functional and evolutionary parameters (9, 19). Our filtering process disproportionately removed candidate LoF variants with a higher predicted probability of haploinsufficiency, P(HI), consistent with the majority of putative LoF variants in highly functionally constrained genes being artifactual (Fig. 2A). High-confidence LoF variants remaining after filtering have significantly lower P(HI) than variants discarded by our filters (P = 2.1 × 10−16) or known haploinsufficient genes (P = 1.8 × 10−73).
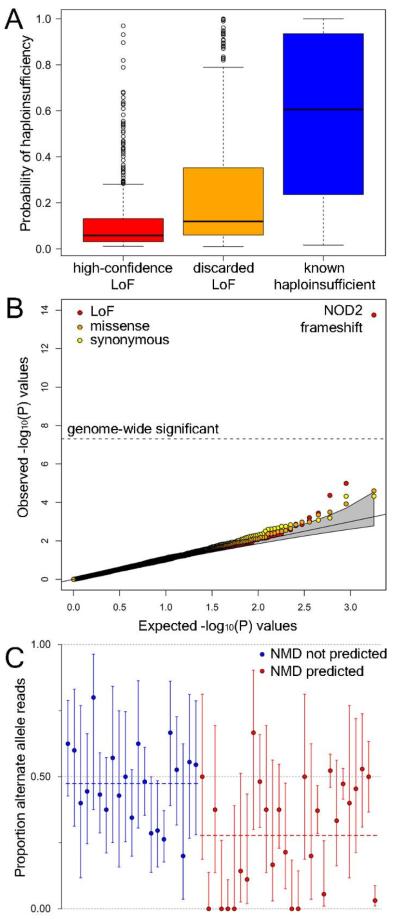
Figure 2.
A. Estimated probability of haploinsufficiency (presence of disease due to heterozygous loss of function), using a model trained using an independent set of LoF deletions as well as a set of known haploinsufficient genes. B. Association of coding variants with complex disease risk. Observed -log10(P) values for disease association in 17,000 individuals from 7 complex disease cohorts and a shared control group, following imputation of variants identified by the 1000 Genomes low-coverage pilot, are plotted against the expected null distribution for all LoF variants and frequency-matched missense and synonymous SNPs. C. Allele-specific expression analysis of nonsense variants, using RNA sequencing data from 119 lymphocyte cell lines. Circles show the proportion of LoF-carrying reads spanning each site across all heterozygous individuals. Variants predicted to cause nonsense-mediated decay (NMD, red) and those predicted to escape NMD (blue) are arbitrarily ordered by genome position within each class. Blue and red dashed horizontal lines indicate mean values in each class. Error bars, 95% CI.
We identified 365 genes with multiple candidate LoF variants. The majority of the genes with three or more independent LoF variants were found to represent systematic sequencing errors: for instance, the CDC27 gene contained 10 separate candidate splice-disrupting variants, all of which were found to represent mapping errors due to an inactive gene copy absent from the human reference sequence. Most of these variants were removed by filtering (Table S3). Of the remaining genes, some likely represent genes drifting towards inactivation in the population: for instance, the VWDE gene contains four separate high-confidence LoF variants, with 42.7% of the sequenced 1000G samples carrying at least one non-functional copy of this gene.
Effects of LoF variants on human phenotypes and disease risk
The high-confidence LoF set includes many known LoF variants reported to have effects on human traits (Table S4). We also found a number of previously uncharacterized LoF variants likely to have phenotypic effects. For instance, we identified three separate LoF variants in PKD1L3 and one in PKD2L1; the protein products of these two genes form a putative sour taste receptor complex (20, 21), so these variants may underlie variation in sour taste sensitivity between humans.
Our high-confidence LoF set includes many variants relevant to severe human disease. We identified 26 known recessive disease-causing mutations in our high-confidence LoF set, including mutations associated with the severe early-onset conditions Leber congenital amaurosis, harlequin ichthyosis, osteogenesis imperfecta and Tay-Sachs disease (Table S5). We also identified 21 strong candidates for novel disease-causing mutations: high-confidence LoF variants affecting all known transcripts of genes in which other null mutations have been convincingly associated with Mendelian disease, including adult-onset muscular dystrophy, Charcot-Marie-Tooth disease and mucolipidosis (Table S6). With one exception (a variant associated with transplant graft-versus-host disease) no individuals were homozygous for the putative disease-causing alleles.
Given the evidence for the presence of known deleterious variants, we hypothesized that LoF variants may also be enriched for association with risk of common, complex diseases. We investigated this hypothesis by imputing genotypes for 417 LoF SNVs and indels into a total of 13,241 patients representing seven complex diseases such as Crohn’s disease and rheumatoid arthritis, along with 2,938 shared controls, who had previously been subjected to genome-wide SNP genotyping (22). We confirmed a previously known frameshift indel in the NOD2 gene associated with Crohn’s disease, with a genome-wide significant imputed P value of 1.78 × 10−14 (two orders of magnitude more significant than the best tag SNP). However, no other LoF variants achieved genome-wide significance, and there was no overall excess of association signals in LoF variants compared to other coding variants (Fig. 2B). Since our catalogue is expected to contain most genuine LoF variants at greater than 5% frequency this result suggests that common gene-disrupting variants play a minor role in complex disease predisposition.
One explanation for the paucity of common LoF variants associated with complex disease risk is purifying selection, which is expected to prevent most severely deleterious alleles from reaching high population frequencies; this is consistent with the skew towards low frequencies amongst high-confidence LoF variants (Fig. 1A). In addition, genes containing homozygous LoF variants have more gene family members (median 5 vs 3; P = 3.76 × 10−3) and are less conserved between macaque and human (P = 1.87 × 10−4) than genes containing only heterozygous LoF variants, suggesting greater redundancy in genes affected by high-frequency loss of function. Similarly small effects on complex disease risk have previously been noted for large, common copy-number variations, another class of variant with a high prior probability of functional impact (23).
Genotype imputation and case-control association studies have low power to detect associations for low-frequency variants, so further experiments involving direct genotyping of LoF variants in large disease cohorts will be required to characterize the impact of rare LoF variation on human complex disorders.
Effects of nonsense SNVs on gene expression
We examined the impact of validated nonsense SNVs on gene expression using RNA sequencing data generated from lymphoblastoid cell lines of 119 samples from two populations (24, 25). Comparison of the relative expression of the LoF and functional alleles within experimentally genotyped heterozygous individuals (Fig. 2C; Table S7) revealed a statistically significant reduction in expression from the LoF allele in 8/49 (16.3%) of variants with sufficient sequencing depth to be assayed. As expected, this reduction in expression is most common for variants predicted to trigger nonsense-mediated mRNA decay (NMD), a cellular process that degrades premature stop codon-containing transcripts: 7/28 (25.0%) of predicted NMD-triggering variants show significant evidence of decay, compared to 1/21 (4.8%) of predicted NMD-evading variants, and the proportion of reads mapping to the alternate allele was significantly lower for predicted NMD-triggering variants (median 0.352 vs 0.481; P = 0.0023). However, most predicted NMD-triggering variants have no detectable effect on gene expression.
These results provide functional confirmation of true loss of gene function for a minority of LoF variants. In addition, they demonstrate that the most widely-used algorithm for NMD prediction (26) is an imperfect indicator of the effects of nonsense SNVs on RNA expression.
Natural selection on LoF variants
We explored whether LoF variants as a class showed evidence of recent positive selection, as expected under the “less is more” hypothesis of adaptive gene loss proposed by Olson (27). We examined the overlap between high-confidence LoF variants and regions showing potential signatures of positive selection using frequency spectrum and haplotype length-based tests on 1000 Genomes pilot data (2). In contrast to the “less is more” hypothesis, LoF variants overlapped with positively selected regions no more often than frequency-matched synonymous SNVs. However, we have identified 20 high-confidence LoF variants in candidate regions for positive selection that warrant further analysis (Table S10).
In some cases, selection for gene inactivation may act through the accumulation of multiple rare LoF variants rather than increased frequency of a specific LoF allele. We identified one potential example of this: in addition to a relatively common nonsense SNV in the CD36 gene reported to be the _target of positive selection in African populations (28) we identified two rare, novel splice-disrupting SNVs in the same gene. All three of these variants were specific to the Yoruban (YRI) population, suggesting that multiple null alleles for CD36 may be accumulating in African populations under the influence of selection.
Using LoF-tolerant genes to predict the probability of disease causation for novel variants
Homozygous inactivation of a gene can have a range of phenotypic effects: at one end of the spectrum are severe recessive disease genes, while at the other end are genes that can be inactivated without overt clinical impact, referred to here as LoF-tolerant genes. Clinical sequencing projects seeking to identify disease-causing mutations would benefit from improved methods to distinguish where along this spectrum each affected gene lies.
Genes homozygously inactivated in 1000 Genomes Project samples are likely to fall close to the LoF-tolerant end of the spectrum. These genes therefore represent a comparison group that can be used to define the functional and evolutionary characteristics that distinguish these genes from severe recessive disease genes.
We examined the 253 genes containing validated LoF variants that were found to be homozygous in at least one individual. These LoF-tolerant genes are significantly less conserved and have fewer protein-protein interactions than the genome average (Fig. 3A). They are also enriched for functional categories related to chemosensation, largely explained by the enrichment of olfactory receptor genes in this class (13.0% vs 1.4% genome-wide), and depleted for genes involved in embryonic development and cellular metabolism (Table S8).
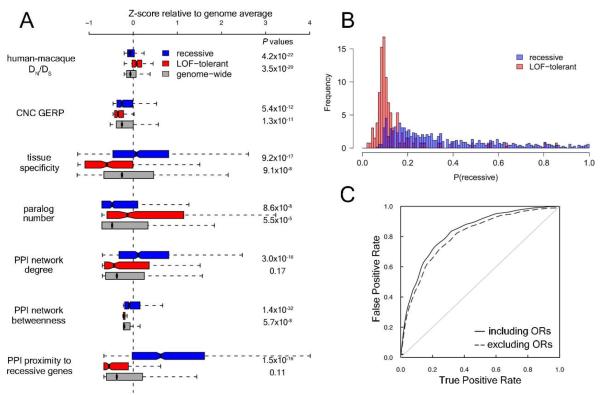
Figure 3.
A. Distribution of selected evolutionary and functional parameters for recessive disease genes (blue) and LoF-tolerant genes (red) compared to all protein-coding genes (grey). Values are transformed to z scores to allow parameters to be plotted together. Boxes show interquartile range with medians indicated with a vertical black line, and whiskers terminate at the most extreme point less than 1.5 times the interquartile range from the box. For each pair of P values, top value refers to the recessive vs LoF-tolerant comparison and bottom refers to the LoF-tolerant vs genome background comparison. As many of the parameters are left-skewed the medians typically fall below zero. B. P value distribution for linear discriminant model (LDM) trained using LoF-tolerant and recessive disease genes, based on human-macaque DN/DS ratio and PPI network proximity to known recessive disease genes. C. Receiver-operating characteristic (ROC) curve for LDM distinguishing between LoF-tolerant and recessive disease genes, both when olfactory receptor genes (ORs) are included (solid line, AUC = 0.831) and excluded (dashed line, AUC = 0.814). DN/DS, ratio of missense to synonymous substitutions; CNC GERP, GERP score for conserved non-coding elements within 50 kb of gene; PPI, protein-protein interaction.
We then identified parameters that could be used to classify candidate genes along the disease/LoF-tolerant spectrum. We first removed olfactory receptors from the LoF-tolerant set, as these genes could be easily excluded as candidates for most severe Mendelian diseases, leaving 213 LoF-tolerant genes to compare with 858 known recessive disease genes. These two gene categories were found to display marked differences in a wide range of properties (Fig. 3A).
We developed a linear discriminant model based on human-macaque conservation and proximity to recessive disease genes in a protein-protein interaction network to classify genes into LoF-tolerant and recessive disease classes (Fig. 3B, 3C). Although insufficient to definitively discriminate between the two classes, this algorithm could be used to prioritize candidates identified by sequencing recessive disease patients for replication and functional follow-up. We have calculated a recessive disease probability score for each protein-coding gene in the genome for use in such analyses (9).
Conclusions
Here we describe a stringently filtered catalogue of variants disrupting the reading frame of human protein-coding genes, including the majority of such variants present at a population frequency of 5% or greater. Because large numbers of candidate LoF variants are present in the genomes of all individuals, but are highly enriched for a variety of sequencing and annotation errors, there is a need for caution in assigning disease-causing status to novel gene-disrupting variants found in patients. More reliable reference gene sets will help: reference sequence and automated gene annotation errors accounted for 44.9% of candidate LoF variants in our deeply characterized individual genome, but most of these have now been corrected as a result of this project and other manual annotation efforts.
Our stringent filtering of the LoF variants found in a single high-quality human genome suggests that a typical “healthy” genome contains ~100 genuine LoF variants, with most of them carried in the heterozygous state. Given that humans (29) and other species (30) have been estimated to carry fewer than 5 recessive lethal alleles per genome, it seems likely that the majority of LoF variants found in an individual genome are common variants in non-essential genes, although these may still have an effect on human phenotypic variation. Nonetheless, the signature of strong purifying selection against high-confidence LoF variants as a class, and the discovery of numerous known and predicted severe recessive disease alleles, indicates that many LoF alleles with large effects on human fitness exist at low frequency in the human population. Large sequencing and genotyping projects will be required to uncover the full spectrum of these variants and their effects on human disease risk.
We have found that LoF-tolerant and recessive disease genes have differing functional and evolutionary properties, allowing us to develop a potential approach for prioritizing novel candidate recessive disease variants identified in patient samples for functional follow-up. As further examples of LoF-tolerant genes are obtained from high-throughput sequencing studies the power of this type of classification approach is likely to grow considerably.
Finally, we note that our catalogue of validated LoF variants comprises a list of naturally occurring “knock-out” alleles for over 1,000 human protein-coding genes, many of which currently have little or no functional annotation attached to them. Identification and systematic phenotyping of individuals homozygous for these variants could provide valuable insight into the function of many poorly characterized human genes.
Supplementary Material
Acknowledgments
T. Shah provided the Pyvoker software used for manual assignment of genotypes based on intensity clusters, S. Edkins was involved in the Sequenom validation, and the genotyping groups at Illumina, the Wellcome Trust Sanger Institute and The Broad Institute of Harvard and MIT provided raw intensity data for the three Illumina arrays used for genotyping validation. The work performed at the Wellcome Trust Sanger Institute was supported by Wellcome Trust grant 098051; DM was supported by a fellowship from the Australian National Health and Medical Research Council; GL by the Wellcome Trust (090532/Z/09/Z); ETD and SBM by the Swiss National Science Foundation, the Louis Jeantet Foundation and the NIH-NIMH GTEx fund; KY by NWO VENI grant 639.021.125; and HZ, YL and JW by a National Basic Research Program of China (973 program no. 2011CB809200), the National Natural Science Foundation of China (30725008; 30890032; 30811130531), the Chinese 863 program (2006AA02A302;2009AA022707), the Shenzhen Municipal Government of China (grants JC200903190767A; JC200903190772A; ZYC200903240076A; CXB200903110066A; ZYC200903240077A; and ZYC200903240080A) and the Ole Rømer grant from the Danish Natural Science Research Council, as well as funding from the Shenzhen Municipal Government and the Local Government of Yantian District of Shenzhen. JKP is on the scientific advisory board of 23andMe and RAG has a shared investment in Life Technologies. Raw sequence data for the 1000 Genomes pilot projects are available from www.1000genomes.org, and a curated list of the loss-of-function variants described in this manuscript is provided in the Supplementary Online Material.
References and notes
- 1.Ng PC, et al. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.1000 Genomes Project Consortium Nature. 2010;467:1061. [Google Scholar]
- 3.Pelak K, et al. PLoS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacArthur DG, Tyler-Smith C. Hum. Mol. Genet. 2010;19:R125. doi: 10.1093/hmg/ddq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian S, et al. Genes Dev. 2011;25:1. doi: 10.1101/gad.1968411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePristo MA, et al. Nat. Genet. 2011;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrow J, et al. Genome Biol. 2006;7(Suppl 1):1. doi: 10.1186/gb-2006-7-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. http://vat.gersteinlab.org/
- 9.See supporting material on Science online.
- 10.Mills RE, et al. Nature. 2011;470:59. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uzumcu A, et al. J. Med. Genet. 2006;43:e5. doi: 10.1136/jmg.2005.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacArthur DG, et al. Nat. Genet. 2007;39:1261. doi: 10.1038/ng2122. [DOI] [PubMed] [Google Scholar]
- 13.Xue Y, et al. Am. J. Hum. Genet. 2006;78:659. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M. Genome Biol. 2010;11:R26. doi: 10.1186/gb-2010-11-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yngvadottir B, et al. Am. J. Hum. Genet. 2009;84:224. doi: 10.1016/j.ajhg.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupski JR, et al. N. Engl. J. Med. 2010;362:1181. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Nature Reviews Genetics. 2007;8:762. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 18.Casola C, Zekonyte U, Phillips AD, Cooper DN, Hahn MW. Genome Res. 2011;22 doi: 10.1101/gr.127738.111. doi:10.1101/gr.127738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang N, Lee I, Marcotte EM, Hurles ME. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimaru Y, et al. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12569. doi: 10.1073/pnas.0602702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang AL, et al. Nature. 2006;442:934. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellcome Trust Case Control Consortium Nature. 2007;447:661. [Google Scholar]
- 23.Conrad DF, et al. Nature. 2010;464:704. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montgomery SB, et al. Nature. 2010;464:773. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickrell JK, et al. Nature. 2010;464:768. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy E, Maquat LE. Trends Biochem. Sci. 1998;23:198. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 27.Olson MV. Am. J. Hum. Genet. 1999;64:18. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fry AE, et al. Hum. Mol. Genet. 2009;18:2683. doi: 10.1093/hmg/ddp192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bittles AH, Neel JV. Nat. Genet. 1994;8:117. doi: 10.1038/ng1094-117. [DOI] [PubMed] [Google Scholar]
- 30.McCune AR, et al. Science. 2002;296:2398. doi: 10.1126/science.1071757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.