Abstract
Progress in understanding the genetic and neurobiological basis of bipolar disorder(s) has come from both human studies and animal model studies. Until recently, the lack of concerted integration between the two approaches has been hindering the pace of discovery, or more exactly, constituted a missed opportunity to accelerate our understanding of this complex and heterogeneous group of disorders. Our group has helped overcome this “lost in translation” barrier by developing an approach called Convergent Functional Genomics (CFG). The approach integrates animal model gene expression data with human genetic linkage/association data, as well as human tissue (postmortem brain, blood) data. This Bayesian strategy for cross-validating findings extracts meaning from large datasets, and prioritizes candidate genes, pathways and mechanisms for subsequent _targeted, hypothesis-driven research. The CFG approach may also be particularly useful for identification of blood biomarkers of the illness.
Keywords: microarray, animal model, convergent functional genomics, bipolar, genes, brain, blood, biomarkers
Introduction
Identifying genes for bipolar disorder through classic human genetic studies has proven arduous, despite some recent successes (as reviewed in (Hayden and Nurnberger 2006), (Kato 2007)). There are at least three possible reasons for this relatively slow pace of discovery. First, bipolar disorder, like other neuropsychiatric disorders, is in all likelihood a complex polygenic disorder, with variable penetrance. Moreover, some (if not all) true illness-causing mutations may be deleterious to reproductive potential and thus be evolutionarily selected against, resulting in minor-frequency alleles that require sample sizes beyond those used to date in order to be able to unequivocally establish statistical association with illness. Second, the phenotypic heterogeneity, overlap and interdependence with other neuropsychiatric disorders (Niculescu et al 2006), (Niculescu 2006) is not fully explored or built into the genetics work carried so far, including the more recent whole-genome association (WGA) efforts. Third, gene-environment interactions and the effect of environmental factors (epigenetic modifications, effects of stress, infections, drugs, medications) on the expression of the phenotype are not fully factored in human genetic linkage studies to date (Tsuang et al 2004), (Abdolmaleky et al 2004), (Crow 2007).
Possible solutions to the above three problems are the following, respectively: 1) carrying out association studies with groups of genes that may work together, rather than with individual genes; 2) analysis of association with discrete quantitative endophenotypes (Hasler et al 2006) (Niculescu et al 2006) rather than broad DSM diagnostic classifications; and 3) use of gene expression studies (in human blood, postmortem brain or animal models), which are a direct reflection of gene-environment interactions, in conjunction with classic genetic studies.
Animal models can provide help with these three potential solutions encountered by classic human genetic research: 1) gene expression studies in animal models can identify groups of genes that change together, and thus may work together, on a homogeneous genetic background, with the signal not masked by the noise generated from the variable genetic background present in human studies; 2) endophenotypes of the disorder can be deliberately mimicked in animal models with pharmacological approaches(Ogden et al 2004), or fortuitously observed in genetic mutants(Roybal et al 2007); and 3) gene-environment interactions are minimized, well defined and well controlled in animal model studies as opposed to human studies.
Our expanded Convergent Functional Genomics approach (Ogden et al 2004),(Bertsch et al 2005),(Rodd et al 2006),(Le-Niculescu et al 2007) embodies an appreciation of the strengths and limitations of animal model data and human data (potential lack of specificity for animal model data, potential lack of sensitivity for human data). It relies on the integration of multiple independent lines of evidence. Each of the lines of evidence is potentially vulnerable to type I or type II errors, but taken together in a Bayesian fashion (Bernardo and Smith 1994), they are less likely to provide false positives or false negatives. By putting together carefully designed animal model experimental data with human genetic and human tissue expression data, the CFG approach provides a comprehensive solution to the challenge of identifying candidate genes, pathways and mechanisms for neuropsychiatric disorders. It has been applied with some success to bipolar disorder (Niculescu et al 2000), (Ogden et al 2004), and more recently to alcoholism (Rodd et al 2006) and schizophrenia (Le-Niculescu et al 2007). Approaches similar to ours (named, alternately, as integrative genomics) have started to be applied widely in various fields of biomedicine (Mootha et al 2003) (Schadt 2006) (Zhu et al 2007).
Convergent Functional Genomics
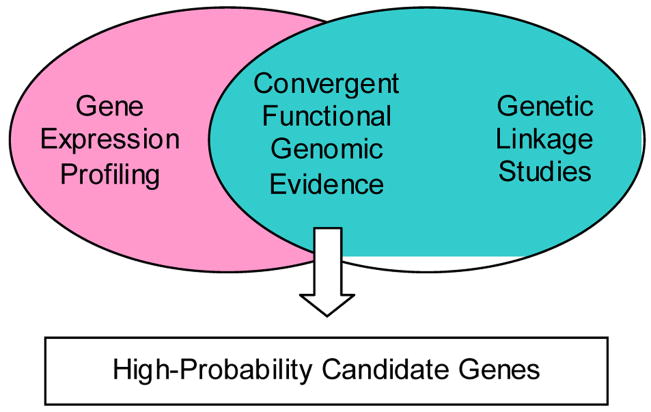
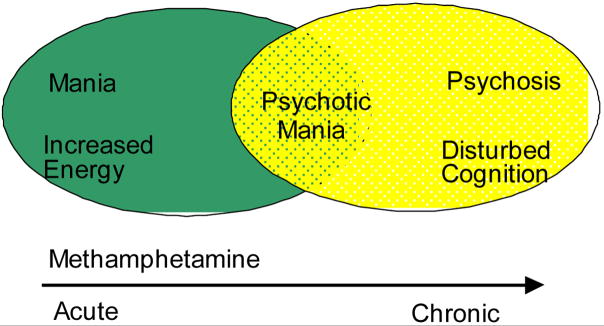
The Convergent Functional Genomics approach was developed initially to integrate gene expression data from a relevant animal model with data from human linkage analyses as a way of cross-validating findings and deriving a short list of high-probability candidate genes that deserve individual scrutiny in a prioritized fashion (Niculescu et al 2000) (Figure 1). For bipolar disorder, one relevant animal model involves the administration of a single dose of methamphetamine. Methamphetamine administration mimics in human and rodents some of the behavioral signs and symptoms of bipolar disorder including manic-like features in the activation phase and depressive-like features in the withdrawal phase. With more chronic escalating-dose binge treatments of methamphetamine, a more complex picture akin to psychosis emerges (Figure 2).
Figure 1.
Initial Iteration of Convergent Functional Genomics
Figure 2.
Candidate genes emerging from our initial application of this strategy have since been actively investigated for a role in bipolar disorder, with some genes such as DBP (which codes for albumin D-box-binding protein) (Sokolov et al 2003), (McFarland et al Submitted 2007) and GRK3 (which codes for G-protein- coupled receptor kinase 3)(Barrett et al 2003; Shaltiel et al 2006) yielding additional evidence for involvement in the disorder that may, upon further replication, warrant distinction as “risk genes” for bipolar disorder.
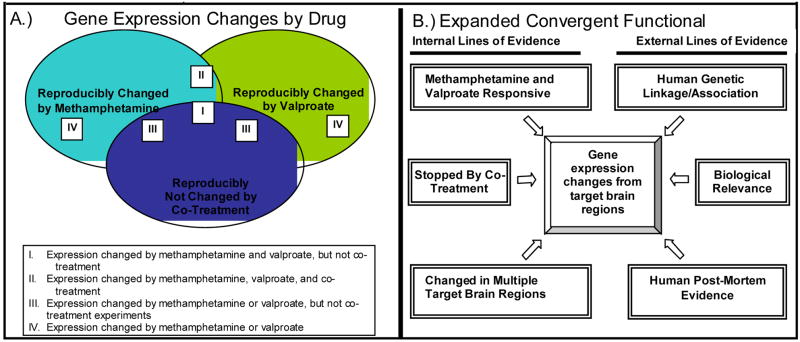
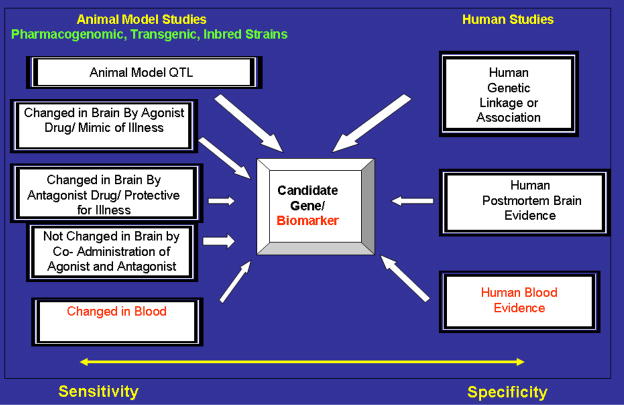
More recently, we have expanded the Convergent Functional Genomics approach to look at the effects of two opposing pharmacological manipulations in mice, using both an agonist of the illness/bipolar disorder-mimicking drug (methamphetamine) and an antagonist of the illness/bipolar disorder-treating drug (valproate) (Ogden et al 2004). In essence, the pharmacogenomic approach is a tool for tagging genes that may have pathophysiological relevance. Additionally, we have extended the convergent cross-validation beyond genetic linkage results to data on biological relevance of genes and postmortem brain changes (Ogden et al 2004). In this application of the Convergent Functional Genomics paradigm, mice received either: 1) the bipolar disorder-mimicking agent methamphetamine; 2) the bipolar disorder-relieving agent valproate; or 3) co-treatment with both drugs.
Comprehensive gene expression analysis of specific _target brain regions that have previously been implicated in bipolar disorder (including prefrontal cortex, amygdala, nucleus accumbens, ventral tegmentum, and caudate-putamen) with oligonucleotide microarrays was carried out in parallel with behavioral studies. As internal cross-validation, genes that were changed by both methamphetamine and valproate were deemed to be higher-probability candidate genes than genes changed by either drug alone (Figure 3A, Categories I and II). Genes that were changed by individual drug treatments but were “nipped in the bud” (showed no change) by co-treatment were also deemed to be of higher probability (Figure 3A, Category III). Lastly, genes that were changed in multiple _target brain regions were deemed to be of higher probability as well, from a detection standpoint if not necessarily from an etio-pathological one. As external cross-validation, gene expression data was cross-referenced to linked loci from human genetic studies of bipolar and related disorders, reports from the literature regarding the biological role of these genes, and reports of changes in post-mortem brain tissue from patients (Figure 3B).
Figure 3.
Expanded Convergent Functional Genomics Approach to Identifying Candidate Genes Involved in Bipolar Disorder and Related Disorders
By integrating multiple internal and external lines of evidence, a prioritized list of candidate genes was generated. Functionally relevant genes represented in this dataset, for some of whom additional evidence has accumulated in the field since our publication (Ogden et al 2004), include: DARPP-32 pathway genes (Meyer-Lindenberg et al 2007), pain related genes (such as TAC1-substance P (Carletti et al 2005), PENK- preproenkephalin (Nieto et al 2005)), circadian clock genes (such as ARNTL1/BMAL1 (Nievergelt et al 2006); (Mansour et al 2006)), apoptosis-related genes (such as BAD (Laeng et al 2004)), G-protein-coupled receptor signaling genes (such as GPR88(Brandish et al 2005), (Conti et al 2007)), intracellular signaling genes (such as GSK3b (Gould et al 2006)), glutamate neurotransmission (such as GRM3 (Fallin et al 2005) transporters (such as DAT1-dopamine transporter (Greenwood et al 2006), neurotrophic/neuronal survival factor genes (such as BDNF -brain-derived neurotrophic factor (Muller et al 2006), and glia/myelin-related genes (such as MOBP, PLP1, CLDN11, PMP22, MAG, GFAP, GMFB). The glia/myelin story is particularly interesting, as similar findings are reported by others (Davis et al 2003; Tkachev et al 2003); (Haroutunian et al 2007) and us (Le-Niculescu et al 2007) in schizophrenia, and alcoholism (Lewohl et al 2005), (Rodd et al 2006). These findings point to the issue of overlap among genes involved in major neuropsychiatric disorders (Niculescu 2006); (Le-Niculescu et al 2007). Moreover, hypomyelination of frontal lobe regions may reflect the incomplete development of brain regions involved in the process of executive control and motivation (Volkow and Li 2005), (Bartzokis 2006),(Sokolov 2007), potentially resulting in cognitive, affective and hedonic dysregulation. As such, this neurobiological abnormality could be a sensitive but non-specific common denominator of mental illness, and an explanation for what are clinically called dual-diagnosis disorders (substance abuse and another psychiatric disorder).
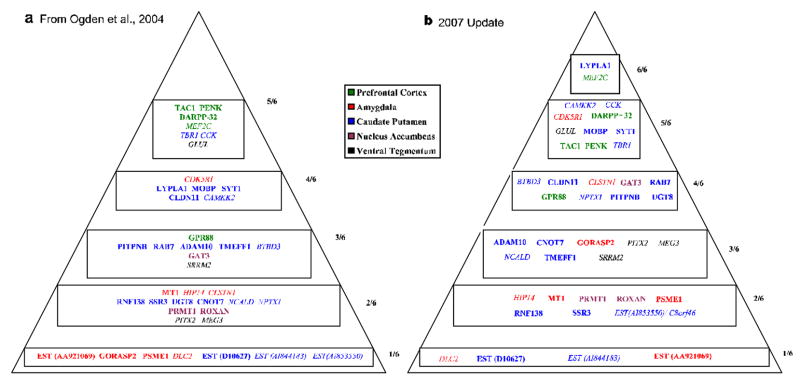
Our dataset also identified candidate genes mapping to linkage loci identified by large-scale meta-analyses for bipolar disorder (McQueen et al 2005; Segurado et al 2003) and schizophrenia (Lewis et al 2003), cross-validated these candidates with human postmortem findings, and revealed other novel genes (Figure 4), pathways and mechanisms that may be of importance in the pathophysiology of bipolar disorder (Ogden et al 2004). These findings are prime starting points for subsequent hypothesis-driven work, such as candidate gene association studies, epistatic interactions testing, and transgenic mouse models generation. Moreover, they provide insights for new pharmacotherapeutic approaches to bipolar disorder.
Figure 4. Top candidate genes for bipolar disorder(s).
Probability pyramid generated by the tabulation of independent convergent lines of evidence (as shown in Figure 3B). Plain text-increased by methamphetamine. Italics-decreased by methamphetamine. Color coding-different _target brain regions in our animal model. (a) from Ogden et al. 2004. (b) updated 2007 with new external lines of evidence published in the field since 2004. Once a gene is on the pyramid, it can only move up in priority as new evidence emerges over time. On the sides of the pyramids is depicted the scoring based on number of lines of evidence.
Future directions: blood gene expression profiling and biomarker research
Objective biomarkers of illness and treatment response would make a significant difference in our ability to diagnose and treat patients with psychiatric disorders, eliminating subjectivity and our reliance on patient’s self-report of symptoms. Lymphocyte gene expression profiling has emerged as a particularly interesting area of research in the search for peripheral biomarkers (Vawter et al 2004), (Tsuang et al 2005), (Segman et al 2005), (Glatt et al 2005; Middleton et al 2005),(Sullivan et al 2006) (Naydenov et al 2007). Most of the studies to date have focused on human blood gene expression profiling, comparison between illness groups and normal controls, and cross-matching with human postmortem brain gene expression data. They suffer from one or both of the following caveats. The first is the widespread use of lymphoblastoid cell lines in lieu of fresh blood. Fresh blood, with quantitative phenotypic state measures gathered at time of harvesting, may be more informative than immortalized lymphocytes, and avoid some of the caveats of Epstein-Barr virus (EBV) immortalization and cell culture passaging. The second potential caveat is that human tissue gene expression studies are susceptible to the issue of being underpowered, due to genetic heterogeneity, the effect of variable environmental exposure (including medications and drugs of abuse) on gene expression, and difficulty of accrual of large sample cohorts, particularly fresh blood samples with state phenotypic information. It is questionable if, by themselves, such studies have sufficient power to identify all the bona fide biomarkers out of large and noisy human blood gene expression datasets, despite a variety of sophisticated statistical methodologies (Glatt et al 2005), (McClintick and Edenberg 2006), (Zapala and Schork 2006), (Li and Horvath 2007) that can be employed.
Summary
The current state of our understanding of the genetic and neurobiological basis for bipolar disorder in general and of peripheral molecular biomarkers of the illness in particular, is still inadequate. Most of the fundamental genetic, environmental, and biological elements needed to delineate the etiology and pathophysiology of bipolar disorder are yet to be completely identified, understood and validated. A rate-limiting step, which we are helping overcome, has been the lack of concerted integration across disciplines and methodologies. The use of a multidisciplinary, integrative research framework such as Convergent Functional Genomics should lead to a reduction in the historically high rate of inferential errors committed in studies of complex diseases like bipolar disorder. Currently emerging whole genome association studies will provide a wealth of information to be mined, made sense of and prioritized with approaches such as ours.
An interesting area of future research is in regards to peripheral biomarkers of the illness. To our knowledge, no one has reported, at the time of writing of this review (early 2007), a comprehensive investigation of blood gene expression profiling in conjunction with brain gene expression studies in an animal model presenting features of bipolar disorder, and cross-validated that data with comprehensive human fresh blood gene expression studies tied to illness state, as well as integrated the findings in the context of the available human genetic linkage/association data, postmortem brain data and information on biological pathways. We suggest that such an expanded Convergent Functional Genomics approach (Figure 5) may be particularly fruitful for biomarker discovery, and overcome the caveats mentioned above. These studies are underway in our laboratories. We anticipate that panels of biomarkers, rather than single biomarkers, are going to emerge as clinically useful tools.
Figure 5.
Expanded convergent Functional Genemonics (2007): Multiple Independent Lines of Evidence for Bayesian cross Validation
Acknowledgments
This work was supported by funds from INGEN (Indiana Genomics Initiative of Indiana University) and INBRAIN (Indiana Biomarker Research Alliance in Neuropsychiatry) to ABN, as well as NIMH R01 MH071912-01 to MTT and ABN. ABN is a NARSAD Mogens Schou Young Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdolmaleky HM, Smith CL, Faraone SV, Shafa R, Stone W, Glatt SJ, et al. Methylomics in psychiatry: Modulation of gene-environment interactions may be through DNA methylation. Am J Med Genet B Neuropsychiatr Genet. 2004;127:51–59. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- Barrett TB, Hauger RL, Kennedy JL, Sadovnick AD, Remick RA, Keck PE, et al. Evidence that a single nucleotide polymorphism in the promoter of the G protein receptor kinase 3 gene is associated with bipolar disorder. Mol Psychiatry. 2003;8:546–557. doi: 10.1038/sj.mp.4001268. [DOI] [PubMed] [Google Scholar]
- Bartzokis G. Acetylcholinesterase Inhibitors May Improve Myelin Integrity. Biol Psychiatry. 2006 doi: 10.1016/j.biopsych.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Bertsch B, Ogden CA, Sidhu K, Le-Niculescu H, Kuczenski R, Niculescu AB. Convergent functional genomics: a Bayesian candidate gene identification approach for complex disorders. Methods. 2005;37:274–279. doi: 10.1016/j.ymeth.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Brandish PE, Su M, Holder DJ, Hodor P, Szumiloski J, Kleinhanz RR, et al. Regulation of gene expression by lithium and depletion of inositol in slices of adult rat cortex. Neuron. 2005;45:861–872. doi: 10.1016/j.neuron.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Carletti R, Corsi M, Melotto S, Caberlotto L. Down-regulation of amygdala preprotachykinin A mRNA but not 3H-SP receptor binding sites in subjects affected by mood disorders and schizophrenia. Eur J Neurosci. 2005;21:1712–1718. doi: 10.1111/j.1460-9568.2005.04002.x. [DOI] [PubMed] [Google Scholar]
- Conti B, Maier R, Barr AM, Morale MC, Lu X, Sanna PP, et al. Region-specific transcriptional changes following the three antidepressant treatments electro convulsive therapy, sleep deprivation and fluoxetine. Mol Psychiatry. 2007;12:167–189. doi: 10.1038/sj.mp.4001897. [DOI] [PubMed] [Google Scholar]
- Crow TJ. How and why genetic linkage has not solved the problem of psychosis: review and hypothesis. Am J Psychiatry. 2007;164:13–21. doi: 10.1176/ajp.2007.164.1.13. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, et al. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77:918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N, et al. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proc Natl Acad Sci U S A. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould TD, Picchini AM, Einat H, Manji HK. _targeting glycogen synthase kinase-3 in the CNS: implications for the development of new treatments for mood disorders. Curr Drug _targets. 2006;7:1399–1409. doi: 10.2174/1389450110607011399. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Schork NJ, Eskin E, Kelsoe JR. Identification of additional variants within the human dopamine transporter gene provides further evidence for an association with bipolar disorder in two independent samples. Mol Psychiatry. 2006;11:125–133. 115. doi: 10.1038/sj.mp.4001764. [DOI] [PubMed] [Google Scholar]
- Haroutunian V, Katsel P, Dracheva S, Stewart DG, Davis KL. Variations in oligodendrocyte-related gene expression across multiple cortical regions: implications for the pathophysiology of schizophrenia. Int J Neuropsychopharmacol. 2007:1–9. doi: 10.1017/S1461145706007310. [DOI] [PubMed] [Google Scholar]
- Hasler G, Drevets WC, Gould TD, Gottesman II, Manji HK. Toward Constructing an Endophenotype Strategy for Bipolar Disorders. Biol Psychiatry. 2006 doi: 10.1016/j.biopsych.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Hayden EP, Nurnberger JI., Jr Molecular genetics of bipolar disorder. Genes Brain Behav. 2006;5:85–95. doi: 10.1111/j.1601-183X.2005.00138.x. [DOI] [PubMed] [Google Scholar]
- Kato T. Molecular genetics of bipolar disorder and depression. Psychiatry Clin Neurosci. 2007;61:3–19. doi: 10.1111/j.1440-1819.2007.01604.x. [DOI] [PubMed] [Google Scholar]
- Laeng P, Pitts RL, Lemire AL, Drabik CE, Weiner A, Tang H, et al. The mood stabilizer valproic acid stimulates GABA neurogenesis from rat forebrain stem cells. J Neurochem. 2004;91:238–251. doi: 10.1111/j.1471-4159.2004.02725.x. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, Jerome RE, et al. Towards understanding the schizophrenia code: An expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:129–158. doi: 10.1002/ajmg.b.30481. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wixey J, Harper CG, Dodd PR. Expression of MBP, PLP, MAG, CNP, and GFAP in the Human Alcoholic Brain. Alcohol Clin Exp Res. 2005;29:1698–1705. doi: 10.1097/01.alc.0000179406.98868.59. [DOI] [PubMed] [Google Scholar]
- Li A, Horvath S. Network neighborhood analysis with the multi-node topological overlap measure. Bioinformatics (Oxford, England) 2007;23:222–231. doi: 10.1093/bioinformatics/btl581. [DOI] [PubMed] [Google Scholar]
- Mansour HA, Wood J, Logue T, Chowdari KV, Dayal M, Kupfer DJ, et al. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 2006;5:150–157. doi: 10.1111/j.1601-183X.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland M, Le-Niculescu H, Balaraman Y, Ogden C, Patel S, Tan J, et al. Convergent Functional Genomics Studies. Mice Lacking The Circadian Clock Gene DBP: A Genetic Animal Model Of Bipolar Disorder 2007 Submitted. [Google Scholar]
- McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, et al. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility Loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77:582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Straub RE, Lipska BK, Verchinski BA, Goldberg T, Callicott JH, et al. Genetic evidence implicating DARPP-32 in human frontostriatal structure, function, and cognition. J Clin Invest. 2007;117:672–682. doi: 10.1172/JCI30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Pato CN, Gentile KL, McGann L, Brown AM, Trauzzi M, et al. Gene expression analysis of peripheral blood leukocytes from discordant sib-pairs with schizophrenia and bipolar disorder reveals points of convergence between genetic and functional genomic approaches. Am J Med Genet B Neuropsychiatr Genet. 2005;136:12–25. doi: 10.1002/ajmg.b.30171. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci U S A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller DJ, de Luca V, Sicard T, King N, Strauss J, Kennedy JL. Brain-derived neurotrophic factor (BDNF) gene and rapid-cycling bipolar disorder: family-based association study. Br J Psychiatry. 2006;189:317–323. doi: 10.1192/bjp.bp.105.010587. [DOI] [PubMed] [Google Scholar]
- Naydenov AV, Macdonald ML, Ongur D, Konradi C. Differences in lymphocyte electron transport gene expression levels between subjects with bipolar disorder and normal controls in response to glucose deprivation stress. Arch Gen Psychiatry. 2007;64:555–564. doi: 10.1001/archpsyc.64.5.555. [DOI] [PubMed] [Google Scholar]
- Niculescu AB., 3rd Polypharmacy in oligopopulations: what psychiatric genetics can teach biological psychiatry. Psychiatr Genet. 2006;16:241–244. doi: 10.1097/01.ypg.0000242195.74268.f9. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, 3rd, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR. Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics. 2000;4:83–91. doi: 10.1152/physiolgenomics.2000.4.1.83. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, Lulow LL, Ogden CA, Le-Niculescu H, Salomon DR, Schork NJ, et al. PhenoChipping of psychotic disorders: A novel approach for deconstructing and quantitating psychiatric phenotypes. Am J Med Genet B Neuropsychiatr Genet. 2006;141:653–662. doi: 10.1002/ajmg.b.30404. [DOI] [PubMed] [Google Scholar]
- Nieto MM, Guen SL, Kieffer BL, Roques BP, Noble F. Physiological control of emotion-related behaviors by endogenous enkephalins involves essentially the delta opioid receptors. Neuroscience. 2005;135:305–313. doi: 10.1016/j.neuroscience.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Nievergelt CM, Kripke DF, Barrett TB, Burg E, Remick RA, Sadovnick AD, et al. Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141:234–241. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, et al. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–1029. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bertsch BA, Strother WN, Le-Niculescu H, Balaraman Y, Hayden E, et al. Candidate genes, pathways and mechanisms for alcoholism: an expanded convergent functional genomics approach. Pharmacogenomics J. 2006 doi: 10.1038/sj.tpj.6500420. [DOI] [PubMed] [Google Scholar]
- Roybal K, Theobold D, Graham A, Dinieri JA, Russo SJ, Krishnan V, et al. From the Cover: Mania-like behavior induced by disruption of CLOCK. Proc Natl Acad Sci U S A. 2007;104:6406–6411. doi: 10.1073/pnas.0609625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE. Novel integrative genomics strategies to identify genes for complex traits. Animal genetics 37 Suppl. 2006;1:18–23. doi: 10.1111/j.1365-2052.2006.01473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segman RH, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev AY. Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry. 2005;10:500–513. 425. doi: 10.1038/sj.mp.4001636. [DOI] [PubMed] [Google Scholar]
- Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI, Jr, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: Bipolar disorder. Am J Hum Genet. 2003;73:49–62. doi: 10.1086/376547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltiel G, Shamir A, Levi I, Bersudsky Y, Agam G. Lymphocyte G-protein receptor kinase (GRK)3 mRNA levels in bipolar disorder. Int J Neuropsychopharmacol. 2006;9:761–766. doi: 10.1017/S146114570500636X. [DOI] [PubMed] [Google Scholar]
- Sokolov BP. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse. Comorbidity, shared traits, or molecular phenocopies? . Int J Neuropsychopharmacol. 2007:1–9. doi: 10.1017/S1461145706007322. [DOI] [PubMed] [Google Scholar]
- Sokolov BP, Polesskaya OO, Uhl GR. Mouse brain gene expression changes after acute and chronic amphetamine. J Neurochem. 2003;84:244–252. doi: 10.1046/j.1471-4159.2003.01523.x. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Bar JL, Stone WS, Faraone SV. Gene-environment interactions in mental disorders. World Psychiatry. 2004;3:73–83. [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, et al. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2005;133:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Ferran E, Galke B, Cooper K, Bunney WE, Byerley W. Microarray screening of lymphocyte gene expression differences in a multiplex schizophrenia pedigree. Schizophr Res. 2004;67:41–52. doi: 10.1016/s0920-9964(03)00151-8. [DOI] [PubMed] [Google Scholar]
- Volkow N, Li TK. The neuroscience of addiction. Nat Neurosci. 2005;8:1429–1430. doi: 10.1038/nn1105-1429. [DOI] [PubMed] [Google Scholar]
- Zapala MA, Schork NJ. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci U S A. 2006;103:19430–19435. doi: 10.1073/pnas.0609333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wiener MC, Zhang C, Fridman A, Minch E, Lum PY, et al. Increasing the Power to Detect Causal Associations by Combining Genotypic and Expression Data in Segregating Populations. PLoS Comput Biol. 2007;3:e69. doi: 10.1371/journal.pcbi.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]