Abstract
Important cellular processes such as inflammation, apoptosis, differentiation, and proliferation confer critical roles in the pathogenesis of human diseases. In the past decade, an emerging process named “autophagy” has generated intense interest in both biomedical research and clinical medicine. Autophagy is a regulated cellular pathway for the turnover of organelles and proteins by lysosomal-dependent processing. Although autophagy was once considered a bulk degradation event, research shows that autophagy selectively degrades specific proteins, organelles, and invading bacteria, a process termed “selective autophagy.” It is increasingly clear that autophagy is directly relevant to clinical disease, including pulmonary disease. This review outlines the principal components of the autophagic process and discusses the importance of autophagy and autophagic proteins in pulmonary diseases from COPD, α1-antitrypsin deficiency, pulmonary hypertension, acute lung injury, and cystic fibrosis to respiratory infection and sepsis. Finally, we examine the dual nature of autophagy in the lung, which has both protective and deleterious effects resulting from adaptive and maladaptive responses, and the challenge this duality poses for designing autophagy-based diagnostic and therapeutic _targets in lung disease.
Autophagy is an evolutionarily conserved lysosomal degradation pathway.1 Often referred to as merely autophagy, macroautophagy is the best-characterized form of autophagy and involves the engulfment of cytoplasmic contents and organelles through a complex reorganization of subcellular membranes to form a new organelle: the autophagosome. The autophagosome then fuses with and delivers its contents to the lysosome (Fig 1A). Lysosomal enzymes subsequently facilitate a degradation process to regenerate metabolic precursor molecules (ie, amino acids, fatty acids) that can be used for anabolic pathways and adenosine-5′-triphosphate production.
Figure 1.
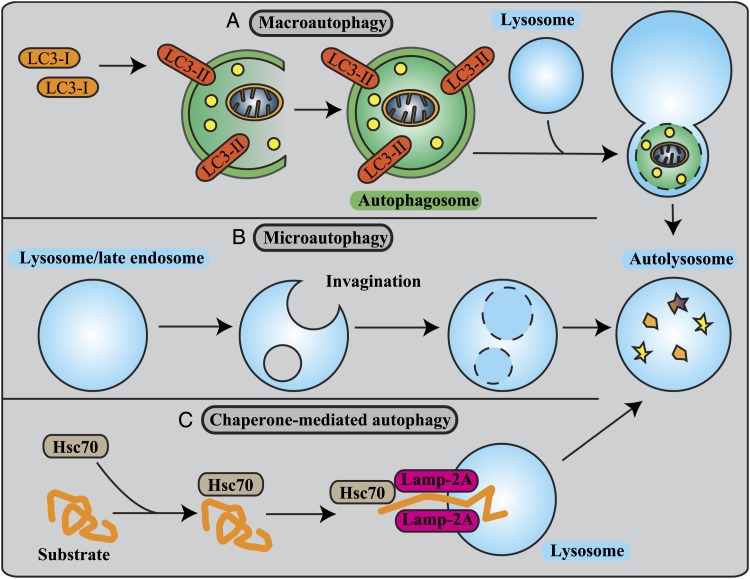
Three different types of autophagy. A, Macroautophagy is a dynamic process involving the rearrangement of subcellular membranes to sequester cytoplasm and organelles for delivery to the lysosome or vacuole where the sequestered cargo is degraded and recycled. LC3 is known to exist on autophagosomes, and hence, LC3 is a widely used marker for autophagosomes. B, In microautophagy, the lysosomal membrane is invaginated to sequester cytosolic constituents into intralysosomal vesicles. C, In chaperone-mediated autophagy, proteins containing the KFERQ motif bind to Hsc70 and cochaperones. These complexes are translocated across the lysosomal membrane after binding with Lamp-2A. Hsc70 = heat shock cognate 70; Lamp-2A = lysosome-associated membrane protein type 2a; LC3 = microtubule-associated protein light 1 chain 3.
Autophagy has been shown to be both protective and injurious in a variety of different models, suggesting that its role in human diseases is complex. Until recently, autophagy was merely deemed a nonspecific homeostatic cellular process; however, mounting evidence suggests that a process termed “selective autophagy” is involved in the delivery of a wide range of autophagic cargo from protein aggregates to whole organelles, such as mitochondria, and even intracellular microbes to the lysosome. In this review, we examine the considerable evidence emerging for the role of autophagy and selective autophagy in cell survival, cell death, and immune and inflammatory responses related to the pathogenesis of complex lung diseases (Table 1). We also discuss the conundrums behind the design of therapeutic agents that either promote autophagy in situations where autophagy is beneficial or that dampen down the autophagic response in situations where autophagy is injurious. A better understanding for the role of this double-edged-sword hypothesis of autophagy in pulmonary diseases will help in the design of personalized therapies for the treatment of pulmonary diseases.
Table 1.
—Pathologic Roles of Autophagy in Pulmonary Diseases
| Author/Year | Diseases | Role of Autophagy |
| Chen et al2,3/2008, 2010 | COPD | Epithelial cell: regulation of cell death |
| Monick et al4/2010 | Macrophage: regulation of infection | |
| Chen et al2/2008 | α1-antitrypsin deficiency | Selective degradation/clearance of aggregated α1-antitrypsin protein |
| Granell et al5/2008 | ||
| Hidvegi et al6/2010 | ||
| Kamimoto et al7/2006 | ||
| Lee et al8/2010 | Pulmonary hypertension | Adult: regulation of proliferation |
| Teng et al9/2012 | Fetal: regulation of angiogenesis | |
| Lee et al10/2011 | Acute lung injury | Regulation of cell death |
| Tanaka et al11/2011 | ||
| Knævelsrud and Simonsen12/2010 | Cystic fibrosis | Clearance of aggregated prone proteins |
| Luciani et al13/2010 | ||
| Intemann et al14/2009 | Respiratory mycobacterial infection | Elimination of mycobacteria |
| Kim et al15/2012 | ||
| Kumar et al16/2010 | ||
| Lam et al17/2012 | ||
| Renna et al18/2011 | ||
| Singh et al19/2006 | ||
| Nakahira et al20/2010 | Sepsis | Regulation of the cytokine production |
| Zhou et al21/2010 |
Macroautophagy, Microautophagy, and Chaperone-Mediated Autophagy
In addition to macroautophagy, other forms of autophagy exist and include microautophagy, which involves the direct invagination of the lysosomal membrane to sequester cytoplasm, and chaperone-mediated autophagy, a dynamic process involving the selective delivery of proteins containing a specific consensus sequence (KFERQ) to the lysosome (Figs 1B, 1C).1 Of the three autophagic pathways, macroautophagy has received the most attention partly because of the easy detection and visualization of autophagosomes, which are relatively large structures that can be visualized using fluorescence or electron microscopy. In the 1990s, genetic studies in yeast identified a series of autophagy-related (Atg) genes shown to be involved in the macroautophagy process.22,23 Among these, Beclin 1 (the mammalian homolog of yeast Atg6) represents a major autophagic regulator.24 Beclin 1 associates with a macromolecular complex that includes the class 3 phosphatidylinositol 3-kinase (Vps34). The Beclin 1 complex produces phosphatidylinositol 3-phosphate, a second messenger that regulates autophagosomal nucleation.25
Microtubule-associated protein light chain (LC3), a homolog of yeast Atg8 is one of the most well-known Atg proteins and is frequently used as a specific marker to monitor macroautophagy in vitro and in vivo.26 The C-terminal fragment of LC3 is cleaved immediately following its synthesis to yield a cytosolic form called LC3-I. Upon the activation of macroautophagy, LC3-II is then conjugated to phosphatidylethanolamine and _targeted to autophagic membranes (Fig 1A).26 Measurement of the conversion of LC3B-I to LC3B-II by western blot is a reliable method to assess whether autophagy is activated in vitro or in vivo. However, although LC3-II expression levels can correlate with autophagosome number, the amount of LC3B-II at a given point in time may not be a true indication of cellular autophagic activity. Increases in LC3C-II may indicate the induction of autophagosome formation or the suppression of autophagosome degradation.27 To distinguish between autophagosome accumulation due to enhanced or inhibited autophagic degradation, autophagic flux assays must be implemented in which substrate flux in cell culture systems can be assessed in the absence and presence of inhibitors of autophagy degradation, such as chloroquine to prevent lysosome acidification; leupeptin to prevent cathepsin B activation; or bafilomycin A, an inhibitor of lysosomal adenosine-5′-triphosphate.27 Our laboratory has also established a reliable method to assess autophagic flux in the lung in vivo by using similar approaches with in vitro autophagic flux assays.28
Autophagy, Cell Survival, and Cell Death
Although basal macroautophagy (herein referred to as autophagy) is generally considered to be protective when it is induced in response to stress, it has also been implicated in cell death.29,30 Autophagy is rarely considered a suicidal mechanism, yet the term “autophagic cell death” has been widely used to indicate instances of cell death that are associated with excessive cytoplasmic vacuolization.31 Autophagic cell death is not considered a form of programmed cell death in its own right; rather, it is seen as cell death with autophagy rather than cell death by autophagy.32 Apoptosis can occur at the same time as autophagy, suggesting a common regulatory mechanism,33-35 and several proapoptotic signaling molecules are known to induce autophagy, including tumor necrosis factor,36 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL),37 Fas-associated protein with death domain (FADD),38 dynamin-related protein-1 (DRP-1), and death-associated protein kinase (DAPK).39 The B-cell lymphoma 2 (BCL-2) proteins are also known to be important in both autophagy and apoptosis signaling.40 Beclin 1 has been shown to interact with BCL-2, resulting in the inhibition of Beclin 1-mediated autophagy in response to starvation,41 and a truncated form of Atg5 (cleaved by calpain 1 and 2) participates in apoptosis regulation and translocates from the cytosol to mitochondria to trigger cytochrome c release and caspase activation.40 Atg3 has also been shown to be activated by caspase-8 cleavage, demonstrating the crosstalk between autophagy and the extrinsic pathway of apoptosis.42
The crosstalk among many cellular processes, including apoptosis, autophagy, translation, energy metabolism, and inflammation, is also controlled by the mammalian _target of rapamycin (mTOR), which is an evolutionarily conserved serine-threonine kinase that acts as a sensor of environmental and cellular nutrition and energy status. A plethora of mitogens, growth factors, and nutrients stimulate the activation of the two mTOR complexes mTORC1 and mTORC2 to regulate diverse functions, such as cell growth, proliferation, development, autophagy, and innate as well as adaptive immune responses. mTORC1 actively suppresses autophagy, and conversely, inhibition of mTORC1 strongly induces autophagy. Atg13 and ULK1 (uncoordinated family member [unc]-51-like kinase) bind to the FIP200 (200-kDa FAK family kinase-interacting protein), a putative ortholog of Atg17, and the mammalian-specific component Atg101. mTOR phosphorylates Atg13 and ULK1 to block autophagosome initiation.43
Selective Autophagy
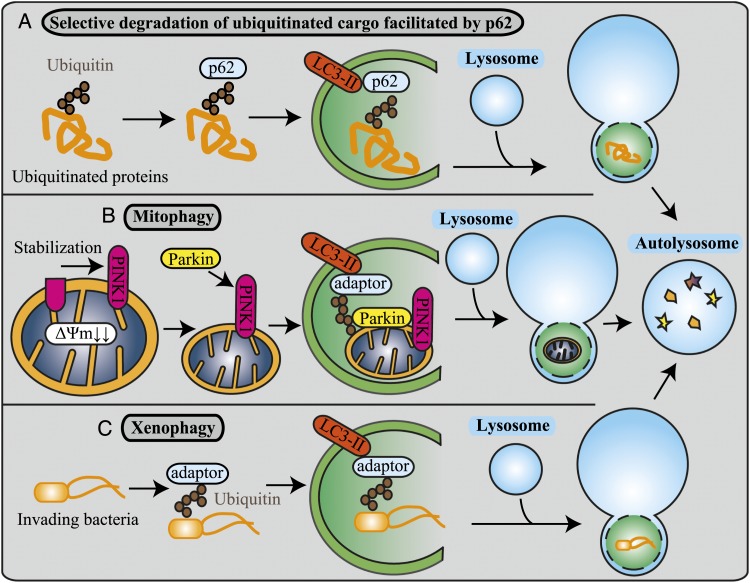
Selective autophagy is involved in the delivery of a wide range of autophagic cargo from protein aggregates to whole organelles and even intracellular microbes to the lysosome for degradation. It is believed that ubiquitin-positive substrates, such as protein aggregates (inclusion bodies), mitochondria, and invading bacteria, not dealt with by the proteasome system are selectively degraded by autophagy.44 Ubiquitin can be conjugated as a monomer on one (monoubiquitination) or more substrate lysines (multiubiquitination) or as a polymer (polyubiquitination) by the sequential addition of further ubiquitins to one another through ubiquitin lysines. Monoubiquitination can regulate DNA repair, viral budding, and gene expression, whereas polyubiquitination generally results in proteasomal degradation and endocytosis.45 Ubiquitination seems to function as a general tag for selective autophagy in mammalian cells.46 Autophagy receptors such as p62 and NBR1 (neighbor of BRCA1 gene 1), that simultaneously bind both monoubiquitinated and polyubiquitinated substrates,47 act as adaptors between ubiquitination and autophagy. An examination of the comprehensive list of selective autophagic processes currently in the literature is beyond the scope of this review; therefore, we focus on the three most documented types of selective autophagy: selective degradation of ubiquitinated cargo facilitated by p62 (disposal of ubiquitinated protein) (Fig 2A), mitophagy (disposal of mitochondria) (Fig 2B), and xenophagy (disposal of intracellular bacteria and viruses) (Fig 2C).
Figure 2.
Selective autophagy. A, In the selective degradation of ubiquitinated cargo facilitated by p62, the p62 substrate interacts with monoubiquitinated or polyubiquitinated proteins through its ubiquitin-associated domain. This complex is selectively sequestered into autophagosomes. B, Mitophagy is the specific autophagic elimination of mitochondria. In response to the loss of the membrane potential, PINK1 accumulates, and parkin translocates to damaged mitochondria. Parkin ubiquitinates outer membrane proteins and induces mitophagy. C, In xenophagy, invading extracellular pathogens are recognized and captured by the autophagic machinery through ubiquitin-dependent and ubiquitin-independent mechanisms. Adaptors mediate autophagic sequestration of the pathogens to restrict their growth. PINK1 = PTEN (phosphatase and tensin homolog)-induced putative kinase 1. See Figure 1 legend for expansion of other abbreviation.
One of the best-characterized substrates of selective autophagy is p62 (Fig 2A). p62 is often used as a general indicator of macroautophagy and autophagic flux and plays an important role in the detection of cytoplasmic proteinaceous inclusion bodies. It is also a hallmark of conformational diseases, such as Alzheimer disease, Parkinson disease, and various chronic liver disorders.46 It has an inherent ability to polymerize or aggregate as well as to specifically recognize substrates.46 The p62 substrate participates in direct protein-protein interactions with LC3 on the isolation membrane and with ubiquitinated proteins through its ubiquitin-associated domain, facilitating the sequestration of ubiquitinated proteins into the autophagosome.1
The selective removal of mitochondria by mitophagy is a process that regulates mitochondrial number to match metabolic demand and to perform quality control to remove damaged mitochondria from the cell (Fig 2B). Loss of the two mitophagy regulatory genes PINK1 (PTEN-induced putative kinase 1) and PARK2 results in progressive mitochondrial damage and dysfunction, which may cause Parkinson disease.48 Damaged depolarized mitochondria stabilize PINK1, which in turn recruits the E3 ubiquitin ligase parkin. Parkin then ubiquitinates various mitochondrial outer membrane proteins (the precise substrate of parkin, which is essential for mitophagy, is still unknown1) and induces mitophagy.48
During infection, autophagy assists in the immune response by providing a mechanism for the selective intracellular degradation of invading pathogens, a process termed “xenophagy” (Fig 2C).49 Invading bacteria are ubiquitinated, and adaptors, including p62, OPTN (optineurin),50 NBR1,51 and NDP52 (nuclear dot protein 52 kDa),52 mediate their autophagic sequestration.25 Defects in xenophagy have been associated with a number of inflammatory disorders, including Crohn’s disease.53 Crohn’s disease is associated with defective xenophagy, where miR-196 (microRNA 196), which is overexpressed in the inflammatory intestinal epithelia of individuals with Crohn’s disease, controls the level of the human immunity-related GTPase M (IRGM), which is critical for both the initiation and the maturation of xenophagy.53 Interestingly, xenophagy may also play a role in host defenses not only by removing intracellular pathogens but also by enhancing immune recognition of infected cells through the generation of antigenic bacterial peptides.54
Autophagy in COPD
COPD remains a major public health problem and is the fourth leading cause of death worldwide.55,56 COPD is characterized by airflow limitation that is not fully reversible and by pathologic changes found in the proximal and peripheral airways, lung parenchyma, and pulmonary vasculature.56,57 Cigarette smoke is the most common identifiable risk factor for COPD, with smokers being known to have a greater COPD mortality rate than nonsmokers.56
We demonstrated a pivotal functional role for autophagic proteins in cigarette smoke-induced emphysema.3 We observed increased autophagosomes and increased expression of LC3B-II in human lung specimens from patients with COPD as well as the increased expression of additional autophagy-associated proteins Atg4, Atg5-Atg12 complex, and Atg7.2 Genetic depletion of the essential autophagy mediators LC3B and Beclin 1 suppressed cigarette smoke extract-induced cell death, and emphysematous airspace enlargement in LC3B-deficient mice was reduced compared with wild-type (WT) mice.3 These data suggest that the stimulation of the autophagic pathway may promote cell death during cigarette smoke exposure. We posit that the observed accumulation in autophagosomes in patients with COPD may be a result of defective autophagic flux. Studies are under way to investigate the effect of cigarette smoke on autophagy activity in lung epithelial cells in vitro and in vivo. In addition, it remains to be determined whether autophagy directly contributes to the alveolar epithelial cell death in COPD or whether other pathways are responsible. Previous studies have also observed defective autophagy in cigarette smoke-exposed macrophages.4 Such a deficit in autophagy was identified in the alveolar macrophages of smokers and was suggested to lead to recurrent infections in persons who smoke because cigarette smoke exposure impairs delivery of bacteria to lysosomes (xenophagy).4
Interestingly, mTOR signaling has also been linked to cigarette-induced COPD and emphysema. RTP801, a DNA damage response gene upregulated in response to cigarette smoke exposure, has been shown to stabilize the assembly of the mTOR inhibitory complex TSC1-TSC2, resulting in the exacerbation of oxidative stress-induced cell death. RTP801 negatively regulates mTOR by dissociating the inhibitory protein 14-3-3 from TSC2, allowing the TSC1-TSC2 complex to block mTOR activity.58 Blocking mTOR may consequently affect autophagy activity. In this study, the mTOR inhibitor rapamycin was protective in WT mice exposed to smoke, reducing alveolar inflammation. On the contrary, basally, rapamycin heightened the number of apoptotic and inflammatory cells in room air control WT mice and abrogated the protective effects of RTP801 knockout in mice exposed to cigarette smoke. These studies highlight that the timing and lung cell _targets of mTOR inhibition might therefore be essential to define its beneficial and pathologic roles in disease.58
Until now, the role of autophagy in COPD has focused on the role of classic nonselective autophagy; however, these data suggest that selective autophagy, such as mitophagy or xenophagy, may play a more important role in this disease. Therapeutic agents that could be beneficial in treating COPD may be those that enhance autophagic clearance and turnover, preventing the accumulation of autophagosomes or agents that affect mTOR signaling. In addition, recently developed chemical chaperones may be used as therapeutic agents to preserve protein function despite the oxidative environment in COPD lungs.59-62 A better understanding of the balance between the cytoprotective and prodeath functions of autophagic proteins in response to cigarette smoke will be required for the therapeutic _targeting of this process in COPD.
Autophagy in α1-Antitrypsin Deficiency
Evidence suggests that selective autophagy contributes to the clearance of aggregated α1-antitrypsin (AAT), a protein that has been implicated in early onset emphysema from the endoplasmic reticulum (ER).63 The most common pathogenic mutation in the AAT gene, termed the “PiZ mutation,” leads to misfolding and retention of AAT aggregates in the ER, which result in the reduced export of the AAT protein from hepatocytes into the bloodstream, leading to unopposed protease activity in the lung.64 Evidence from our group demonstrated that lung biopsy specimens obtained from patients with AAT deficiency have increased ratios of LC3B-II/LC3B-I, suggesting that the regulation of autophagy in the lungs of these patients may be altered.2 In addition, PiZ protein aggregates were shown to colocalize with transfected green fluorescent protein-LC3B protein, and degradation of AAT was impaired by genetic or chemical inhibition of autophagy in COS7 cells (African Green Monkey Cercopithecus aethiops fibroblast-like kidney cells).7 Conversely, polymerized mutant AAT is found concentrated in inclusion bodies that are associated with the ER, but these structures do not colocalize with the autophagosome marker LC3B.5 However, Hidvegi et al6 reported that administering high doses of carbamazepine, a drug known to stimulate autophagic activity, reversed the liver pathology observed in AAT PiZ overexpressing transgenic mice. The implication of these results suggest that the autophagy pathway might be exploited to arrest or even reverse the disease phenotype associated with mutant AAT protein, at least in the liver. Moreover, mutant PiZ-AAT protein aggregates can be detected in alveolar macrophages as well as in BAL fluid and lung biopsy specimens of patients with AAT deficiency with emphysema.65,66 It is tempting to speculate that the clearance of these aggregates by autophagy within alveolar macrophages might be impaired, particularly in patients who smoke. It is anticipated that continued investigations into the role of autophagy in AAT deficiency may lead to a better understanding of emphysema development in this disorder and perhaps improved therapy.
Autophagy in Pulmonary Hypertension
Pulmonary arterial hypertension is a sustained elevation of pulmonary arterial pressure to >25 mm Hg at rest or to >30 mm Hg with exercise, with a mean pulmonary capillary wedge pressure and left ventricular end-diastolic pressure of <15 mm Hg.67 Hypoxia results in secondary pulmonary hypertension (PH), and hypoxic PH is a progressive and often fatal complication of chronic lung disease.68
We have shown the elevated occurrence of autophagy in lung tissue from patients with PH.8 To determine the involvement of autophagic proteins in PH, we subjected mice genetically deficient in LC3B to chronic hypoxia and measured indices of PH. Hypoxic lung tissues showed an increase in autophagic vacuoles compared with lung tissue from normoxic mice. After chronic hypoxia, LC3B−/− mice demonstrated increased indices of PH, right ventricular systolic pressure, and Fulton index relative to WT mice.8 These results suggest that the autophagic protein LC3B may exert a protective function during the pathogenesis of PH.
On the other hand, previous studies have reported a role for autophagy in the impaired angiogenesis observed in developing lungs.9 Although pulmonary artery endothelial cells from fetal lambs with persistent PH showed impaired angiogenesis, inhibitors of autophagy and knockdown of the autophagic protein Beclin 1 (Atg6) improved angiogenesis. These data indicate that in developing lungs, autophagy might contribute to the disease progression of PH. From a clinical perspective, the period of intervention might therefore be important for the treatment of PH. Further studies are needed to clarify the functional relevance of autophagy and how it contributes to the pathogenesis of PH.
Autophagy in Acute Lung Injury
Acute lung injury (ALI) is the clinical syndrome of rapid-onset bilateral pulmonary infiltrates and hypoxemia of noncardiac origin.69 ALI is associated with sepsis, hyperoxia, trauma, pharmaceutical or xenobiotic exposure, and mechanical ventilation. To manage patients with severe respiratory failure, mechanical ventilation with high concentrations of oxygen are required. However, prolonged exposure to hyperoxia can result in lung injury. We have reported that LC3B regulates hyperoxia-induced apoptotic pathways in lung epithelial cells.11 Hyperoxia induces autophagosome formation and conversion of LC3B-I to the active LC3B-II form. Cell viability after hyperoxic conditions was lower in LC3B siRNA transfected cells compared with control siRNA. In addition, LC3B was found to interact with the Fas death-inducing signaling complex-dependent apoptotic pathway under hyperoxia. These data suggest that LC3B plays a protective role against hyperoxia-induced cell death by potentially interacting with and inhibiting apoptotic pathways.11 Other autophagic proteins may also have dynamic interactions with the Fas-dependent pathway. Specifically, the interaction of p62, LC3B, and the caspase-8 substrate Atg342 may provide further complexity in the mechanism by which LC3B regulates Fas.
We have suggested a novel therapeutic intervention using low concentrations of carbon monoxide (CO) in a model of hyperoxic stress.10 CO increased the expression of LC3B in mouse lung and in cultured human alveolar or human bronchial epithelial cells as well as increased autophagosome formation in epithelial cells as seen by electron microscopy and green fluorescent protein-LC3 puncta assays. This increase in autophagy activity was inhibited by N-acetyl-l-cysteine and the mitochondria-_targeting antioxidant Mito-TEMPO, suggesting that CO promotes the autophagic process through mitochondrial reactive oxygen species (ROS) generation.10 The protection afforded by CO against hyperoxia-induced cell death was also compromised on treatment with siRNA against LC3B. These findings suggest that LC3B plays an important role in the cytoprotection afforded by CO through the production of mitochondrial ROS. These studies lend further support to the concept that autophagic proteins, such as LC3B, participate in signaling networks that maintain a dynamic equilibrium, which determines the fate of cells under stress. Thus, autophagic proteins may be selected as potential therapeutic _targets to modulate ALI to hyperoxia. A further understanding of these processes must be obtained before _targeting the autophagic pathway in human disease.70
Autophagy in Cystic Fibrosis
Cystic fibrosis (CF) is the most common fatal autosomal recessive disease among white populations and is caused by a mutation in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR). CF is characterized by abnormally viscid mucous that obstructs organ passages, resulting in recurrent pulmonary infections. It is caused by a mutation in a single large gene on chromosome 7, with the most common CFTR mutation being a deletion of phenylalanine at position 508 (CFTRF508del).71-73 Cells with CFTRF508del display accumulated polyubiquitinated proteins, defective autophagy, and decreased clearance of aggresomes.74 Defective CFTR also results in increased ROS production and upregulation of tissue transglutaminase (TG2), an important factor of the inflammatory response in CF.13 TG2 results in the crosslinking and inactivation of Beclin 1, leading to sequestration of PI3K (phosphoinositide 3-kinase) complex III and accumulation of p62, which regulates aggresome formation.13 Restoring Beclin 1 or knocking down p62 rescues the trafficking of CFTRF508del to the cell surface. In addition to these data linking defective CFTR to autophagy deficiency, the autophagy-stimulating agent rapamycin markedly decreases Burkholderia cenocepacia infection in the lungs of CF mice and drastically reduces signs of lung inflammation. Infection by B cenocepacia is a particularly lethal threat to patients with CF because it causes severe and persistent lung inflammation and is resistant to nearly all available antibiotics.75 On the other hand, azithromycin, a potent macrolide antibiotic used in patients with CF, is associated with increased infection with nontuberculous mycobacteria. Interestingly, azithromycin has been shown to block autophagosome clearance by preventing lysosomal acidification, impairing autophagic and phagosomal degradation in primary human macrophages and thereby inhibiting the intracellular killing of mycobacteria within macrophages in vivo.18 These studies highlight the role of maintaining regulated autophagy activity in patients with CF. If efficiently activated, autophagy can aid in the removal of polyubiquitinated proteins and control infection. However, inadvertent genetic or pharmacologic blockade of autophagy puts patients at risk for the accumulation of protein inclusions and aggresomes and infection from drug-resistant pathogens. Therapies that restore autophagy or enhance autophagosome clearance or activity offer great therapeutic value in the treatment of CF.
Autophagy in Respiratory Infection and Sepsis
As previously mentioned, xenophagy has been shown to be disrupted in lung alveolar macrophages exposed to cigarette smoke and may be disrupted in patients with COPD.4 Autophagy also plays other important roles in the response of the lung to bacterial, viral, and protozoan infections25; however, to provide a comprehensive summation of the role of autophagy in each of these respiratory infections is beyond the scope of this review. The role for autophagy in respiratory mycobacterial infection, including Mycobacterium tuberculosis (MTb) is well known.25 In particular, polymorphisms in the IRGM gene are linked to increased susceptibility to MTb infection, and IRGM has been shown to induce autophagy to eliminate intracellular mycobacteria,14,19 A genome-wide analysis of host genes that regulate survival of MTb identified a number of genes involved in the regulation of autophagy,16 and autophagy-defective, Atg7 mutant Drosophila infected with Mycobacterium marinum could not be rescued by antimycobacterial treatment, exhibiting decreased survival rates. Moreover, activation of autophagy by antibiotic (isoniazid) treatment dampened MTb-induced proinflammatory responses in macrophages.15 A number of therapies that involve enhancing autophagy activity have been shown to be effective against MTb infection. The antiprotozoal drug nitazoxanide and its active metabolite tizoxanide strongly stimulate autophagy and inhibit mTORC1 signaling and intracellular proliferation of MTb,17 and vitamin D has shown therapeutic benefits in persons with HIV and MTb infection through the activation of autophagy.76 After HIV, TB is the leading infectious cause of death worldwide. Interestingly, HIV also manipulates autophagy to improve viral infectivity,77 and a common _target of RNA viruses is IRGM.77
Together, these findings underscore the importance of host autophagy in orchestrating successful antimicrobial responses to mycobacteria. Reports have identified cases of totally drug-resistant TB in India with fears that totally drug-resistant TB will spread worldwide.78 New therapeutic agents that have different mechanisms of action from conventional anti-TB drugs are needed to prevent the development of drug resistance. Although agents that promote autophagy may improve the efficacy of conventional anti-TB drugs as an adjunctive treatment, some of these drugs have undesirable immunosuppressive effects (eg, rapamycin). Other candidates with better side effect profiles, such as metformin or carbamazepin, may be beneficial.6,79
Finally, autophagy has been implicated in the regulation of inflammation, particularly regulation of the inflammasome pathway.21 Along with Zhou et al,21 we have identified a role for autophagy in NALP3 (NOD-like receptor 3)-dependent inflammation, where autophagy is believed to preserve mitochondrial integrity and segregate damaged ROS-producing mitochondria away from inflammasome complexes.20 In this study, we identified a role for autophagy in severe sepsis using endotoxemic mice, which are commonly used as a model of septic shock, and cecal ligation and puncture, a more clinically relevant polymicrobial sepsis model.20 Mice genetically deficient in LC3B were found to be more susceptible to both lipopolysaccharide (LPS)- and CLP-induced lethality than control mice. Serum concentrations of IL-1β and IL-18 were significantly higher in endotoxemic LC3B−/− mice than in their WT littermates. The data suggest that autophagy may regulate the cytokine production commonly associated with severe sepsis. More importantly, the findings are consistent with the role of IL-1β and IL-18 in patients with sepsis in the medical ICU.20,80 Again, these findings underscore the importance of host autophagy in orchestrating successful cytokine responses to infection. Agents that enhance autophagy activity in vivo may have beneficial effects for treating sepsis. Hepcidin has been shown to protect against LPS-induced liver injury through autophagy in a mouse model of obstructive jaundice.81 Low doses of CO have been shown to prevent LPS-induced organ damage most likely through the ability of CO to activate autophagy,82 and ES-62, an immunomodulator secreted by the filarial nematode Acanthocheilonema viteae, protected mice against endotoxic and polymicrobial septic shock by Toll-like receptor 4-mediated induction of autophagy.83 Given that the treatments for septic shock at present are inadequate, the autophagy-dependent mechanism of action by new and existing therapies might form the basis for urgently needed therapeutic intervention against this life-threatening condition.
Conclusions
Accumulating evidence demonstrates that autophagy plays a previously unforeseen role in the pathogenesis of pulmonary diseases. It is now clear that autophagy plays a complex role in the lung where it can have both protective and injurious effects on the progression of lung disease. When autophagy is activated correctly and there is proper turnover of autophagosomes to the lysosomes (good flux), autophagy promotes the survival of the cell. Autophagy activity is responsible for the clearance of injurious protein aggregates as well as for promoting bacterial and viral clearance by xenophagy and removing damaged organelles, such as mitochondria, by mitophagy. On the other hand, if autophagosome turnover or flux is defective, autophagy becomes injurious to the cell; protein- and infection-laden autophagosomes accumulate inside the cell and may promote cell death pathways. The precise mechanisms by which autophagy directly promotes apoptotic cell death or autophagic cell death is not clearly understood. Identification of cellular and molecular _targets of the autophagy machinery to promote efficient autophagosome formation and clearance of cellular debris or damaged proteins or organelles will be invaluable not only for a wide range of pulmonary diseases but also for a plethora of other human disorders and diseases. Further research into other autophagic pathways in the lung, such as selective autophagy, chaperone-mediated autophagy, and microautophagy, will also allow for new therapeutic interventions.
Acknowledgments
Financial/nonfinancial disclosures: The authors have reported to CHEST that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or in the preparation of the manuscript.
Abbreviations
- AAT
α1-antitrypsin
- ALI
acute lung injury
- Atg
autophagy related
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- CLP
cecal ligation and puncture
- CO
carbon monoxide
- ER
endoplasmic reticulum
- IRGM
immunity-related GTPase M
- LC3
microtubule-associated protein light 1 chain 3
- LPS
lipopolysaccharide
- MTb
Mycobacterium tuberculosis
- mTOR
mammalian _target of rapamycin
- PH
pulmonary hypertension
- ROS
reactive oxygen species
- TG2
tissue transglutaminase 2
- WT
wild type
Footnotes
Funding/Support: The research reviewed here was supported by grants from the National Institutes of Health [R01-HL060234, R01-HL055330, 2R01-HL079904] and a Flight Attendant Medical Research Institute Clinical Innovator Award to Dr Choi.
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians. See online for more details.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728-741 [DOI] [PubMed] [Google Scholar]
- 2.Chen ZH, Kim HP, Sciurba FC, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS ONE. 2008;3(10):e3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen ZH, Lam HC, Jin Y, et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A. 2010;107(44):18880-18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monick MM, Powers LS, Walters K, et al. Identification of an autophagy defect in smokers’ alveolar macrophages. J Immunol. 2010;185(9):5425-5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granell S, Baldini G, Mohammad S, et al. Sequestration of mutated alpha1-antitrypsin into inclusion bodies is a cell-protective mechanism to maintain endoplasmic reticulum function. Mol Biol Cell. 2008;19(2):572-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidvegi T, Ewing M, Hale P, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329(5988):229-232 [DOI] [PubMed] [Google Scholar]
- 7.Kamimoto T, Shoji S, Hidvegi T, et al. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281(7):4467-4476 [DOI] [PubMed] [Google Scholar]
- 8.Lee SJ, Smith A, Guo L, et al. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am J Respir Crit Care Med. 2010;183(5):649-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teng RJ, Du J, Welak S, et al. Cross-talk between NADPH oxidase and autophagy in pulmonary artery endothelial cells with intrauterine persistent pulmonary hypertension. 2012;302(7):L651-L663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SJ, Ryter SW, Xu JF, et al. Carbon monoxide activates autophagy via mitochondrial reactive oxygen species formation. Am J Respir Cell Mol Biol. 2011;45(4):867-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka A, Jin Y, Lee SJ, et al. Hyperoxia induced LC3B interacts with the Fas apoptotic pathway in epithelial cell death. Am J Respir Cell Mol Biol. 2011;46(4):507-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knævelsrud H, Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584(12):2635-2645 [DOI] [PubMed] [Google Scholar]
- 13.Luciani A, Villella VR, Esposito S, et al. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat Cell Biol. 2010;12(9):863-875 [DOI] [PubMed] [Google Scholar]
- 14.Intemann CD, Thye T, Niemann S, et al. Autophagy gene variant IRGM -261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5(9):e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JJ, Lee HM, Shin DM, et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe. 2012;11(5):457-468 [DOI] [PubMed] [Google Scholar]
- 16.Kumar D, Nath L, Kamal MA, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140(5):731-743 [DOI] [PubMed] [Google Scholar]
- 17.Lam KK, Zheng X, Forestieri R, et al. Nitazoxanide stimulates autophagy and inhibits mTORC1 signaling and intracellular proliferation of Mycobacterium tuberculosis. PLoS Pathog. 2012;8(5):e1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renna M, Schaffner C, Brown K, et al. Azithromycin blocks autophagy and may predispose cystic fibrosis patients to mycobacterial infection. J Clin Invest. 2011;121(9):3554-3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313(5792):1438-1441 [DOI] [PubMed] [Google Scholar]
- 20.Nakahira K, Haspel JA, Rathinam VAK, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2010;12(3):222-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2010;469(7329):221-225 [DOI] [PubMed] [Google Scholar]
- 22.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1-2):169-174 [DOI] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Cregg JM, Dunn WA, Jr, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539-545 [DOI] [PubMed] [Google Scholar]
- 24.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by Beclin 1. Nature. 1999;402(6762):672-676 [DOI] [PubMed] [Google Scholar]
- 25.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117(pt 13):2805-2812 [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313-326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haspel J, Shaik RS, Ifedigbo E, et al. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy. 2011;7(6):629-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu S, Kanaseki T, Mizushima N, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6(12):1221-1228 [DOI] [PubMed] [Google Scholar]
- 30.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131(6):1137-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galluzzi L, Vitale I, Abrams JM, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19(1):107-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9(12):1004-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4(5):600-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorski SM, Chittaranjan S, Pleasance ED, et al. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol. 2003;13(4):358-363 [DOI] [PubMed] [Google Scholar]
- 35.Jin Z, Li Y, Pitti R, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137(4):721-735 [DOI] [PubMed] [Google Scholar]
- 36.Jia L, Dourmashkin RR, Allen PD, Gray AB, Newland AC, Kelsey SM. Inhibition of autophagy abrogates tumour necrosis factor alpha induced apoptosis in human T-lymphoblastic leukaemic cells. Br J Haematol. 1997;98(3):673-685 [DOI] [PubMed] [Google Scholar]
- 37.Herrero-Martín G, Høyer-Hansen M, García-García C, et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28(6):677-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyo JO, Jang MH, Kwon YK, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280(21):20722-20729 [DOI] [PubMed] [Google Scholar]
- 39.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157(3):455-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou F, Yang Y, Xing D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011;278(3):403-413 [DOI] [PubMed] [Google Scholar]
- 41.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927-939 [DOI] [PubMed] [Google Scholar]
- 42.Oral O, Oz-Arslan D, Itah Z, et al. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis. 2012;17(8):810-820 [DOI] [PubMed] [Google Scholar]
- 43.Weichhart T. Mammalian _target of rapamycin: a signaling kinase for every aspect of cellular life. Methods Mol Biol. 2012;821:1-14 [DOI] [PubMed] [Google Scholar]
- 44.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259-269 [DOI] [PubMed] [Google Scholar]
- 45.Sadowski M, Sarcevic B. Mechanisms of mono- and poly-ubiquitination: ubiquitination specificity depends on compatibility between the E2 catalytic core and amino acid residues proximal to the lysine. Cell Div. 2010;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ichimura Y, Komatsu M. Selective degradation of p62 by autophagy. Semin Immunopathol. 2010;32(4):431-436 [DOI] [PubMed] [Google Scholar]
- 47.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105(52):20567-20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120(2):159-162 [DOI] [PubMed] [Google Scholar]
- 50.Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33(4):505-516 [DOI] [PubMed] [Google Scholar]
- 52.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10(11):1215-1221 [DOI] [PubMed] [Google Scholar]
- 53.Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43(3):242-245 [DOI] [PubMed] [Google Scholar]
- 54.Yano T, Kurata S. Intracellular recognition of pathogens and autophagy as an innate immune host defense. J Biochem. 2011;150(2):143-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dal-Ré R. Worldwide clinical interventional studies on leading causes of death: a descriptive analysis. Ann Epidemiol. 2011;21(10):727-731 [DOI] [PubMed] [Google Scholar]
- 56.Rabe KF, Hurd S, Anzueto A, et al. ; Global Initiative for Chronic Obstructive Lung Disease Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176(6):532-555 [DOI] [PubMed] [Google Scholar]
- 57.Hogg JC. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. Lancet. 2004;364(9435):709-721 [DOI] [PubMed] [Google Scholar]
- 58.Yoshida T, Mett I, Bhunia AK, et al. Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat Med. 2010;16(7):767-773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanojkovic I, Kotur-Stevuljevic J, Milenkovic B, et al. Pulmonary function, oxidative stress and inflammatory markers in severe COPD exacerbation. Respir Med. 2011;105(suppl 1):S31-S37 [DOI] [PubMed] [Google Scholar]
- 60.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968):905-909 [DOI] [PubMed] [Google Scholar]
- 61.Mu TW, Ong DS, Wang YJ, et al. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134(5):769-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ozcan U, Yilmaz E, Ozcan L, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137-1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marciniak SJ, Lomas DA. Alpha1-antitrypsin deficiency and autophagy. N Engl J Med. 2010;363(19):1863-1864 [DOI] [PubMed] [Google Scholar]
- 64.Ranes J, Stoller JK. A review of alpha-1 antitrypsin deficiency. Semin Respir Crit Care Med. 2005;26(2):154-166 [DOI] [PubMed] [Google Scholar]
- 65.Mornex JF, Chytil-Weir A, Martinet Y, Courtney M, LeCocq JP, Crystal RG. Expression of the alpha-1-antitrypsin gene in mononuclear phagocytes of normal and alpha-1-antitrypsin-deficient individuals. J Clin Invest. 1986;77(6):1952-1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elliott PR, Bilton D, Lomas DA. Lung polymers in Z alpha1-antitrypsin deficiency-related emphysema. Am J Respir Cell Mol Biol. 1998;18(5):670-674 [DOI] [PubMed] [Google Scholar]
- 67.Farber HW, Loscalzo J. Pulmonary arterial hypertension. N Engl J Med. 2004;351(16):1655-1665 [DOI] [PubMed] [Google Scholar]
- 68.Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. 2011;365(6):537-547 [DOI] [PubMed] [Google Scholar]
- 69.Herridge MS, Angus DC. Acute lung injury—affecting many lives. N Engl J Med. 2005;353(16):1736-1738 [DOI] [PubMed] [Google Scholar]
- 70.Jin Y, Tanaka A, Choi AM, Ryter SW. Autophagic proteins: New facets of the oxygen paradox. Autophagy. 2012;8(3):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rommens JM, Iannuzzi MC, Kerem B, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245(4922):1059-1065 [DOI] [PubMed] [Google Scholar]
- 72.Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245(4922):1066-1073 [DOI] [PubMed] [Google Scholar]
- 73.Kerem B, Rommens JM, Buchanan JA, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245(4922):1073-1080 [DOI] [PubMed] [Google Scholar]
- 74.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552-1555 [DOI] [PubMed] [Google Scholar]
- 75.Abdulrahman BA, Khweek AA, Akhter A, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by Burkholderia cenocepacia in a model of cystic fibrosis. Autophagy. 2011;7(11):1359-1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campbell GR, Spector SA, Vitamin D. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8(5):e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grégoire IP, Richetta C, Meyniel-Schicklin L, et al. IRGM is a common _target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7(12):e1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Udwadia ZF, Amale RA, Ajbani KK, Rodrigues C. Totally drug-resistant tuberculosis in India. Clin Infect Dis. 2012;54(4):579-581 [DOI] [PubMed] [Google Scholar]
- 79.Buzzai M, Jones RG, Amaravadi RK, et al. Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 2007;67(14):6745-6752 [DOI] [PubMed] [Google Scholar]
- 80.Fahy RJ, Exline MC, Gavrilin MA, et al. Inflammasome mRNA expression in human monocytes during early septic shock. Am J Respir Crit Care Med. 2008;177(9):983-988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang YH, Yang YL, Tiao MM, Kuo HC, Huang LT, Chuang JH. Hepcidin protects against lipopolysaccharide-induced liver injury in a mouse model of obstructive jaundice. Peptides. 2012;35(2):212-217 [DOI] [PubMed] [Google Scholar]
- 82.Constantin M, Choi AJ, Cloonan SM, Ryter SW. doi: 10.1155/2012/859235. Therapeutic potential of heme oxygenase-1/carbon monoxide in lung disease. Int J Hypertens. 2012; 2012:859235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Puneet P, McGrath MA, Tay HK, et al. The helminth product ES-62 protects against septic shock via Toll-like receptor 4-dependent autophagosomal degradation of the adaptor MyD88. Nat Immunol. 2011;12(4):344-351 [DOI] [PubMed] [Google Scholar]