Abstract
Induction of epithelial-to-mesenchymal transition (EMT) by TGF-β1 requires Ras signaling. We recently identified the transcriptional repressor Blimp-1 (PRDM1) as a downstream effector of the NF-κB, RelB/Bcl-2/Ras-driven pathway that promotes breast cancer cell migration. As the RelB/Blimp-1 pathway similarly required Ras signaling activation, we tested whether Blimp-1 plays a role in TGF-β1-mediated EMT. Here, TGF-β1 treatment of untransformed NMuMG mammary epithelial and MDA-MB-231 breast cancer cells was shown to induce Blimp-1 expression, which promoted an EMT signature and cell migration. TGFB1 and BLIMP1 RNA levels were correlated in patient breast tumors. BLIMP1 gene transcription was activated by TGF-β1 via a c-Raf (RAF1) to AP-1 pathway. Blimp-1 induced expression of the EMT master regulator Snail (SNAI1) via repressing BMP-5, which inhibited Snail expression upon TGF-β1 treatment. Interestingly, a similar cascade was observed during postnatal mouse mammary gland development. RelB expression was detected early in pregnancy followed progressively by Blimp-1 and then Snail; whereas, BMP-5 levels were high in nulliparous and regressing glands. Finally, lower BMP5 RNA levels were detected in patient breast tumors versus normal tissues, and correlated with cancer recurrence. Thus, the Ras effector Blimp-1 plays an essential role in TGF-β1-induced EMT via repression of BMP-5 in breast cancer.
Keywords: RelB NF-κB, Blimp-1, TGF-β1, BMP-5, AP-1, breast cancer, EMT, mammary gland development
INTRODUCTION
Aberrant constitutive expression of NF-κB subunits, reported in over 90% of breast cancers (1, 2) and multiple other malignancies, play a pivotal role in tumorigenesis (3, 4). Notably, the RelB NF-κB subunit is more highly expressed in more aggressive estrogen receptor (ER)α negative breast tumors versus ERα positive ones, and promotes their invasive phenotype (5). Similarly, nuclear RelB expression in prostate cancer correlates directly with Gleason score (6) and plays a radioprotective role in aggressive prostate cancer cells (7). In breast cancer cells, RelB enhances migration and invasion via induction of the BCL2 gene (5). The resulting Bcl-2 protein recruits Ras to the mitochondria, where it is activated (8). The zinc-finger B-lymphocyte induced maturation protein (Blimp-1) was identified as a pivotal downstream mediator of the migratory phenotype regulated by RelB and Ras (8). Blimp-1, which was not previously known to be expressed in breast cancer, reduced ERα expression by directly repressing ERa (ESR1) gene transcription (8). Consistently, higher BLIMP1 RNA expression was detected in ERα negative breast tumors. Blimp-1 was originally identified as a silencer of IFNB gene transcription (9), and subsequently as a master regulator essential for the differentiation of B and T lymphocytes (10, 11). Blimp-1 also plays crucial roles in early embryonic development, including the specification and migration of primordial germ cells (12, 13). Consistently, we demonstrated that Blimp-1 promotes breast cancer cell migration (8).
Transforming growth factor (TGF)-β, which suppresses growth of normal cells, functions as a tumor promoter in the presence of active Ras signaling. TGF-β1 induces epithelial-to-mesenchymal transition (EMT) and promotes the invasive phenotype of breast cancer cells that are signaling via a Ras to Erk cascade (14). TGF-β1 treatment of the ERα negative untransformed NMuMG mouse mammary epithelial line (15, 16) or the MDA-MB-231 human breast cancer cells promotes EMT as judged by enhanced cell migration and wound closure in vitro (17), and metastasis to bone (18) and lung (19) in mouse models. As Blimp-1 is induced downstream of Ras signaling, we hypothesized a role for this master regulator in TGF-β1-mediated EMT. Here, we elucidate a new EMT pathway mediated by Blimp-1 that leads to Snail induction via the repression of bone morphogenetic protein 5 (BMP-5) expression, which has been previously implicated in certain features of TGF-β-induced EMT in renal epithelial cells (20). Consistently, lower BMP5 RNA levels are detected in breast tumors versus normal tissues, and correlate with disease recurrence in patients, suggesting the clinical relevance of this pathway.
MATERIALS AND METHODS
Cell culture and treatment conditions
ERα negative MDA-MB-231, NMuMG and Hs578T and ERα positive MCF-7 and ZR-75 cells were purchased from the American Type Culture Collection (ATCC), grown in medium recommended by ATCC and vials frozen immediately. Fresh vials were used approximately every 6 weeks. Cells were confirmed to be free of mycoplasma contamination using PCR (VenorGeM Mycoplasma Detection Kit, Sigma). The identity of the MDA-MB-231 line was authenticated using short tandem repeat analysis (Genetica DNA Laboratories), which demonstrated 100% identity with the MDA-MB-231 cell line of ATCC. Hs578T pRELB-shRNA (RelB shRNA) or pRELB-sense (Control) cell lines were established as described (5) and grown in the presence of 1 μg/ml puromycin (Sigma). NMuMG and MDA-MB-231 cells were treated with 2 and 5 ng/ml TGF-β1, respectively or with 100 ng/ml BMP-5 (R&D Systems). For both reagents, the equivalent volume of vehicle, 4 mM HCl containing 1 mg/ml BSA in PBS, was used as control. Plasmids, transfection analysis and wound healing assays are described in the Supplementary Materials.
RNA and immunoblot analyses
RNA isolation, RT-PCR and real-time quantitative Q-PCR analyses were performed as detailed in Supplemental Data. Whole-cell extracts (WCEs) and nuclear extracts (NEs) were prepared and subjected to immunoblotting using the antibodies described in the Supplementary Materials. Protein extracts were prepared from the 4th mammary gland of FVB/N mice following removal of the lymph node as previously described (21), see Supplementary Materials.
ChIP assay
The ChIP assay for AP-1 binding was performed using Hs578T cells as previously described (8), see Supplementary Materials. The Blimp-1 ChIP assay for BMP5 promoter was performed using a pcDNA4/Blimp1-V5-tag expression vector and the EZ-ChIP kit (Millipore Corporation) (Supplementary Materials).
Gene expression and statistical analysis
Individual cancer datasets were downloaded from Oncomine (Compendia Bioscience), as previously described (5), see Supplemental Data. The prognostic value of BMP5 in recurrence of breast cancer was assessed with Kaplan-Meier Plotter (www.kmplot.com), which used gene expression microarray data and survival information of 2324 breast cancer patients downloaded from GEO (Affymetrix HGU133A and HGU133+2). To compare the different conditions used in wound healing assays, the Student’s t test was performed and a P-value < 0.05 was considered statistically significant.
RESULTS
Blimp-1 is required for TGF-β1-induced EMT
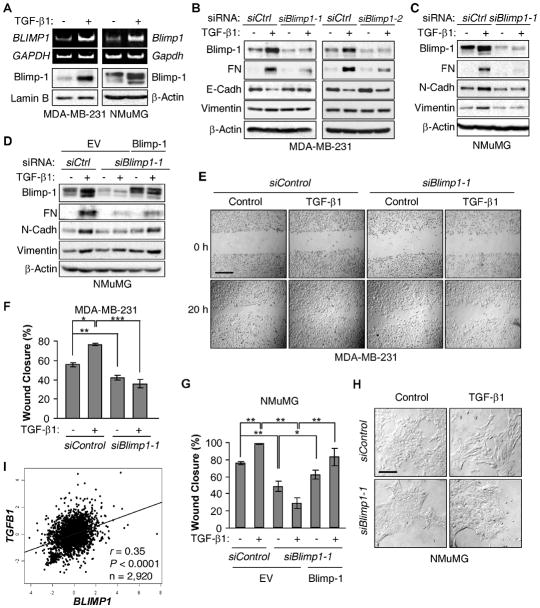
Incubation of MDA-MB-231 or NMuMG cells with TGF-β1 resulted in robust increases in Blimp1 RNA and Blimp-1 protein levels (Fig. 1A). Using two different Blimp-1 siRNAs to knockdown Blimp-1 levels in MDA-MB-231 cells, the normally observed induction of Fibronectin caused by TGF-β1 treatment was attenuated (17) and the decrease in basal E-Cadherin levels was prevented (Fig. 1B). MDA-MB-231 cells consist of two subpopulations with different levels of E-Cadherin, one negative and one positive (22); the mesenchymal marker N-Cadherin is not detectable (23). No significant increase in the high basal levels of Vimentin was noted. TGF-β1 treatment of NMuMG cells has been shown to lead to EMT, as judged by the upregulation of mesenchymal markers Fibronectin, N-Cadherin and Vimentin (15). Knockdown of Blimp-1 overrode the characteristic upregulation of all three mesenchymal makers by TGF-β1 (Fig. 1C). Notably, ectopic expression of Blimp-1 largely rescued the effects of Blimp-1 knockdown in these cells, confirming the specificity of the Blimp-1 siRNA (Fig. 1D). These results were recapitulated in another ERα negative breast cancer line Hs578T using both knockdown and dominant negative strategies (Supplementary Fig. S1).
Figure 1. Blimp-1 is induced by TGF-β1 and required for TGF-β1-induced EMT.
A, MDA-MB-231 and NMuMG cells were treated with TGF-β1. RNAs were subjected to RT-PCR analysis, and NEs or WCEs to immunoblotting. B, MDA-MB-231 cells were transiently transfected with two siRNAs specific for Blimp1 or with Control (siCtrl) siRNA. After 24 h, cells were incubated with TGF-β1 or vehicle (−) and WCEs analyzed. C, NMuMG cells were transfected with siCtrl or siBlimp1-1 and processed as in part B. D, NMuMG were transfected with Blimp-1 expressing vector or empty vector (EV). After 24 h, cells were transfected with siRNAs and processed as in part C. E-F, MDA-MB-231 cells were transfected with siCtrl or siBlimp1-1 and subjected to a wound healing assay with TGF-β1 or vehicle. (E) Cultures were photographed at 10X magnification right after the scratch and 20 h later. Bar, 200 μm. (F) The percentage of wound closure (mean ± S.D.) from 3 independent experiments is shown. * P = 1.3×10−4, ** P = 2.3×10−3, *** P = 1.5×10−4. G, NMuMG cells were transfected as in part D and subjected to a wound healing assay with TGF-β1 or vehicle. The percentage of wound closure (mean ± S.D.) from 3 independent experiments is shown. * P < 0.05, ** P < 0.005. H, NMuMG cells were transfected with siControl or siBlimp1-1. After 24 h, cells were incubated with TGF-β1 and photographed at 20X magnification. Bar, 100 μm. I, The Pearson’s pairwise correlation plot depicts the expression of TGFB1 and BLIMP1 from datasets (bc-GenExMiner Database) showing a positive correlation in human breast tumors.
Another measure of EMT is increased cell migration (16). TGF-β1 treatment led to more rapid wound closure by both MDA-MB-231 and NMuMG cells. Blimp-1 knockdown reduced the basal level of wound closure and prevented its acceleration by TGF-β1 in MDA-MB-231 (Figs. 1E and 1F and Supplementary Fig. S2) and NMuMG cells (Fig. 1G). Furthermore, ectopic Blimp-1 expression in NMuMG cells partially rescued the decreased migration seen upon Blimp-1 knockdown (Fig. 1G). Consistently, the acquisition of a fibroblastoid phenotype by NMuMG cells upon TGF-β1 treatment was prevented by knockdown of Blimp-1 (Fig. 1H). Thus, TGF-β1 treatment induces Blimp-1 expression, which is essential for the acquisition of a mesenchymal phenotype.
Increased TGF-β1 production has been observed in breast carcinomas compared to normal tissues (24), and associated with disease progression in human breast cancer (25). Using the bc-GenExMiner database of 2,920 patients with breast cancer, a significant correlation was observed between TGFB1 and BLIMP1 RNA levels (r = 0.35, P < 0.0001) (Fig. 1I), which was also observed when one stratified by ERα status (Supplementary Fig. S3), thus extending the clinical relevance of these findings to human disease.
Blimp-1 expression in breast cancer cells is mediated via a c-Raf to AP-1 pathway
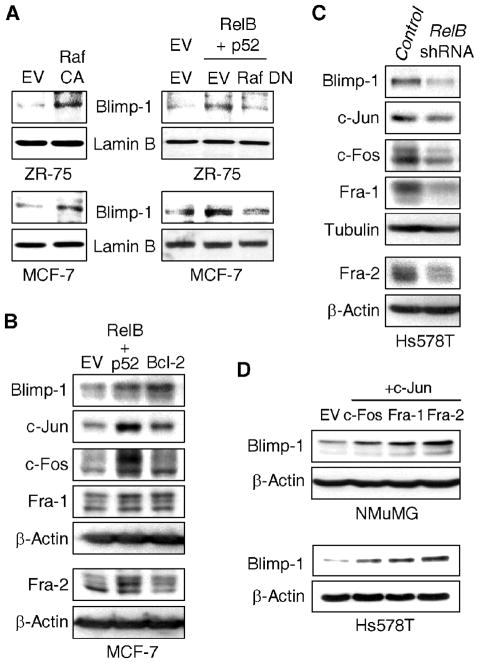
To address the mechanism of Blimp-1 activation, we focused on Ras signaling, as it is implicated downstream of RelB signaling (8) and also required for the transforming activity of TGF-β1 (14, 26). As c-Raf is an effector of Ras-mediated migration (27), its involvement in Blimp-1 induction was tested. Expression of a constitutively active form of c-Raf (Raf CA) into ERα positive ZR-75 and MCF-7 breast cancer cells, which contain low levels of Blimp-1, led to increased Blimp-1 levels (Fig. 2A). Conversely, a dominant negative variant of c-Raf (Raf DN) prevented Blimp-1 induction by ectopic expression of RelB and its binding partner p52 (Fig. 2A). Consistent with a c-Raf to Erk pathway downstream of RelB and Bcl-2, we confirmed the ability of RelB and Bcl-2 to activate Erk1/2 (Supplementary Figs. S4A and S4B), and showed that their effects on Blimp-1 induction could be abrogated with the MEK inhibitor PD98059 (Supplementary Fig. S4C). Thus, c-Raf and Erk1/2 are important downstream effectors of the Ras pathway leading to induction of Blimp-1 expression in breast cancer cells.
Figure 2. Blimp-1 is induced by the RelB/Bcl-2/c-Raf to AP-1 pathway.
A, ZR-75 and MCF-7 cells were transfected with a constitutively active c-Raf (Raf CA) vector or with RelB + p52 expression vectors in the presence of a dominant negative c-Raf expression (Raf DN) vector for 48 h. NEs were subjected to immunoblotting. B, WCEs from MCF-7 cells transfected with the indicated vectors for 48 h were subjected to immunoblotting. C, WCEs from stable mixed populations of Hs578T cells expressing RelB or Control shRNA were subjected to immunoblotting. D, WCEs from NMuMG or Hs578T cells transfected for 48 h with EV or indicated AP-1 vector were subjected to immunoblotting.
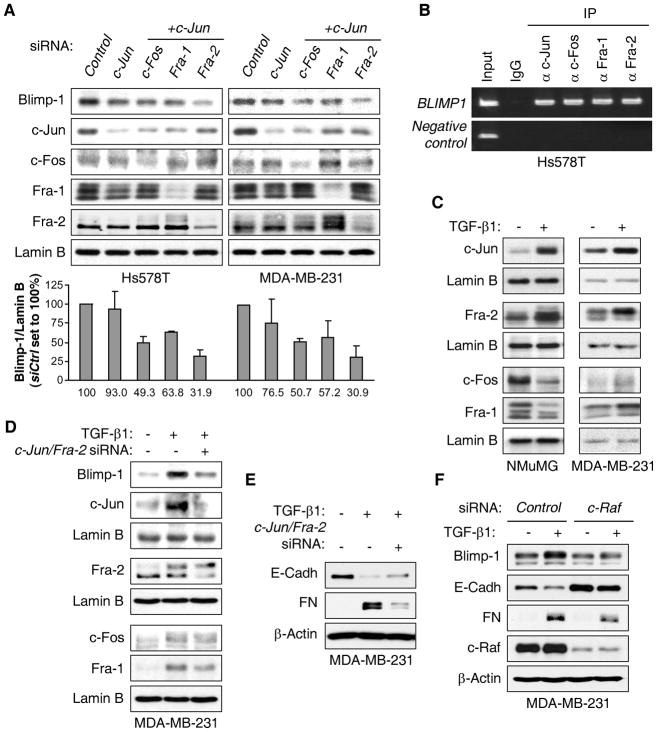
Dimeric AP-1 factors are important mediators of transformation by the Ras/c-Raf/Erk1/2 pathway and have been implicated in TGF-β1-mediated transformation (28). AP-1 elements have been involved in the transcriptional control of the BLIMP1 gene although different subunits were identified, suggesting cell-type specificity (29). Following ectopic expression of RelB/p52 or Bcl-2 in MCF-7 cells, activation of c-Jun, c-Fos, Fra-1 and Fra-2 AP-1 subunits was observed, as judged by an increase in their levels and/or relative phosphorylation status (Fig. 2B). Consistently, Blimp-1 expression was induced. Conversely, knockdown of RelB in Hs578T cells led to decreased levels of these same AP-1 subunits as well as of endogenous Blimp-1 expression (Fig. 2C). Co-transfection of vectors expressing c-Jun with c-Fos, Fra-1 or Fra-2 induced Blimp1 promoter activity (Supplementary Fig. S4D). Expression of these AP-1 subunits increased endogenous levels of Blimp-1 protein (Fig. 2D). Complexes of c-Jun/Fra-2 resulted in the largest induction in NMuMG and Hs578T cells (2.4-fold and 7.8-fold, respectively), with the latter line transfecting with a much higher efficiency. Consistently, knockdown of c-Jun and Fra-2 in MDA-MB-231 and Hs578T cells led to the most robust decreases in Blimp-1 levels (reduction of ~70% with this combination of siRNAs) (Fig. 3A). Knockdown of c-Jun/c-Fos or c-Jun/Fra-1 also led to decreased endogenous Blimp-1 levels, but to a lesser extent. Two nearby AP-1 binding sites have been identified in the BLIMP1 promoter in B cells (29). We focused on the −1647 bp site as it showed more binding in an initial test (data not shown). ChIP assays found endogenous c-Jun, c-Fos, Fra-1 and Fra-2 protein bound to the AP-1 site in BLIMP1 promoter in Hs578T cells (Fig. 3B), suggesting that they all contribute to induction of BLIMP1 transcription. Thus, activation of AP-1 signaling potently induces Blimp-1 expression in breast cancer cells.
Figure 3. TGF-β1 induces Blimp-1 expression via c-Raf/AP-1 signaling.
A, NEs from Hs578T and MDA-MB-231 cells transfected with the indicated siRNA for 48 h were subjected to immunoblot analysis. Quantification of Blimp-1 normalized to Lamin B from 3 independent experiments is given below relative to Control siRNA condition. B, Chromatin prepared from Hs578T cells was immunoprecipitated with antibodies against (α) the indicated proteins or control IgG. The region of the BLIMP1 promoter containing an AP-1 binding site (−1647 bp) was amplified by PCR, while a region of this promoter that does not carry any AP-1 site (−5480 bp) was used as negative control. C, NEs from NMuMG and MDA-MB-231 cells treated as in Fig. 1A were subjected to immunoblotting. D–E, MDA-MB-231 cells were transfected for 24 h with Control siRNA (−) or c-Jun/Fra-2 siRNAs (+) and treated with TGF-β1 for another 24 h. Immunoblotting was performed on NEs (E) or WCEs (F). F, MDA-MB-231 cells were transfected for 24 h with Control or c-Raf siRNA and treated with TGF-β1 for another 24 h. WCEs were subjected to immunoblotting.
TGF-β1 induces Blimp-1 via a c-Raf to AP-1 pathway
We next addressed whether increased AP-1 activity by TGF-β1 was responsible for Blimp-1 induction. TGF-β1 treatment increased nuclear levels of c-Jun and Fra-2 in both NMuMG and MDA-MB-231 cells (Fig. 3C). In NMuMG cells, c-Fos and Fra-1 were downregulated, while they were modestly increased in MDA-MB-231 cells. The role of c-Jun/Fra-2 activation in TGF-β1-mediated induction of Blimp-1 was tested. Knockdown of both c-Jun and Fra-2 greatly reduced the induction of Blimp-1 in MDA-MB-231 cells treated by TGF-β1 (Fig. 3D), which is consistent with the partial rescue of E-Cadherin expression and the decrease in Fibronectin induction seen upon TGF-β1 treatment (Fig. 3E). The remaining Blimp-1 expression may be due to the increase in c-Fos and Fra-1 seen in these cells upon TGF-β1 treatment (Fig. 3C). Lastly, c-Raf knockdown reduced basal Blimp-1 expression and virtually completely prevented its induction by TGF-β1, which led to the rescue of E-Cadherin expression and the attenuation of Fibronectin induction by TGF-β1 (Fig. 3F), confirming the importance of the Ras/c-Raf signaling axis. Together we conclude that TGF-β1 activates AP-1, thereby inducing Blimp-1 expression, which promotes EMT.
Blimp-1 is required for the induction of Snail by TGF-β1
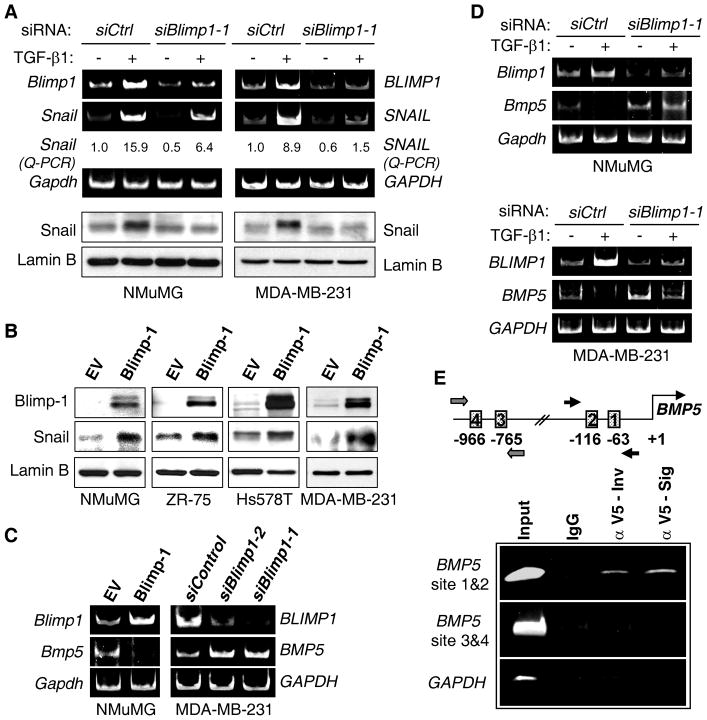
Blimp-1 was shown to promote EMT through the direct repression of ERa gene expression in ERα positive breast cancer cells (8). However, as NMuMG and MDA-MB-231 cells are ERα negative and its re-expression was not observed upon Blimp-1 knockdown (data not shown), we sought to identify the TGF-β1-induced EMT mediator downstream of Blimp-1. Snail and Slug are two key transcription factors that regulate the TGF-β-induced EMT molecular program during tumor progression (30). Blimp-1 knockdown greatly reduced the induction of Snail/SNAIL RNA and Snail protein resulting from TGF-β1 treatment (Fig. 4A), whereas the induction of Slug was unaffected (data not shown). Moreover, ectopic Blimp-1 expression was sufficient to increase levels of Snail in NMuMG and multiple breast cancer lines (Fig. 4B). Thus, Blimp-1 promotes the induction of Snail expression and its knockdown appears to profoundly decrease Snail protein levels following treatment with TGF-β1.
Figure 4. Blimp-1 is required for Snail induction by TGF-β1 and repression of BMP5 gene transcription.
A, NMuMG and MDA-MB-231 cells were transfected with Control (siCtrl) or Blimp1-1 siRNA and treated with TGF-β1 as in Fig. 1B. RNA or NEs were subjected to RT-PCR and real-time Q-PCR, or immunoblotting. B, The indicated cells were transfected with EV or Blimp-1 expression vector for 48 h, and NEs immunoblotted. C, Blimp-1 was either ectopically expressed in NMuMG cells or knocked down in MDA-MB-231 cells using 2 siRNAs for 48 h. RNA was subjected to RT-PCR. D, Cells were transiently transfected with Control or Blimp1-1 siRNA for 24 h and treated with TGF-β1 for 6 h. RNA was subjected to RT-PCR. E, Upper panel: Schematic diagram of the BMP5 promoter region with the two sets of putative Blimp-1 binding elements and their respective primers indicated. Lower panel: Chromatin prepared from MDA-MB-231 cells expressing a V5-tagged Blimp-1 was immunoprecipitated with antibodies against (α) V5 from Invitrogen (Inv) or Sigma (Sig) or with control IgG. The two BMP5 promoter regions containing putative Blimp-1 binding sites were amplified by PCR, while a region of the GAPDH promoter that does not carry any Blimp-1 site was used as negative control.
Blimp-1 repression of the BMP5 gene relieves inhibition of Snail expression by BMP-5 and promotes EMT by TGF-β1
We next sought to elucidate the mechanism whereby Blimp-1, which is a zinc finger protein only known to repress transcription, induces Snail expression. As Blimp-1 did not appear to affect the canonical TGF-β1 pathway mediated by Smad signaling (see Discussion), we hypothesized that Blimp-1 reduced the expression of an inhibitor of Snail expression. Microarray analysis was performed to identify genes controlled by Blimp-1 in the MDA-MB-231 line. Cells were reverse transfected with the two Blimp-1 siRNAs used above for 48 h. The quality of RNA from two independent experiments was confirmed using an Agilent RNA 6000 Pico kit and effective Blimp-1 knockdown using western blot (data not shown). The RNA was analyzed using Affymetrix U133A 2.0 arrays, representing 14,500 human genes. Sixty eight genes were significantly upregulated with Blimp-1 siRNAs, which represent potential direct Blimp-1 _targets. Bone morphogenetic protein 5 (BMP5) was found upregulated 2.75-fold (P = 5×10−4) with the two Blimp-1 siRNAs compared to Control siRNA, which was confirmed by RT-PCR (Fig. 4C). Interestingly, both TGF-β1 and BMP-5 belong to the TGF-β superfamily and their signaling pathways display complex and frequently antagonistic crosstalk during organogenesis and tissue homeostasis (31). To confirm the effects of Blimp-1 on BMP5 expression, Blimp-1 was ectopically expressed in NMuMG cells and led to a profound repression of Bmp5 RNA (Fig. 4C). Importantly, TGF-β1 treatment resulted in almost complete loss of BMP5 expression; whereas, BMP5 levels were substantially maintained upon knockdown of Blimp-1 in NMuMG (Fig. 4D) and MDA-MB-231 cells (Fig. 4D and data not shown). Using Transfac analysis, four putative Blimp-1 binding sites were identified in the BMP5 promoter (Fig. 4E). As there are currently no commercially available Blimp-1 antibodies for ChIP analysis, V5-tagged Blimp-1 protein was ectopically expressed in MDA-MB-231 cells. Blimp-1 binding to the region containing sites 1 and 2 was demonstrated using two different V5 antibodies (Fig. 4E). Together these findings identify BMP5 as a direct _target of Blimp-1 repression.
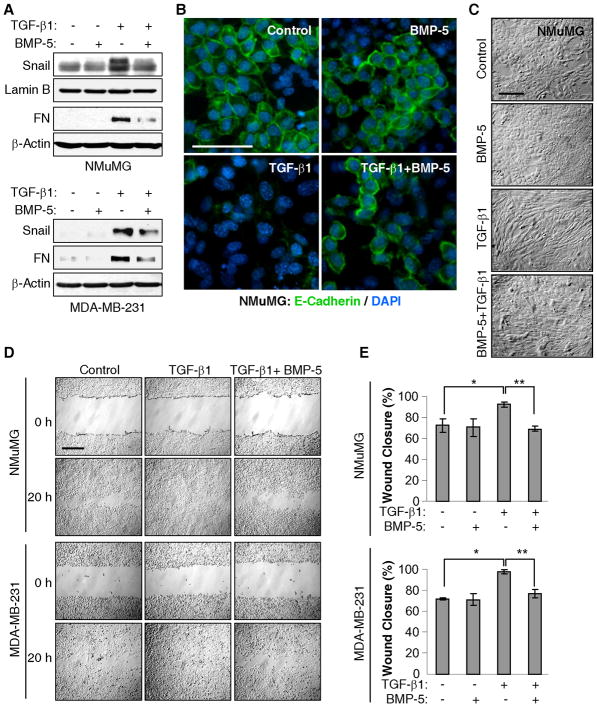
To assess the role of BMP-5 in Snail induction by TGF-β1, NMuMG and MDA-MB-231 cells were treated with TGF-β1 or BMP-5 alone, or the two in combination. Notably, addition of recombinant BMP-5 significantly attenuated Snail induction by TGF-β1 and reduced the induction of Fibronectin in both lines (Fig. 5A). Moreover, the loss of E-Cadherin expression on the surface of NMuMG cells in response to TGF-β1 was largely overridden by BMP-5 (Fig. 5B) and the fibroblastoid phenotype was also substantially reversed (Fig. 5C). Lastly, the acceleration of wound closure seen with TGF-β1 treatment was abrogated with addition of BMP-5 (Figs. 5D and 5E). Together, the data indicate that repression of BMP-5 levels by TGF-β1 contributes to the induction of EMT.
Figure 5. BMP-5 attenuates TGF-β1-induced Snail expression and cell migration.
A, NMuMG and MDA-MB-231 cells were treated for 24 h with TGF-β1 and BMP-5 alone or in combination. NEs and WCEs were assessed by immunoblot. B, NMuMG cells, treated for 48 h as in part A, were subjected to immunofluorescence microscopy for E-Cadherin (green) and their nuclei stained with DAPI (blue). Bar, 50 μm. C, NMuMG cells, treated as in part A, were photographed at 20X magnification. Bar, 100 μm. D–E, Cells were subjected to a wound healing assay in the absence or presence of TGF-β1 or BMP-5 alone or in combination. (D) Cultures were photographed right after the scratch and 20 h later. Bar, 200 μm. (E) Wound healing assays were measured as in Fig. 1F. The percentage of wound closure is shown. NMuMG: * P = 0.009, ** P = 4×10−4. MDA-MB-231: * P = 3.3×10−5, ** P = 1.6×10−3.
The RelB/Blimp-1/Snail pathway is active during mammary gland development
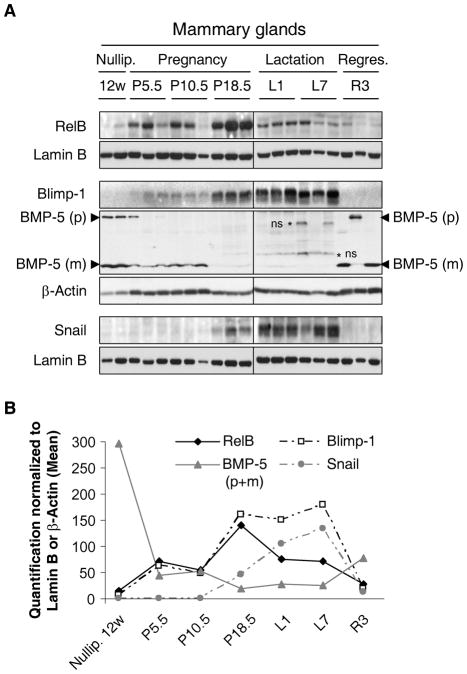
Previously, we detected RelB expression in the developing mouse mammary gland and found this subunit involved in ductal branching during pregnancy (21). As pathways utilized in malignant cells are often derived from normal development, we examined the expression of RelB and Blimp-1 in the mouse mammary gland. For each time point during pregnancy, nuclear and whole-cell extracts were prepared from three individual female mice. As seen previously, expression of RelB was detected early during pregnancy and increased between days 10.5 and 18.5 (Fig. 6A). During lactation and regression, RelB levels continued to decline. Blimp-1 expression followed RelB, with amounts increasing during late pregnancy and peaking during lactation (Fig. 6A), as judged by quantification of the 3 samples per time point (Fig. 6B). By comparison, Snail expression was quite low during pregnancy and increased following Blimp-1 induction. Expression of Blimp-1 and Snail dropped precipitously during regression (Fig. 6). Thus, the relative patterns of Blimp-1 and Snail were consistent with signaling downstream of RelB. Notably we could detect the precursor and mature forms of BMP-5 in nulliparous mice and during regression. Mature BMP-5 expression decreased progressively during pregnancy until it disappeared at the latest time point of pregnancy (P18.5) and during lactation, which is consistent with our findings identifying BMP-5 as a _target of Blimp-1 repression and involved in Snail repression, suggesting a similar cascade occurs during adult mouse mammary gland development.
Figure 6. Blimp-1/BMP5/Snail axis is active in vivo during adult mammary gland development.
A, Mammary glands were harvested from 12-week (12w) old nulliparous mice, and from mice at different days of pregnancy (P5.5, P10.5, P18.5), lactation (L1, L7) and regression (R3). NEs were immunoblotted for RelB, Snail and Lamin B, and WCEs for Blimp-1, precursor (p) and mature (m) forms of BMP-5, and β-Actin, respectively. * non-specific (ns) bands. B, Data were quantified, normalized to Lamin B or β-Actin as appropriate. The mean values of the multiple samples taken at the same time points are shown.
Breast cancer and poor prognosis are associated with low BMP5 expression
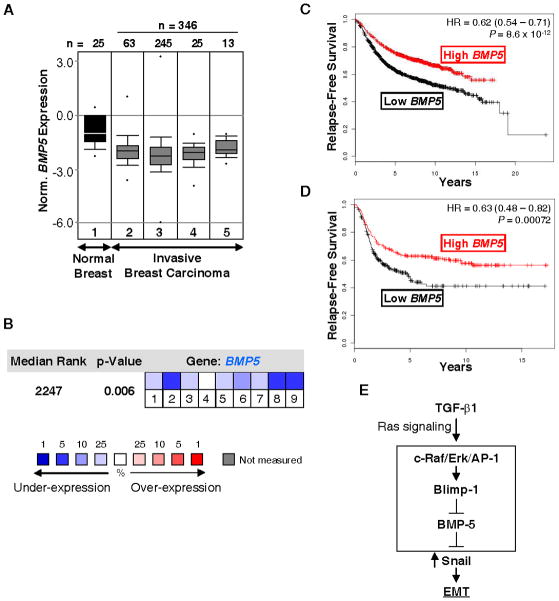
To assess the role of BMP-5 in breast cancer, we used the Oncomine database to evaluate their levels of BMP5 expression. The TCGA_Breast study was selected initially, as this dataset includes a substantial number of normal breast tissues (n = 25), as well as a large number of invasive breast carcinoma samples (n=346) (Fig. 7A). BMP5 RNA was significantly lower in overall invasive tumors versus normal breast tissues (see legend for individual P-values). Data from other studies that include fewer normal tissue samples showed similar results (Supplemental Figs. S5A and S5B). Overall, when all the studies containing informative BMP5 expression levels were combined, BMP5 was identified as one of the most downregulated genes in breast cancer compared to normal tissue (Figure 7B). Furthermore, numerous BMPs were found to be aberrantly downregulated in breast cancer, whereas TGFB1 was upregulated when compared to normal tissues (Supplemental Fig. S5C). Finally, the prognostic value of BMP5 was assessed using Kaplan-Meier Plotter, an online tool to correlate survival with gene expression, based upon microarray data from 2,324 breast cancer patients. Low BMP5 RNA levels were significantly correlated with lower disease free survival (Figure 7C), particularly in patients with ERα negative cancers (Figure 7D).
Figure 7. BMP5 expression is downregulated in breast cancer and low BMP5 levels correlate with breast cancer recurrence.
A, The TCGA_Breast microarray dataset was accessed using Oncomine database (www.oncomine.com), and includes normal breast tissues (1), invasive (2, P = 4.55×10−11), invasive ductal (3, P = 6.98×10−10), invasive lobular (4, P = 2.67×10−10) and invasive mixed breast carcinomas (5, P = 3.87×10−7). n = sample number. Box plots depict the distribution of BMP5 expression within each group. The P-values were obtained using a Student’s t-test for the comparison of BMP5 expression between the normal breast tissue and each breast carcinoma group. B, The median BMP5 rank was assessed across 9 analyses comparing the normal tissue group to each breast carcinoma group found in Gluck_Breast (1. invasive ductal breast carcinoma), Radvanyi_Breast (2. ductal breast carcinoma in situ, 3. invasive ductal breast carcinoma, 4. invasive lobular breast carcinoma, 5. invasive mixed breast carcinoma) and TCGA_Breast (6. invasive breast carcinoma, 7. invasive ductal breast carcinoma, 8. invasive lobular breast carcinoma, 9. invasive mixed breast carcinoma) datasets. The P-value is given for the median-ranked analysis. C–D, The Kaplan-Meier survival plots were obtained using Kaplan-Meier Plotter (www.kmplot.com) and display (C) the probability of relapse-free survival of 2,324 breast cancer patients or (D) 494 ERα negative breast cancer patients grouped according to BMP5 median expression. Hazard ratio (HR) with 95% confidence intervals (CI) and P-value for the comparison of the low and high BMP5 groups, are shown on the plot. E, Summary scheme. In the presence of Ras signaling, TGF-β1 induces Snail, which promotes EMT. Here a novel mechanism of Snail induction is elucidated in mammary epithelial cells and breast cancer cells. TGF-β1 induces Blimp-1 expression via a c-Raf/Erk/AP-1 pathway. Blimp-1 is required for TGF-β1-induced Snail expression and repression of BMP-5. BMP-5 is an inhibitor of the induction of Snail expression by TGF-β1.
DISCUSSION
These studies elucidate a new pathway of transformation by TGF-β1 via induction of Snail. Blimp-1 was induced by TGF-β1 via a c-Raf/AP-1 pathway, and directly inhibited the BMP5 gene (see scheme in Figure 7E). The resulting decrease in BMP-5 levels led to a de-repression of Snail expression. Thus Blimp-1 and BMP-5 are crucial intermediates between the Ras/c-Raf/AP-1 signaling axes and the induction of Snail and EMT by TGF-β1 in breast epithelial and cancer cells.
The transcriptional master regulator Snail is induced in virtually all EMT processes and notably by TGF-β in a broad spectrum of cell types (30). Snail expression has been correlated with mesenchymal phenotype and metastasis of breast cancer (32). Abnormal production of TGF-β1 in breast cancer is associated with disease progression (25). Interrogation of microarray datasets demonstrated a significant correlation between TGFB1 and BLIMP1 RNA levels in 2,920 breast cancer patients, extending our findings to human disease. The induction of Snail by Blimp-1 is mediated via release of repression by BMP-5, and consistently Blimp-1 is shown here to directly bind to and repress the BMP5 gene. To our knowledge, this is the first study demonstrating the direct binding of a transcription factor to the mammalian BMP5 promoter leading to its transcriptional regulation. Previous studies of the BMP5 promoter identified putative regulatory elements (i.e., GATA-1, engrailed and long-range cis regulatory elements), but binding studies or more fine-tuned mapping was not performed (33, 34). In untransformed renal epithelial cells, BMP-5 expression was decreased by TGF-β and its addition attenuated TGF-β-induced EMT, as judged by levels of smooth muscle cell actin and ZO-1 (20); although the _target(s) of BMP-5 was not elucidated in these cells. TGF-β and BMP signaling pathways display complex (positive and negative) crosstalk during organogenesis and homeostasis of many tissues (31). In mammary cells, BMP-5 and TGF-β1 had antagonistic activities on Snail expression. Blimp-1 tilted the balance toward TGF-β1-induced EMT by repressing BMP-5 expression, which led to the de-repression of Snail.
Bmp5 was identified as the gene mutated in the short-ear mouse model, which displays various skeletal defects (35). Interestingly, these mice also have a 2-fold increase in skin tumor susceptibility (36). Decreased BMP-5 expression was observed in a number of primary tumors or cancer cell lines. In pancreatic cancers, which are typified by active Ras signaling, low expression of BMP5 RNA was detected in sixteen tumor cell lines compared to four normal samples (37). BMP-5 expression was also significantly lower in adrenocortical carcinomas and tumor cell lines compared with normal adrenal glands (38), and in malignant schwannoma versus benign lesions (39). We observed that BMP5 RNA is one of the most under-expressed transcripts in breast tumors compared to normal tissue, and its decreased expression correlated with disease recurrence in breast cancer patients. Reduced BMP5 levels also correlated with more invasive tumors in these patients. Previously, Blimp-1 was found more highly expressed in ERα negative vs positive breast tumors (5). Consistently, the prognostic value of BMP-5 was restricted to ERα-negative breast tumors, validating the biological relevance of this inhibitory pathway. Interestingly, the Blimp-1/BMP-5/Snail axis appeared active in vivo during adult mammary gland development. In contrast to BMP-5, Blimp-1 and Snail were not expressed in nulliparous or regressing mammary glands, and only detected during pregnancy and lactation. This suggests that after pregnancy, Blimp-1 expression might need to be shut down in mammary tissue in order to reactivate BMP-5, decreasing Snail levels and allowing the gland to go back to a resting stage.
AP-1 complexes drive the oncogenic capacity of TGF-β at the transcriptional level (40). Here, AP-1 dimers of c-Jun and Fra-2 were identified as critical downstream _targets required for Blimp-1 induction in response to TGF-β1. Transcriptional activation of other TGF-β-induced genes involved in tumor progression, such as Collagen, MMP2, Laminin α3a, and the autoinduction of TGF-β1 itself is similarly mediated by the direct promoter binding of AP-1 complexes (40). Interestingly, overexpression of c-Jun in MCF-7 cells strongly promoted their migratory and invasive properties and allowed for tumor formation independent of estrogen (41). Fra-1 and Fra-2 were implicated in the malignant behavior of breast cancer cells and the progression of mammary carcinoma (8, 42).
This study identifies Blimp-1 as a critical node in the cooperation between the Ras pathway and TGF-β1 signaling in favor of Snail expression and induction of EMT. Blimp-1 did not appear to affect the canonical TGF-β1 pathway mediated by Smad signaling as its knockdown did not affect either levels of Smad2/3 and Smad4 in the nucleus, or RNA levels of Smad6 and Smad7 (data not shown). The antagonistic effects of BMP-5 on TGF-β1-induced EMT provide a rationale for the prognostic value of BMP-5 observed in patients with invasive breast cancer. Interestingly addition of BMP-5 did not recapitulate all the EMT marker modulations seen with Blimp-1 knockdown upon TGF-β1, suggesting that Blimp-1 regulates genes other than BMP5 and SNAIL to promote certain features of TGF-β1-induced EMT in ERα negative cells. Studies are in progress to identify these additional _target genes and pathways.
Supplementary Material
Acknowledgments
Grant Support: These studies were supported by NIH grants P01 ES11624 (D.C.S. and G.E.S.) and R01 CA129129 (G.E.S.), and by the DOD postdoctoral fellowship award W81XWH-10-1-1003 (M.R.).
We thank Pat Hogan for assistance with mammary gland preparations and Drs. Nuria Sánchez-Morgan and Albert Tai for help with the immunofluorescence microscopy.
Footnotes
The authors have no conflict of interest to declare.
References
- 1.Sovak MA, Bellas RE, Kim DW, et al. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. The Journal of clinical investigation. 1997;100(12):2952–60. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS., Jr Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000;19(9):1123–31. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 3.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18(49):6938–47. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 4.Gilmore TD, Garbati MR. Inhibition of NF-kappaB signaling as a strategy in disease therapy. Current topics in microbiology and immunology. 2011;349:245–63. doi: 10.1007/82_2010_105. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Belguise K, Kersual N, et al. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its _target BCL2. Nature cell biology. 2007;9(4):470–8. doi: 10.1038/ncb1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lessard L, Begin LR, Gleave ME, Mes-Masson AM, Saad F. Nuclear localisation of nuclear factor-kappaB transcription factors in prostate cancer: an immunohistochemical study. British journal of cancer. 2005;93(9):1019–23. doi: 10.1038/sj.bjc.6602796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Josson S, Xu Y, Fang F, Dhar SK, St Clair DK, St Clair WH. RelB regulates manganese superoxide dismutase gene and resistance to ionizing radiation of prostate cancer cells. Oncogene. 2006;25(10):1554–9. doi: 10.1038/sj.onc.1209186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Belguise K, O’Neill CF, et al. RelB NF-kappaB represses estrogen receptor alpha expression via induction of the zinc finger protein Blimp1. Molecular and cellular biology. 2009;29(14):3832–44. doi: 10.1128/MCB.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5(5):868–79. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer AL, Lin KI, Kuo TC, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17(1):51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 11.Martins GA, Cimmino L, Shapiro-Shelef M, et al. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7(5):457–65. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 12.Ohinata Y, Payer B, O’Carroll D, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–13. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 13.Vincent SD, Dunn NR, Sciammas R, et al. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development (Cambridge, England) 2005;132(6):1315–25. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 14.Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6(5):603–10. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J Cell Biol. 1994;127(6 Pt 2):2021–36. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by deltaEF1 proteins in epithelial mesenchymal transition induced by TGF-beta. Molecular biology of the cell. 2007;18(9):3533–44. doi: 10.1091/mbc.E07-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115(Pt 15):3193–206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 18.Yin JJ, Selander K, Chirgwin JM, et al. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. The Journal of clinical investigation. 1999;103(2):197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padua D, Zhang XH, Wang Q, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133(1):66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bramlage CP, Muller GA, Tampe B, et al. The role of bone morphogenetic protein-5 (BMP-5) in human nephrosclerosis. Journal of nephrology. 2011;24(5):647–55. doi: 10.5301/JN.2011.6330. [DOI] [PubMed] [Google Scholar]
- 21.Demicco EG, Kavanagh KT, Romieu-Mourez R, et al. RelB/p52 NF-kappaB complexes rescue an early delay in mammary gland development in transgenic mice with _targeted superrepressor IkappaB-alpha expression and promote carcinogenesis of the mammary gland. Molecular and cellular biology. 2005;25(22):10136–47. doi: 10.1128/MCB.25.22.10136-10147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lombaerts M, van Wezel T, Philippo K, et al. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. British journal of cancer. 2006;94(5):661–71. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147(3):631–44. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Travers MT, Barrett-Lee PJ, Berger U, et al. Growth factor expression in normal, benign, and malignant breast tissue. British medical journal (Clinical research ed) 1988;296(6637):1621–4. doi: 10.1136/bmj.296.6637.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorsch SM, Memoli VA, Stukel TA, Gold LI, Arrick BA. Immunohistochemical staining for transforming growth factor beta 1 associates with disease progression in human breast cancer. Cancer research. 1992;52(24):6949–52. [PubMed] [Google Scholar]
- 26.Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of Ras signaling in the induction of snail by transforming growth factor-beta. The Journal of biological chemistry. 2009;284(1):245–53. doi: 10.1074/jbc.M804777200. [DOI] [PubMed] [Google Scholar]
- 27.McCubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochimica et biophysica acta. 2007;1773(8):1263–84. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies M, Robinson M, Smith E, Huntley S, Prime S, Paterson I. Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-beta1 involves MAPK, Smad and AP-1 signalling pathways. Journal of cellular biochemistry. 2005;95(5):918–31. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 29.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169(4):1922–9. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 30.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Bubnoff A, Cho KW. Intracellular BMP signaling regulation in vertebrates: pathway or network? Developmental biology. 2001;239(1):1–14. doi: 10.1006/dbio.2001.0388. [DOI] [PubMed] [Google Scholar]
- 32.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development (Cambridge, England) 2005;132(14):3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 33.Sakaue M, Kitazawa S, Nishida K, Kitazawa R, Maeda S. Molecular cloning and characterization of human bone morphogenic protein (BMP)-5 gene promoter. Biochemical and biophysical research communications. 1996;221(3):768–72. doi: 10.1006/bbrc.1996.0671. [DOI] [PubMed] [Google Scholar]
- 34.Pregizer S, Mortlock DP. Control of BMP gene expression by long-range regulatory elements. Cytokine & growth factor reviews. 2009;20(5–6):509–15. doi: 10.1016/j.cytogfr.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kingsley DM, Bland AE, Grubber JM, et al. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell. 1992;71(3):399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- 36.Kangsamaksin T, Morris RJ. Bone morphogenetic protein 5 regulates the number of keratinocyte stem cells from the skin of mice. The Journal of investigative dermatology. 2011;131(3):580–5. doi: 10.1038/jid.2010.378. [DOI] [PubMed] [Google Scholar]
- 37.Virtanen S, Alarmo EL, Sandstrom S, Ampuja M, Kallioniemi A. Bone morphogenetic protein -4 and -5 in pancreatic cancer--novel bidirectional players. Experimental cell research. 2011;317(15):2136–46. doi: 10.1016/j.yexcr.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Johnsen IK, Kappler R, Auernhammer CJ, Beuschlein F. Bone morphogenetic proteins 2 and 5 are down-regulated in adrenocortical carcinoma and modulate adrenal cell proliferation and steroidogenesis. Cancer research. 2009;69(14):5784–92. doi: 10.1158/0008-5472.CAN-08-4428. [DOI] [PubMed] [Google Scholar]
- 39.Jin Y, Lu HB, Liong E, Lau TY, Tipoe GL. Transcriptional mRNA of BMP-2, 3, 4 and 5 in trigeminal nerve, benign and malignant peripheral nerve sheath tumors. Histology and histopathology. 2001;16(4):1013–9. doi: 10.14670/HH-16.1013. [DOI] [PubMed] [Google Scholar]
- 40.Mulder KM. Role of Ras and Mapks in TGFbeta signaling. Cytokine & growth factor reviews. 2000;11(1–2):23–35. doi: 10.1016/s1359-6101(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 41.Smith LM, Wise SC, Hendricks DT, et al. cJun overexpression in MCF-7 breast cancer cells produces a tumorigenic, invasive and hormone resistant phenotype. Oncogene. 1999;18(44):6063–70. doi: 10.1038/sj.onc.1202989. [DOI] [PubMed] [Google Scholar]
- 42.Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene. 2005;24(8):1434–44. doi: 10.1038/sj.onc.1208312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.