Abstract
Secretory and transmembrane proteins enter the endoplasmic reticulum (ER) as unfolded proteins and exit as either folded proteins in transit to their _target organelles or as misfolded proteins _targeted for degradation. The unfolded protein response (UPR) maintains the protein-folding homeostasis within the ER, ensuring that the protein-folding capacity of the ER meets the load of client proteins. Activation of the UPR depends on three ER stress sensor proteins, Ire1, PERK, and ATF6. Although the consequences of activation are well understood, how these sensors detect ER stress remains unclear. Recent evidence suggests that yeast Ire1 directly binds to unfolded proteins, which induces its oligomerization and activation. BiP dissociation from Ire1 regulates this oligomeric equilibrium, ultimately modulating Ire1’s sensitivity and duration of activation. The mechanistic principles of ER stress sensing are the focus of this review.
Activation of the UPR depends on three stress sensor proteins: Ire1, PERK, and ATF6. In yeast, Ire1 appears to bind directly to unfolded proteins.
Secreted and transmembrane proteins enter the secretory pathway by translocation into the endoplasmic reticulum (ER). These newly synthesized polypeptides must properly fold before being transported to their _target organelles. Proteins that do not properly fold within a certain time are _targeted for ER-associated degradation (ERAD), which efficiently retro-translocates them from the ER into the cytosol for degradation via the ubiquitin-proteasome system (Smith et al. 2011). To support proper protein folding and prevent aggregation in the ER lumen, an environment with a high protein concentration (∼100 mg/mL), numerous ER-resident chaperones, and folding enzymes assist maturation by signal-peptide cleavage, glycosylation, and disulfide bond formation (Araki and Nagata 2011). Chaperones in particular are involved in every aspect of ER quality control. The most abundant and best-characterized ER-resident chaperone is BiP/Grp78 (immunoglobulin heavy chain binding protein/glucose-regulated protein 78), an Hsp70 family ATPase involved in numerous functions, including translocating nascent polypeptides, facilitating de novo protein folding and assembly, _targeting misfolded proteins to ERAD machinery, and maintaining calcium homeostasis (Hendershot 2004; Otero et al. 2010). To sustain protein-folding homeostasis in the ER, the cell must balance the ER protein folding load with sufficient ER protein folding machinery, particularly chaperones such as BiP.
Multiple physiological and pathological conditions can interfere with ER quality control and lead to an accumulation of misfolded proteins in the ER. Such an increase of unfolded proteins is termed “ER stress” and can have deleterious consequences for the cell. To cope with ER stress and maintain protein homeostasis, eukaryotic cells have evolved the unfolded protein response (UPR). The UPR coordinates the increase in ER-folding capacity through a broad transcriptional up-regulation of ER folding, lipid biosynthesis, and ERAD machinery (Travers et al. 2000) with a decrease in folding load through selective mRNA degradation and translational repression (Harding et al. 1999; Hollien and Weissman 2006; Hollien et al. 2009). The UPR is therefore cytoprotective, allowing cells to adapt to developmental and environmental conditions that impinge on ER protein folding. During severe and prolonged ER stress, however, the UPR can become cytotoxic rather than cytoprotective, inducing apoptosis (Lin et al. 2007). ER stress-induced apoptosis is an important pathogenic factor in a number of widespread diseases, including diabetes, neurodegenerative diseases, atherosclerosis, and renal disease (Tabas and Ron 2011). Because of the UPR’s central role in determining cell fate, there have been multiple studies to identify small molecules modulators to exploit the UPR for therapeutic benefit (Fribley et al. 2011; Papandreou et al. 2011; Volkmann et al. 2011; Cross et al. 2012; Mimura et al. 2012).
THREE ER STRESS SENSORS INITIATE THE UPR
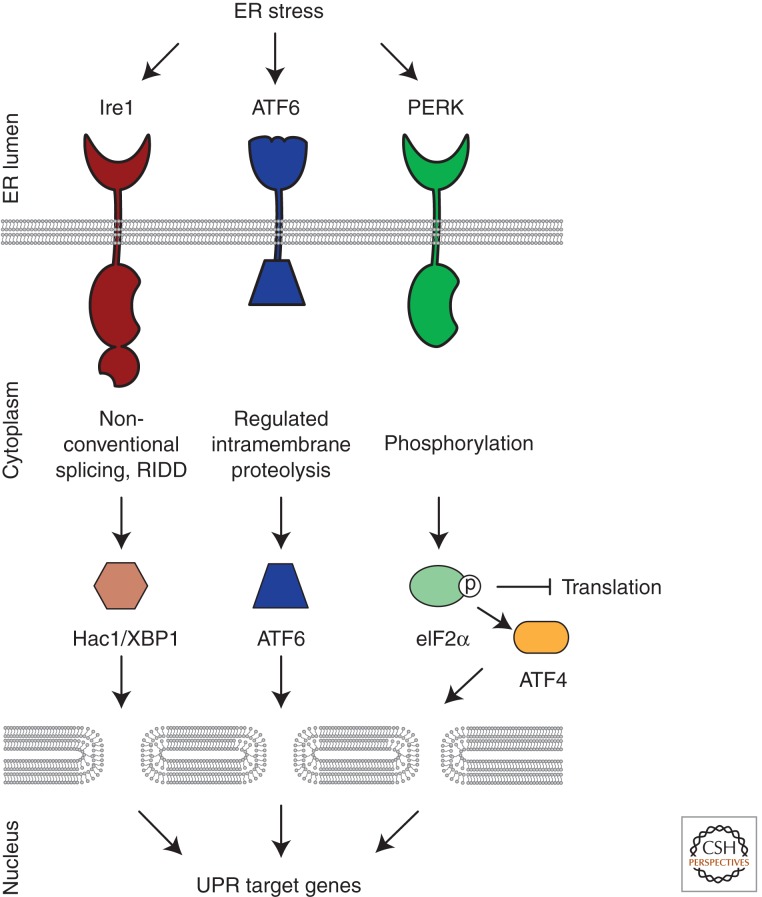
In metazoans, three parallel pathways employing unique signal transduction mechanisms collectively comprise the UPR. In each branch, an ER-resident integral membrane protein, Ire1 (inositol requiring enzyme 1), PERK (protein kinase RNA (PKR)-like ER kinase), or ATF6 (activating transcription factor 6) senses abnormal conditions in the ER lumen and transmits the information across the membrane into the cytosol where a series of transcription factors carry information to the nucleus (Fig. 1) (Walter and Ron 2011). The three branches collaborate to up-regulate protein folding machinery and decrease the burden of unfolded proteins. In this review, we highlight recent mechanistic insights into how ER stress is detected in yeast and subsequently discuss the implications for ER stress sensing in metazoan cells.
Figure 1.
In metazoans, three parallel signaling pathways comprise the UPR. ER-resident transmembrane sensor proteins, Ire1, PERK, and ATF6, activate signaling in each UPR branch. Upon activation, each sensor elicits unique downstream responses. Ire1’s cytosolic RNase domain cleaves an intron out of an mRNA, leading to the production of a potent transcriptional activator that induces genes to increase the folding capacity of the ER (XBP1 in metazoans, Hac1 in budding yeast). The active RNase also cleaves ER-localized messages, leading to their degradation, to reduce the load of unfolded proteins entering the ER. Active ATF6 translocates from the ER to the Golgi where it is proteolytically processed to release its amino-terminal transcriptional activator domain that induces genes to increase the folding capacity in the ER. PERK’s cytosolic kinase domain phosphorylates the translation initiation factor eIF2α, thereby inhibiting global translation and reducing the load of newly synthesized proteins entering the ER. Although generally inhibiting translation, eIF2α phosphorylation allows messages with inhibitory leader sequences to be preferentially translated. One of these messages encodes ATF4, a transcriptional activator that induces a cascade that ultimately produces proapoptotic factors.
Ire1
Ire1 is the only ER stress sensor present in all eukaryotes and therefore reflects the most ancient and most conserved branch of the UPR (Mori 2009). As a type I transmembrane protein, Ire1 contains an amino-terminal ER lumenal domain and carboxy-terminal cytoplasmic kinase and RNase domains. In the presence of ER stress, Ire1 forms higher-order oligomeric assemblies triggered by self-association of the ER lumenal domain (Ire1-LD). ER stress-dependent Ire1 oligomerization can be visualized microscopically as foci in vivo (Kimata et al. 2007; Aragon et al. 2009; Li et al. 2010; Pincus et al. 2010) and is required for downstream activation of its cytosolic kinase and RNase.
Although oligomerization driven trans-autophosphorylation is a common stereotype of cell signaling, Ire1 does not signal downstream via phosphorylation. Rather, a conformational change that occurs upon nucleotide binding acts as a molecular switch to activate Ire1’s RNase domain (Papa et al. 2003; Aragon et al. 2009; Korennykh et al. 2009). Once activated, Ire1’s RNase specifically cleaves its mRNA substrate, HAC1 mRNA (homolog of ATF/CREB1) in yeast or XBP1 mRNA (X-box binding protein 1) (Cox and Walter 1996; Yoshida et al. 2001) in metazoan cells to initiate an unconventional splicing reaction. After Ire1 removes the intron, the severed exons are ligated by tRNA ligase in yeast and an unknown ligase in metazoan cells, and the mature mRNA is translated to produce a potent bZIP transcription factor that up-regulates genes encoding ER quality control components (Sidrauski et al. 1996; Travers et al. 2000; Yoshida et al. 2001).
The enzymatic activity of the purified kinase/RNase domain of Ire1 is highly cooperative, indicating that full RNase activity is only obtained on assembly of more than two Ire1 molecules (Korennykh et al. 2009; Li et al. 2010). Crystal structures of the yeast kinase/RNase domains suggest that Ire1 first associates into back-to-back dimers, followed by assembly into higher-order oligomers that stabilize the RNase active site through oligomer-specific protein/protein interfaces (Lee et al. 2008; Korennykh et al. 2009). This higher-order assembly is facilitated by both the oligomerization initiated by the lumenal domain and the active, nucleotide-bound conformation of the kinase. Ire1 mutants impaired in nucleotide binding lose RNase activity, whereas mutants impaired in phosphotransfer, but retaining nucleotide binding, maintain RNase activity (Shamu and Walter 1996; Chawla et al. 2011; Rubio et al. 2011).
To date, Ire1 remains the only validated substrate of its kinase domain. Although only nucleotide binding and not phosphorylation is required for HAC1 mRNA cleavage, the absence of phosphorylation does affect the kinetics of the UPR, leading to prolonged HAC1 mRNA splicing and delayed disassembly of Ire1 foci (Chawla et al. 2011; Rubio et al. 2011). These results suggest that phosphorylation plays a role in deactivation of Ire1 by destabilizing Ire1 oligomers, perhaps through electrostatic repulsion from the high local concentration of negatively charged phosphate groups (Rubio et al. 2011). Initial phosphorylation events, however, may promote oligomer formation (Lee et al. 2008). The deactivation of Ire1 signaling is important for physiology because sustained HAC1 mRNA splicing impairs cell survival (Chawla et al. 2011; Rubio et al. 2011). Phosphorylation may play an expanded role in metazoan cells in which adaptor protein docking to phosphorylation sites may mediate branching of the signaling pathway. For example, interactions between phosphorylated IRE1 and the adaptor protein TRAF2 ultimately lead to JNK (Jun amino-terminal kinase) activation; sustained JNK activity induces proapoptotic signaling (Urano et al. 2000).
Although HAC1 mRNA is the only known RNA substrate of yeast Ire1 (Niwa et al. 2005), metazoan IRE1 not only mediates XBP1 mRNA splicing but also the rapid degradation of a subset of mRNAs (Hollien and Weissman 2006). This pathway, termed Regulated Ire1-Dependent Decay (RIDD), cleaves mRNAs encoding membrane and secreted proteins and may consequently decrease the protein influx into the ER. Substrate specificity of this more promiscuous Ire1-mediated cleavage and decay appears to be mediated by both localization of the mRNA to the ER membrane and low stringency consensus sites. Intriguingly, in vivo experiments show that the specific XBP1 mRNA cleavage and RIDD modes of RNase activity can be uncoupled. In mammalian cells, an ATP competitive analog (1NM-PP1) can specifically activate a drug-sensitized Ire1 mutant, but this method of activation only induces XBP1 mRNA splicing but not RIDD (Han et al. 2009; Hollien et al. 2009). It remains to be shown whether and how Ire1 switches between XBP1-specific and RIDD modes of activity. The state of oligomerization may determine the stringency of the RNase for substrate recognition.
PERK
PERK is a type I transmembrane protein present in metazoans that partially resembles Ire1. PERK’s lumenal stress-sensing domain is structurally and functionally related to Ire1’s, though the sequence identity is low, and PERK also contains a cytosolic kinase domain that undergoes trans-autophosphorylation in response to ER stress. Unlike Ire1, however, PERK also phosphorylates the eukaryotic translation initiation factor eIF2α. Phosphorylation of eIF2α results in a reduction of general protein synthesis and thus a decrease in the load of proteins entering the ER (Harding et al. 1999). Under such conditions, mRNAs containing inhibitory upstream open reading frames in their 5′-untranslated region are preferentially translated (Jackson et al. 2010). One such mRNA encodes the transcription factor ATF4 that activates downstream UPR _target genes, including GADD34 (growth arrest and DNA damage-inducible 34) and CHOP (transcription factor C/EBP homologous protein) (Harding et al. 2000; Scheuner et al. 2001). GADD34 encodes the regulatory subunit of the protein phosphatase PP1C complex that dephosphorylates eIF2α (Novoa et al. 2001), comprising a negative feedback loop to reverse the translational attenuation mediated by PERK. The downstream transcription factor CHOP activates genes involved in apoptosis (Wang et al. 1998; Zinszner et al. 1998). Thus, the PERK branch first mediates a prosurvival response, which switches into a proapoptotic response on prolonged ER stress (Walter and Ron 2011).
ATF6
ATF6 is an additional sensor in metazoan cells responsible for ER stress-induced ER expansion and up-regulation of chaperones, foldases, and components of the ERAD pathway (Adachi et al. 2008; Bommiasamy et al. 2009). Unlike Ire1 and PERK, ATF6 is a type II transmembrane protein with a carboxy-terminal stress-sensing lumenal domain and an amino-terminal bZip transcription factor domain. In response to ER stress, ATF6 transits from the ER to the Golgi, in which it is proteolyzed sequentially by the site-1 and site-2 proteases (S1P and S2P) to release its amino-terminal transcription factor domain from the membrane (Haze et al. 1999). The released amino terminus (ATF6-N) translocates to the nucleus where it binds to the ER stress response element (ERSE) and activates transcription of UPR _target genes (Yoshida et al. 1998).
THE CHALLENGE OF DETECTING ER STRESS
Conceptually there are many possibilities for how these three sensors could monitor ER stress: redox potential, calcium homeostasis, membrane aberrancy, concentration of unfolded proteins, or availability of folding machinery. In all cases, however, ER stress sensing must be delicately tuned to avoid hypersensitivity to small fluctuations in normal conditions but also respond quickly to legitimate ER stress. In this review, we highlight recent mechanistic insights into how yeast Ire1 detects ER stress and subsequently discuss the implications for ER stress sensing in metazoans.
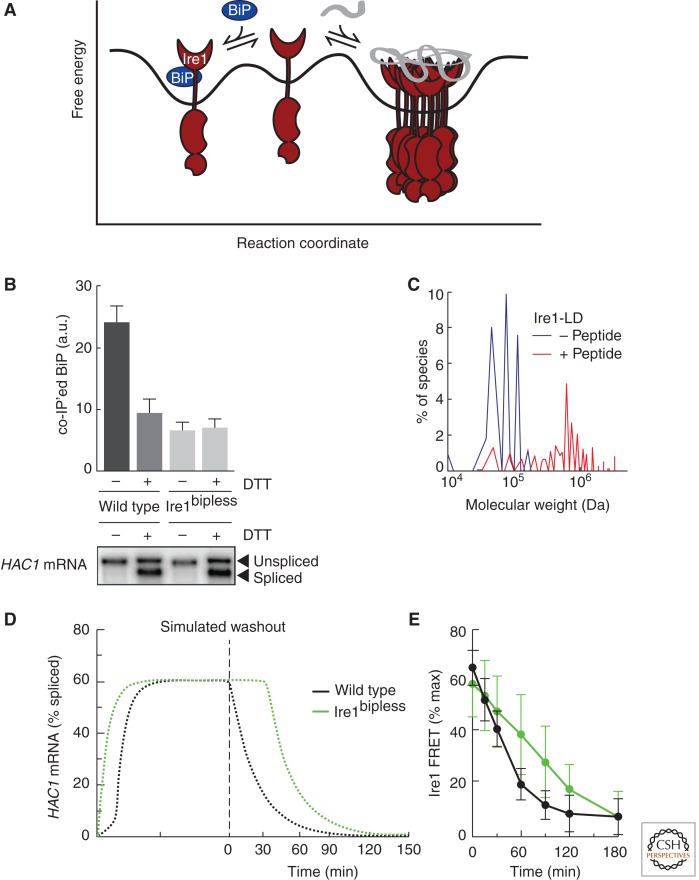
The UPR was first proposed to explain the observation that overexpression of a misfolded protein led to up-regulation of the chaperones BiP and Grp94 (Kozutsumi et al. 1988). Because ER stress-dependent BiP up-regulation operationally defined the UPR, BiP dissociation from Ire1 was originally proposed to be the primary regulatory step for UPR activation. In this model, BiP controls its own expression, and ER stress is monitored by the concentration of free chaperone (Ng et al. 1992; Shamu et al. 1994). Recent evidence, however, favors an alternative mechanism for ER stress sensing: direct binding to unfolded proteins. The crystal structure of Ire1 lumenal domain accompanied by biochemical evidence that Ire1 binds misfolded proteins in vivo and short peptide proxies in vitro strongly suggests that Ire1 can sense ER stress by directly monitoring the concentration of unfolded proteins (Credle et al. 2005; Zhou et al. 2006; Gardner and Walter 2011). Therefore, the higher-order oligomerization of Ire1, which is required for downstream activation, is influenced by two factors: binding to unfolded proteins, which shifts Ire1 toward an active, oligomeric state, and binding to BiP, which stabilizes Ire1 in the inactive, monomeric state (Fig. 2A). Although BiP buffers UPR activity and helps turn Ire1 off, direct binding to unfolded proteins switches Ire1 on.
Figure 2.
Unfolded proteins are the switch that activates Ire1, while BiP binding regulates the sensitivity and duration of the response. (A) Ire1 is in equilibrium between monomeric and oligomeric states. Binding to BiP stabilizes the monomeric state by providing a sink to buffer the amount of free Ire1. Binding to unfolded proteins stabilizes the oligomeric state by overcoming the activation barrier for Ire1 oligomerization. (B) BiP dissociation from Ire1 is not sufficient to activate Ire1 (panel B is from Pincus et al. 2010; reprinted, with permission, from the authors). (Upper panel) Quantification of coimmunoprecipitation experiments showing that BiP dissociates from wild-type Ire1 in the presence of ER stress. A mutant of Ire1 lacking the ER juxtamembrane segment (Ire1bipless) binds to BiP in the absence of stress only to the degree that wild-type Ire1 binds to BiP in the presence of stress. In ER stress conditions, Ire1bipless shows no change in its association with BiP. (Lower panel) Northern blot measuring splicing of HAC1 mRNA showing that, despite its lack of ER-stress-dependent BiP dissociation, the RNase of Ire1bipless is not constitutively active and remains ER-stress inducible. (C) Peptide binding triggers Ire1-LD oligomerization in vitro (panel C is from Gardner and Walter 2011; reprinted, with permission, from the authors). Velocity sedimentation analysis of recombinant Ire1-LD in the presence and absence of a peptide proxy for an unfolded protein shows that Ire1-LD redistributes into large oligomeric assemblies in the presence of peptide. (D) Mathematical model simulation predicts that Ire1bipless would display delayed deactivation kinetics compared to wild-type Ire1 once ER stress is removed (panel D is from Pincus et al. 2010; reprinted, with permission, from the author). (E) Experimental verification that Ire1bipless deactivates less efficiently than wild-type Ire1 once ER stress is removed (panel E is from Pincus et al. 2010; reprinted, with permission, from the authors). FRET measurements between Ire1 molecules reveals that deoligomerization is impaired in the Ire1bipless mutant.
BIP DISSOCIATION IS NOT THE SWITCH FOR IRE1 ACTIVATION
Three observations led to the original proposal that Ire1 monitors levels of free BiP in the ER and induces the UPR as a feedback mechanism to restore BiP levels during ER stress. First, BiP overexpression diminished UPR signaling; second, decreasing the concentration of BiP in the ER activated the UPR; and third, accumulated misfolded proteins in the ER that do not interact with BiP failed to induce the UPR (Hardwick et al. 1990; Dorner et al. 1992; Ng et al. 1992; Kohno et al. 1993). Analogous to how cytoplasmic chaperone hsp70 regulates Heat Shock Factor (HSF1) in eukaryotic cells—on heat shock hsp70 is titrated away from HSF1—it was proposed that BiP binding sequesters Ire1 in an inactive state (Abravaya et al. 1992; Shamu et al. 1994). In ER stress conditions, accumulated unfolded proteins in the ER would saturate the free pool of chaperones, titrating BiP away from Ire1 to activate UPR signaling.
BiP titration as a switch of Ire1 activation gained further support when it was shown that BiP binds to Ire1, PERK, and ATF6 in unstressed cells and dissociates from the UPR sensors during acute ER stress (Bertolotti et al. 2000; Okamura et al. 2000; Shen et al. 2002). The effect of BiP mutations on Ire1 activity were also consistent with a role for BiP as a negative regulator of Ire1 activity. BiP mutants that constitutively bound Ire1 hindered Ire1 activation during ER stress, suggesting that BiP release is required for activation. BiP mutants defective in Ire1 binding either showed constitutive activation of Ire1 (Kimata et al. 2003) or showed an inability to negatively regulate Ire1 (Todd-Corlett et al. 2007). However, because BiP is essential for protein translocation and folding in the ER, BiP mutants are plagued with pleiotropic effects and cause varying degrees of constitutive ER stress. Thus, it is difficult to decipher whether the effect of BiP mutants on Ire1 activation results from the change in association with Ire1 or altered conditions in the ER.
By contrast, a mutational analysis of Ire1’s lumenal domain in yeast mapped the BiP binding site to the juxtamembrane segment (Kimata et al. 2004). Although removal of this region abolished ER stress-regulated BiP binding, the Ire1 mutant remained inactive in the absence of ER stress and retained its stress-dependent activation (Fig. 2B). Though this Ire1 mutant was hyper-responsive to heat shock and ethanol, it became evident that BiP is not the primary regulator of Ire1 activity.
STRUCTURAL HINTS OF DIRECT BINDING TO UNFOLDED PROTEINS
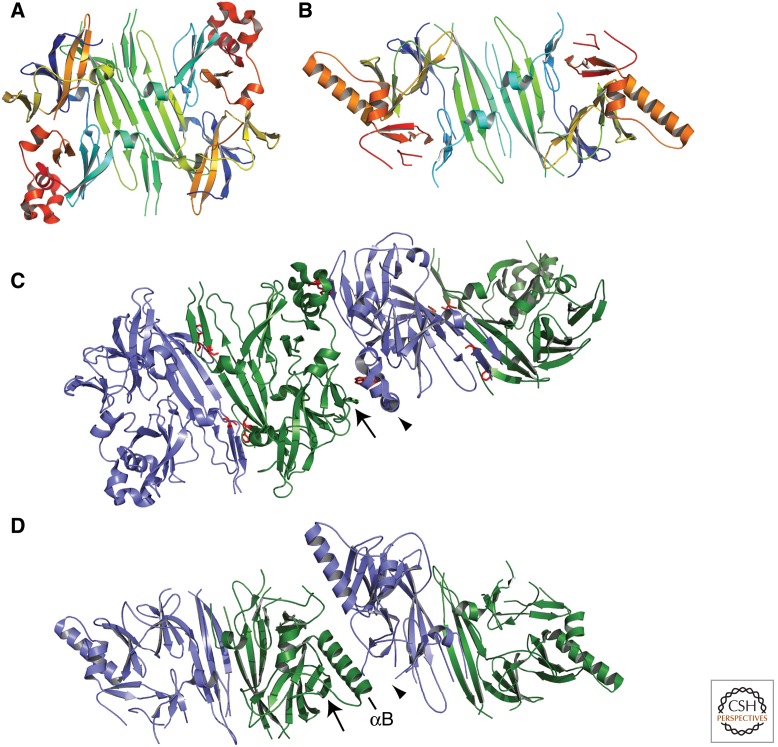
An alternative view of Ire1 activation posits that a ligand produced by the accumulation of unfolded proteins directly binds and activates Ire1 (Shamu et al. 1994). There was little evidence for ligand mediated Ire1 activation until the crystal structure of the lumenal domain of Ire1 (Ire1-LD) from Saccharomyces cerevisiae revealed that the dimer of Ire1-LD forms a deep groove, suggesting a ligand-binding site (Fig. 3A) (Credle et al. 2005). The architecture of this groove, a floor of β-sheets with α-helical walls, resembles the peptide-binding groove of major histocompatibility complexes (MHC) (Fig. 4A,C). Because of this similarity, it was proposed that unfolded proteins are Ire1-activating ligands.
Figure 3.
Crystal structure of yeast and human Ire1-LD indicate how Ire1 oligomerizes. (A) The S. cerevisiae Ire1-LD dimer formed at Interface 1, creating a putative ligand-binding groove (PDB: 2BE1). Each monomer is colored red to blue from amino to carboxyl terminus. Compare to Figure 4A for a surface representation. (B) The H. sapiens Ire1-LD dimer formed at Interface 1 has similar architecture to yeast Ire1-LD, though the α-helical walls narrow the putative ligand binding groove (PDB: 2HZ6). Each monomer is colored red to blue from amino to carboxyl terminus. Compare to Figure 4B for a surface representation. (C) Two groove-forming dimers of yeast Ire1-LD oligomerize through Interface 2. An α-helical turn region (arrowhead) interacts with a β-sheet (arrow) to mediate this interaction. Residues that have been mutated and shown to impair UPR activation are colored red. (D) Two groove-forming dimers of human Ire1-LD interact through a novel Interface 2. The interaction between the β-sheet (arrow) and the α-helical turn region, unstructured here (arrowhead), that formed Interface 2 in the yeast Ire1-LD is now interrupted by the long helix αB. Figures were made using PyMOL (PyMOL Molecular Graphics System).
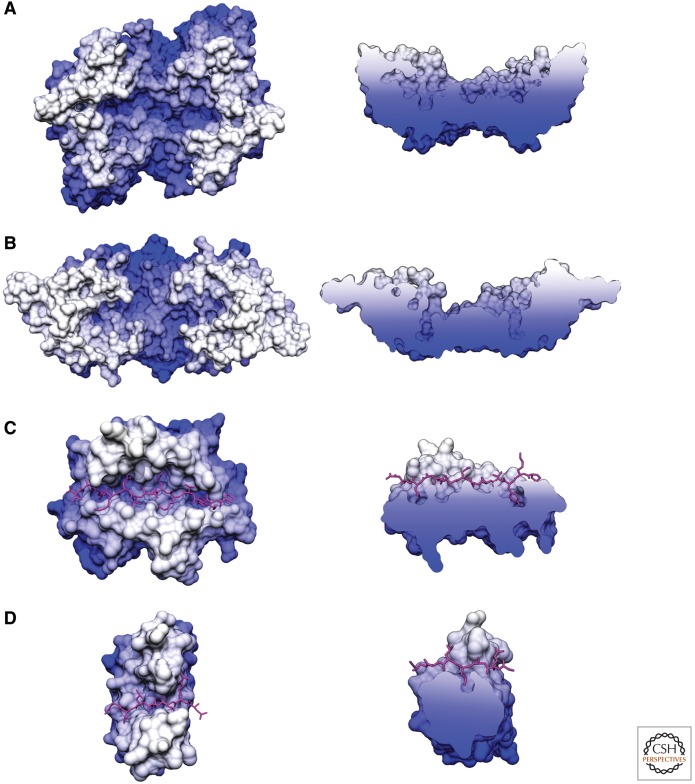
Figure 4.
Crystal structures of Ire1-LD reveal a putative ligand-binding groove similar to those of MHC and DnaK. Surface models of the structures of (A) yeast Ire1-LD, (B) human Ire1-LD, (C) MHCII peptide binding domain crystallized with a peptide ligand (magenta; PDB: 1DLH), and (D) DnaK substrate binding domain crystallized with a peptide ligand (magenta; PDB: 1DKX). All structures are colored white to blue by depth looking down on the proposed or proven ligand-binding groove. Capped side views show a cross section of the ligand-binding groove to illustrate the pockets available to bind anchor residues.
Although no ligand was cocrystallized with yeast Ire1-LD, this structure featuring an open and obvious groove is thought to represent the active form of Ire1 (Fig. 4A). In support of this notion, Ire1 forms oligomers at two interfaces within the crystal lattice (Fig. 3C) (Credle et al. 2005). Interface 1 buries 2380 Å2 of solvent-accessible surface and is formed by hydrogen bonding and hydrophobic interactions between two central, antiparallel β-sheets. The dimer created at Interface 1 forms the putative peptide-binding groove (Figs. 3A, 4A). Interface 2 relates these dimers in a back-to-back fashion, burying 2117 Å2 (Fig. 3C). Mutations within either of these interfaces prevent Ire1 foci formation, HAC1 mRNA splicing, and activation of UPR _target genes, indicating the functional importance of oligomerization through these interfaces in vivo (Credle et al. 2005; Aragon et al. 2009). Ligand binding across the proposed groove would directly stabilize the dimer formed at Interface 1, as well as the conformation required for oligomerization through Interface 2.
BIOCHEMICAL EVIDENCE OF UNFOLDED PROTEIN BINDING TO IRE1
There is increasing biochemical evidence to support the structure-based view that unfolded proteins are Ire1-activating ligands. Purified yeast Ire1-LD inhibits the aggregation of unfolded proteins in vitro, potentially by binding and shielding regions that would otherwise cause aggregation (Kimata et al. 2007). Ire1 also coimmunoprecipitates with a model misfolded protein known to activate the UPR (carboxypeptidase Y, CPY*) (Gardner and Walter 2011). Further coimmunoprecipitation experiments suggest that the interaction between Ire1 and CPY* depends on the formation of the proposed ligand-binding groove: mutations disrupting Interface 1 and residues within the groove abolish the interaction between Ire1 and CPY*, while mutations in Interface 2, which only disrupt oligomerization, do not affect the interaction between Ire1 and CPY* (Promlek et al. 2011).
Furthermore, analysis of Ire1-LD binding to peptides derived from CPY* showed that there are discrete Ire1 binding sites within CPY* (Gardner and Walter 2011). These results suggest that Ire1 ligands are unstructured proteins with sufficient flexibility to sample the binding groove and that multiple ligands can be displayed as part of a single unfolded protein. Moreover, Ire1 discriminates among peptide sequences and displays clear amino acid preferences. Systematic mutational analysis of a binding peptide revealed that yeast Ire1-LD preferentially binds to regions containing basic and hydrophobic residues. Although similar sequence preferences are also displayed by some chaperones, a comparison of peptides bound by BiP and Ire1 revealed that they each bind a separate, but overlapping set of peptides (Gardner and Walter 2011). This indicates that BiP and Ire1 do not always compete for ligands and that BiP saturation is not required for Ire1 activation.
INSIGHTS INTO THE MECHANISM OF LIGAND BINDING
As of yet, no experiments verify the groove as the ligand-binding site within Ire1-LD. Notably, however, residues on the β-sheet floor with side-chains that point into the groove are more conserved than adjacent ones that point away. Mutations of three of these conserved residues, M229A, F285A, and Y301A, inhibit the activation of Ire1 in vivo, prevent coimmunoprecipitation with CPY*, and diminish binding to unfolded proteins and peptides in solution, strongly indicating that the groove is important for ligand binding (Credle et al. 2005; Kimata et al. 2007; Gardner and Walter 2011; Promlek et al. 2011). Moreover, in silico docking shows that a peptide ligand seeks out the groove as a highly preferred binding site in the yeast Ire1-LD crystal structure (BM Gardner, A Korennykh, and P Walter, unpubl.).
Similar to DnaK and MHC molecules, Ire1 must bind a variety of ligands while remaining selective for unfolded proteins (DnaK, Ire1) or extended peptides (MHC). DnaK and MHCII, which have both been crystallized with ligands (Fig. 4C,D), may provide insight into how Ire1 binds a variety of unfolded proteins within the same binding pocket. Sidechains of DnaK and MHCII form extensive hydrogen bond networks with the backbone of the ligand, ensuring binding to extended polypeptides with little stringency for ligand character. The limited specificity for ligand residues arises from pockets with variable volume, charge, and available stacking interactions. Deeper pockets specific to large hydrophobic side-chains anchor the peptide, whereas shallower pockets can accommodate a wider range of residues (Stern et al. 1994; Murthy and Stern 1997).
Ire1 could combine all three of these characteristics to bind unfolded proteins: hydrogen bonding to the polypeptide backbone, electrostatic interactions, and the burial of anchor residues in deep pockets. Indeed, Ire1 displays side-chains across the groove capable of hydrogen bonding to the polypeptide backbone. Ire1’s preference for basic residues may be mediated by two acidic residues (E172, D176) along the sides of the groove that are highly conserved among fungi or through acidic residues in an unstructured loop (aa 380–387). Finally, the proposed ligand-binding groove of Ire1 contains deep pockets on either end that could bind specific side-chains and anchor ligand-binding (Fig. 4A). These pockets are particularly deep and contain several chambers that may select for specific adjacent side-chains, perhaps basic and hydrophobic residues.
LIGAND-BINDING TRIGGERS IRE1 OLIGOMERIZATION
As mentioned previously, the oligomerization of Ire1-LD activates the cytosolic kinase domain and highly cooperative RNase domain (Korennykh et al. 2009). Analytical ultracentrifugation experiments in the presence of a short peptide with a high affinity for Ire1-LD revealed that peptide binding alters the oligomeric state of Ire1-LD, causing higher-order oligomerization (Fig. 2C). By mutating Interface 2, the peptide-induced oligomerization can be limited to dimerization (Gardner and Walter 2011). These experiments show that binding to a short peptide, a proxy for an unfolded protein, stabilizes the dimer of Ire1 in an oligomerization competent state, therefore driving Ire1 activation.
THE EQUILIBRIUM MODEL OF UPR ACTIVATION
The regulatory contribution of BiP binding and unfolded protein binding can be viewed as opposing forces on Ire1’s oligomeric equilibrium (Fig. 2A). Although ligand-induced oligomerization activates Ire1, BiP association stabilizes the inactive, monomeric form of Ire1 preventing Ire1 from hyper-responding to low levels of ER stress (Korennykh et al. 2009; Pincus et al. 2010; Gardner and Walter 2011). Computational simulations of this equilibrium model remarkably recapitulate the observed dynamics of the UPR in yeast and illustrate the importance of BiP regulation. In the absence of BiP binding, Ire1 molecules are poised closer to the activation threshold, but are not constitutively active without further stimulus. Moreover, as predicted by the computational model (Fig. 2D), in the absence of BiP binding, Ire1 displayed delayed deactivation and deoligomerization kinetics when ER stress was removed (Fig. 2E) (Pincus et al. 2010). Further refinement of the model takes into account that the activity of BiP is dynamically modified by posttranslational modification (Chambers et al. 2012). Therefore, the combination of buffering by BiP and cooperative activation of Ire1 achieves an appropriate sensitivity to ER stress while maintaining a robust and speedy response.
OTHER REGULATION OF IRE1 ACTIVATION
BiP and unfolded proteins may not be the only factors to influence Ire1 activation. Additional layers of regulation could arise through alternate inputs detected either by Ire1-LD or other domains. Several studies of Ire1 in which the LD was removed and replaced with a leucine zipper show that Ire1 still responds to ER stress, although the response is delayed and less robust (Liu et al. 2000; Promlek et al. 2011). Although the mechanism of this activation is unclear, it has been proposed that Ire1’s transmembrane domain is sensitive to aberrations in the ER membrane. Furthermore, biochemical and structural studies of the yeast cytoplasmic domain of Ire1 identified a small ligand-binding pocket on Ire1’s cytosolic face that binds quercetin, a naturally occurring flavonol, and stabilizes the active RNase dimer (Wiseman et al. 2010). Presumably any mechanism of stabilizing Ire1 oligomers, including limited membrane diffusion or increased affinity at cytoplasmic interfaces, could contribute to Ire1 activation.
ER STRESS SENSING BY METAZOAN UPR SENSORS
ER stress sensing by the three metazoan sensors, IRE1, PERK, and ATF6, is less understood. During ER stress, all three sensor proteins undergo a change in oligomerization state, which is accompanied by BiP dissociation. IRE1 and PERK oligomerize to activate their cytosolic domains, whereas ATF6 deoligomerizes in response to ER stress. We predict that the principles of yeast Ire1 activation are maintained for the homologous metazoan sensors, IRE1 and PERK. The mechanism of ATF6 activation remains elusive, and although it may also be buffered by BiP binding, its downstream activation is more analogous to that of SREBP than Ire1 or PERK.
IRE1 and PERK
The secondary structure homology between yeast Ire1 and metazoan IRE1 and PERK indicates that they may have similar activation mechanisms in which they directly bind unfolded proteins but are also regulated by BiP association. Indeed, PERK and IRE1 lumenal domains from different species are experimentally interchangeable and can substitute for yeast Ire1-LD (Bertolotti et al. 2000; Liu et al. 2000). Similar to yeast Ire1, PERK and IRE1 residues required for disulfide bond formation and glycosylation are dispensable for ER stress sensing (Ma et al. 2002; Liu et al. 2003; Oikawa et al. 2005, 2009). BiP binds to both IRE1 and PERK in the juxtamembrane region and dissociates during ER stress when IRE1 and PERK form higher-order complexes (Bertolotti et al. 2000; Liu et al. 2000). For IRE1, this higher-order oligomerization has been observed in vivo as foci formation, and the cooperativity of the endoribonuclease of human IRE1 indicates that it is required for robust Ire1 activation (Li et al. 2010). Mutants of IRE1 and PERK in which the BiP interaction is impaired show higher basal level activation (Ma et al. 2002; Oikawa et al. 2009). In the case of IRE1, the BiP-binding impaired mutant retained significant stress-dependent induction, indicating that BiP binding and dissociation is not the only layer of IRE1 regulation. The importance of BiP dissociation from endogenous IRE1 PERK requires further characterization.
The crystal structure of human IRE1-LD is remarkably similar to that of yeast Ire1-LD with a few key differences (Fig. 3B) (Zhou et al. 2006). The core structure of a dimer forming a putative ligand-binding groove with a β-sheet platform and α-helical walls is conserved. However, although a groove is still present, it is not as deep as the yeast counterpart because the α-helices are closer together and block the continuous, deep groove across the dimer (Figs. 3B, 4B). Because of this, it was suggested that human IRE1-LD is unlikely to bind unfolded proteins (Zhou et al. 2006), even though the remaining shallow groove contains deep pockets that could still bind anchor residues as a ligand arches over the helices.
Given the overall conservation of the Ire1-LD structure, we find it more likely that these differences represent distinct conformational steps in the same pathway of Ire1 activation. Perhaps the yeast structure is an open, oligomeric, active conformation, and the human structure is a closed, dimeric, inactive conformation. Indeed, Interface 2, which is required for oligomerization and activity of yeast Ire1-LD, is not observed in the human structure, consistent with it being an inactive conformation. The formation of Interface 2 between a β-sheet and an α-helical turn region (arrows, Fig. 3C,D) is sterically hindered by the long helix αB in the human structure, which forms a new, smaller interface that has not been tested in vivo. The αB-helix corresponds to an unresolved region in the yeast LD structure, whereas an α-helical turn region involved in Interface 2 is alternately unstructured in the human structure. These changes may suggest the conformational change observed on ligand binding.
If PERK and IRE1 do indeed bind unfolded proteins, it will be interesting to see how the altering characteristics of residues within the groove may have redefined their ligand preference. Specialization of the recognition code illustrated by yeast Ire1 would allow some sensors to be “blind” to certain unfolded proteins, yet acutely sensitive to others. On the other hand, sensors could have evolved to be particularly sensitive to certain markers of ER stress or “canary” proteins. Hypothetically, pathogens or developmental programs could also exploit this binding specificity in higher organisms.
ATF6
The lumenal domain of ATF6 bears no sequence homology to those of Ire1 or PERK, and its activation is strikingly different, relying on a change in subcellular localization and intramembrane proteolysis. Analogous to Ire1 and PERK, ATF6 activation is associated with both a change in oligomerization state and BiP dissociation. Unlike Ire1 and PERK, ATF6 is an oligomer in the ER of unstressed cells (Nadanaka et al. 2007). Conversion from an oligomer to a monomer may be important for activation as the oligomer is not found in the Golgi and, when compared to the monomer, is a poor substrate for S1P cleavage. Deoligomerization is not sufficient for ATF6 activation, however, because mutation of both conserved cysteines, which mediate oligomerization, did not lead to constitutive trafficking of ATF6 to the Golgi (Nadanaka et al. 2007). During ER stress, BiP dissociates from the lumenal domain of ATF6 (Shen et al. 2002, 2005). BiP dissociation is not sufficient for activation as deletion of most of the lumenal domain, and therefore BiP binding, did not lead to constitutive trafficking of ATF6.
Mechanistically, the activation of ATF6 in response to ER stress is more analogous to sterol response element binding protein (SREBP), which also transits to the Golgi where its amino-terminal transcription factor is released by S1P and S2P-mediated proteolysis. SREBP’s transit to the ER is regulated by the interaction between two binding proteins, SCAP (SREBP cleavage-activating protein), which acts as a positive transport factor, and INSIG (insulin-induced gene), which acts as a retention factor by masking the COPII sorting signal in SCAP (Sun et al. 2007). The interaction between SCAP and INSIG is regulated by cholesterol—the absence of cholesterol induces a conformational change in SCAP leading to its dissociation from INSIG (Adams et al. 2004; Feramisco et al. 2005). Therefore, it is SCAP and not SREBP that senses cholesterol levels in the membrane. Although SCAP is not required (Ye et al. 2000), it remains to be seen if activation of ATF6 during ER stress requires a specific interacting partner or whether ATF6 lumenal domain functions as an intrinsic stress sensor.
CONCLUDING REMARKS
In conclusion, there is increasing evidence that direct binding to unfolded proteins triggers Ire1 activation in S. cerevisiae by inducing a conformational change to promote oligomerization. BiP dissociation from Ire1 fine-tunes this oligomeric equilibrium, regulating the sensitivity and duration of UPR activation. The mechanics of Ire1 binding to unfolded proteins requires further research to detail both ligand specificity and conformational changes. Structural homology suggests that the mechanism of IRE1 and PERK activation have been conserved, though they may have specialized to detect different types of unfolded proteins or integrate other characteristics of ER stress. We anticipate that the ER stress-sensing mechanisms of metazoan cells are precisely tuned to accommodate the specialized needs of the organism. A detailed understanding of these mechanisms could direct the design of small molecule modulators to specifically modulate branches of the UPR for therapeutic benefit.
ACKNOWLEDGMENTS
P.W. is an investigator of the Howard Hughes Medical Institute. Molecular graphics and analyses were performed either with The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC or with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the National Institutes of Health (National Center for Research Resources grant 2P41RR001081 and National Institute of General Medical Sciences grant 9P41GM103311).
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
- Abravaya K, Myers MP, Murphy SP, Morimoto RI 1992. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev 6: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K 2008. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct 33: 75–89 [DOI] [PubMed] [Google Scholar]
- Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL 2004. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and insigs. J Biol Chem 279: 52772–52780 [DOI] [PubMed] [Google Scholar]
- Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P 2009. Messenger RNA _targeting to endoplasmic reticulum stress signalling sites. Nature 457: 736–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki K, Nagata K 2011. Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol 3: a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2: 326–332 [DOI] [PubMed] [Google Scholar]
- Bommiasamy H, Back SH, Fagone P, Lee K, Meshinchi S, Vink E, Sriburi R, Frank M, Jackowski S, Kaufman RJ, et al. 2009. ATF6 α induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci 122: 1626–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D 2012. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol 198: 371–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Chakrabarti S, Ghosh G, Niwa M 2011. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J Cell Biol 193: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404 [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci 102: 18773–18784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross BC, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, et al. 2012. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci 109: E869–E878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, Kaufman RJ 1992. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J 11: 1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feramisco JD, Radhakrishnan A, Ikeda Y, Reitz J, Brown MS, Goldstein JL 2005. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc Natl Acad Sci 102: 3242–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribley AM, Cruz PG, Miller JR, Callaghan MU, Cai P, Narula N, Neubig RR, Showalter HD, Larsen SD, Kirchhoff PD, et al. 2011. Complementary cell-based high-throughput screens identify novel modulators of the unfolded protein response. J Biomol Screen 16: 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Walter P 2011. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333: 1891–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR 2009. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell 138: 562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang YH, Ron D 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274 [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Lewis MJ, Semenza J, Dean N, Pelham HR 1990. ERD1, a yeast gene required for the retention of luminal endoplasmic reticulum proteins, affects glycoprotein processing in the Golgi apparatus. EMBO J 9: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10: 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM 2004. The ER function BiP is a master regulator of ER function. Mt Sinai J Med 71: 289–297 [PubMed] [Google Scholar]
- Hollien J, Weissman JS 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107 [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K 2003. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell 14: 2559–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K 2004. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol 167: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K 2007. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol 179: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol 13: 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P 2009. The unfolded protein response signals through high-order assembly of Ire1. Nature 457: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332: 462–464 [DOI] [PubMed] [Google Scholar]
- Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F 2008. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P 2010. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci 107: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science 318: 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem 275: 24881–24885 [DOI] [PubMed] [Google Scholar]
- Liu CY, Xu Z, Kaufman RJ 2003. Structure and intermolecular interactions of the luminal dimerization domain of human IRE1α. J Biol Chem 278: 17680–17687 [DOI] [PubMed] [Google Scholar]
- Ma K, Vattem KM, Wek RC 2002. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J Biol Chem 277: 18728–18735 [DOI] [PubMed] [Google Scholar]
- Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, Santo L, Hu Y, Fabre C, Minami J, Ohguchi H, et al. 2012. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood 119: 5772–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K 2009. Signalling pathways in the unfolded protein response: Development from yeast to mammals. J Biochem 146: 743–750 [DOI] [PubMed] [Google Scholar]
- Murthy VL, Stern LJ 1997. The class II MHC protein HLA-DR1 in complex with an endogenous peptide: Implications for the structural basis of the specificity of peptide binding. Structure 5: 1385–1396 [DOI] [PubMed] [Google Scholar]
- Nadanaka S, Okada T, Yoshida H, Mori K 2007. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol 27: 1027–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Watowich SS, Lamb RA 1992. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol Biol Cell 3: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Patil CK, DeRisi J, Walter P 2005. Genome-scale approaches for discovering novel nonconventional splicing substrates of the Ire1 nuclease. Genome Biol 6: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J Cell Biol 153: 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Takeuchi M, Kohno K 2005. An essential dimer-forming subregion of the endoplasmic reticulum stress sensor Ire1. Biochem J 391: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T 2009. Activation of mammalian IRE1α upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res 315: 2496–2504 [DOI] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K 2000. Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279: 445–450 [DOI] [PubMed] [Google Scholar]
- Otero JH, Lizak B, Hendershot LM 2010. Life and death of a BiP substrate. Semin Cell Dev Biol 21: 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa FR, Zhang C, Shokat K, Walter P 2003. Bypassing a kinase activity with an ATP-competitive drug. Science 302: 1533–1537 [DOI] [PubMed] [Google Scholar]
- Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, et al. 2011. Identification of an Ire1α endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood 117: 1311–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P 2010. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promlek T, Ishiwata-Kimata Y, Shido M, Sakuramoto M, Kohno K, Kimata Y 2011. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell 22: 3520–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio C, Pincus D, Korennykh A, Schuck S, El-Samad H, Walter P 2011. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J Cell Biol 193: 171–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P 1996. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J 15: 3028–3039 [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Cox JS, Walter P 1994. The unfolded-protein-response pathway in yeast. Trends Cell Biol 4: 56–60 [DOI] [PubMed] [Google Scholar]
- Shen JS, Chen X, Hendershot L, Prywes R 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of golgi localization signals. Dev Cell 3: 99–111 [DOI] [PubMed] [Google Scholar]
- Shen JS, Snapp EL, Lippincott-Schwartz J, Prywes R 2005. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol 25: 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P 1996. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87: 405–413 [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS 2011. Road to ruin: _targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC 1994. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368: 215–221 [DOI] [PubMed] [Google Scholar]
- Sun LP, Seemann J, Goldstein JL, Brown MS 2007. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci 104: 6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D 2011. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol 13: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.5.0.4, Schrödinger, LLC [Google Scholar]
- Todd-Corlett A, Jones E, Seghers C, Gething MJ 2007. Lobe IB of the ATPase domain of Kar2p/BiP interacts with Ire1p to negatively regulate the unfolded protein response in Saccharomyces cerevisiae. J Mol Biol 367: 770–787 [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287: 664–666 [DOI] [PubMed] [Google Scholar]
- Volkmann K, Lucas JL, Vuga D, Wang X, Brumm D, Stiles C, Kriebel D, Der-Sarkissian A, Krishnan K, Schweitzer C, et al. 2011. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem 286: 12743–12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D 2011. The unfolded protein response: From stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Wang XZ, Kuroda M, Sok J, Batchvarova N, Kimmel R, Chung P, Zinszner H, Ron D 1998. Identification of novel stress-induced genes downstream of chop. EMBO J 17: 3619–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RL, Zhang YH, Lee KPK, Harding HP, Haynes CM, Price J, Sicheri F, Ron D 2010. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell 38: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6: 1355–1364 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins—Involvement of basic leucine zipper transcription factors. J Biol Chem 273: 33741–33749 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891 [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ 2006. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci 103: 14343–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev 12: 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]