Summary
Activating mutations in BRAF are the most common genetic alterations in melanoma. Inhibition of BRAF by small molecule inhibitors leads to cell cycle arrest and apoptosis. We show here that BRAF inhibition also induces an oxidative phosphorylation gene program, mitochondrial biogenesis, and the increased expression of the mitochondrial master regulator, PGC1α. We further show that a _target of BRAF, the melanocyte lineage factor MITF, directly regulates the expression of PGC1α. Melanomas with activation of the BRAF/MAPK pathway have suppressed levels of MITF and PGC1α, and decreased oxidative metabolism. Conversely, treatment of BRAF mutated melanomas with BRAF inhibitors renders them addicted to oxidative phosphorylation. Our data thus identify an adaptive metabolic program that limits the efficacy of BRAF inhibitors.
Keywords: BRAF, melanoma, vemurafenib, metabolism
Introduction
Activating mutations in the BRAF protein kinase are the most common genetic alterations in melanoma, found in ~50% of tumors (Davies et al., 2002; Curtin et al., 2005). The most frequent BRAF mutation is the substitution of valine at position 600 by glutamic acid (BRAF V600E) that results in the constitutive activation of its serine/threonine kinase activity and sustained activation of MAP kinase signal transduction pathway (Davies et al., 2002; Wan et al., 2004). BRAF directly phosphorylates the dual-specificity kinases MEK1 and MEK2, which in turn phosphorylate and activate the mitogen-activated protein kinases, ERK1 and ERK2. BRAF has been shown by overexpression and knockdown experiments to be a critical mediator of melanomagenesis. In mice, activation of BRAF in combination with deletion of the tumor suppressor genes PTEN or INK4A leads to melanoma with complete penetrance (Dankort et al., 2009; Dhomen et al., 2009). Conversely, treatment of BRAF mutant melanomas in vitro with chemical inhibitors of BRAF or MEK1/2 promotes cell cycle arrest and apoptosis (Hingorani et al., 2003; Karasarides et al., 2004; Hoeflich, 2006; Wellbrock et al., 2008). Moreover, the BRAF inhibitor vemurafenib (PLX4032) leads to tumor regression and improved overall survival in patients whose melanomas have the BRAF(V600E) mutation, leading to its approval as a treatment for patients with metastatic melanoma (Flaherty et al., 2010; Chapman et al., 2011; Sosman et al., 2012). Despite the promise and dramatic initial effects of BRAF inhibitors in the clinic, patients eventually relapse within several months, suggesting that combination therapies may be needed to overcome intrinsic or acquired resistance (Gray-Schopfer et al., 2007; Poulikakos and Rosen, 2011).
Although melanomas with BRAF mutations have constitutively active growth signals, how they sustain their growth in the setting of nutrient scarcity is not well understood. In 1930, Otto Warburg proposed that cancer cells have a high rate of glycolysis as compared to oxidative metabolism even under conditions of high oxygen, a phenomenon known as the Warburg effect (Warburg, 1956; Vander Heiden et al., 2009). Oxidative phosphorylation depends on the ability of functionally intact mitochondria to metabolize oxygen, whereas glycolysis can occur independently of mitochondria. Warburg theorized that this metabolic switch facilitated the uptake and incorporation of nutrients that were required for cellular proliferation. Although poorly understood in melanoma, the molecular mechanisms of metabolic reprogramming in cancer have been described in other tumor types. TP53-deficient tumor cells have diminished levels of the genes TIGAR and SCO2 which regulate glycolysis and assembly of the mitochondrial cytochrome c oxidase complex respectively (Bensaad et al., 2006; Matoba, 2006). Similarly, the dysregulation of the proto-oncogene MYC leads to profound effects on tumor metabolism through multiple mechanisms (reviewed in Dang, 2012).
These observations have raised the possibility of _targeting key metabolic pathways to inhibit cancer growth. Yun et al. demonstrated that several colorectal cancers with KRAS or BRAF mutations have increased glucose uptake and glycolysis and survived better in low glucose conditions compared to non-mutated cell lines (Yun et al., 2009). Suppression of glycolysis with 3-bromopyruvate (a non-active intermediary in the glycolysis pathway) reactivates mitochondrial metabolism in tumor cells, induces their selective killing and suppresses cancer growth. Similarly, suppression of glycolysis by inhibiting conversion of pyruvate to lactate enhances oxidative phosphorylation and suppressed the growth of breast cancer cell lines (Fantin et al., 2006; Bonnet et al., 2007). Thus, while metabolic reprogramming is commonly found in cancer, the mechanism and details of the metabolic alterations in melanoma are unknown.
Mitochondrial biogenesis and oxidative phosphorylation are well known to be controlled by the members of the peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1) family of transcriptional coactivators (PGC1α, PGC1β, PPRC1) (Lin et al., 2005a; Finck, 2006; Handschin, 2009; Leone and Kelly, 2011). The best-studied PGC-1 family member, PGC1α, potently activates coordinated gene expression programs by interacting with transcription factors, the basal transcriptional machinery, histone-modifying enzymes, and the RNA splicing machinery. PGC1α drives mitochondrial biogenesis in multiple contexts, including brown and white adiocytes (Wu et al., 1999; Uldry et al., 2006), skeletal muscle (Lin et al., 2002) and heart (Lehman et al., 2000). PGC1α mRNA expression is sensitive to numerous signaling inputs that are often implicated in cancer biology (Herzig et al., 2001; Yoon et al., 2001; Chinsomboon et al., 2009; Arany et al., 2008). PGC1β shares significant sequence homology and functional overlap with PGC1α, including the activation of mitochondrial biogenesis and oxidative phosphorylation, but also has several distinct functions in different tissues (Lin et al., 2005b; Uldry et al., 2006; Wolfrum and Stoffel, 2006). The regulation of PGC1β has been less extensively studied. Although little is known about the role of the PGCs in melanoma, they have recently been implicated in metabolic shifts in breast cancer cells and colon cancers (Eichner et al., 2010; Bhalla et al., 2011; Sahin et al., 2011; Wang and Moraes, 2011; Girnun, 2012; Klimcakova et al., 2012). Here we comprehensively evaluate the effect of BRAF pathway activation on metabolic gene expression and function in melanoma.
Results
BRAF regulates metabolic reprogramming of melanomas
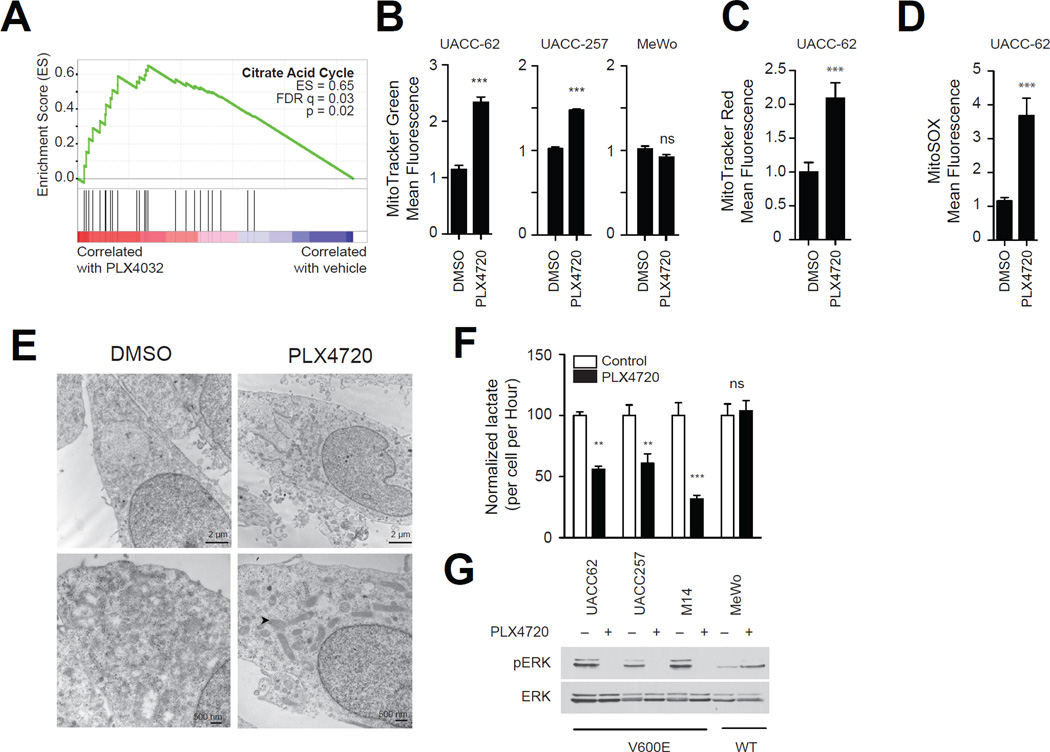
The mechanisms by which oncogenic BRAF promotes oncogenesis are incompletely understood. Gene Set Enrichment Analysis (GSEA) provides a bioinformatics approach to identify gene signatures among microarray datasets that are induced or suppressed as small coordinated changes in individual genes (Mootha et al., 2003). In order to identify gene expression programs altered by oncogenic BRAF, we evaluated previously published gene expression profiles of BRAF mutant melanomas treated with the BRAF inhibitor vemurafenib (Joseph et al., 2010). As shown in Figure 1a, treatment of BRAF mutant melanomas with vemurafenib resulted in significant increases in the expression the citric acid cycle gene set (Figure 1a) as well as multiple oxidative phosphorylation and ATP synthesis gene sets (Table S1). Similarly, melanoma cells treated with PD0325901, a pre-clinical inhibitor of the MEK1/2 protein kinases (Pratilas et al., 2009; Joseph et al., 2010), also exhibited a trend towards induction of the citric acid cycle and oxidative phosphorylation gene sets (Table S2). In contrast, we did not find enrichment of oxidative phosphorylation, or citric acid cycle gene sets in BRAF-mutant non-melanomas treated with PD0325901 (Table S3). We validated the effects of vemurafenib on OXPHOS gene expression in three melanoma cell lines by quantitative PCR (Figure S1a,b,c).
Figure 1.
BRAF inhibitors induce mitochondrial biogenesis and oxidative metabolism. (A) Gene Set Enrichment Analysis plot of melanoma cells treated with vemurafenib showing the most significantly changed gene set. FDR, false-discovery rate; ES, enrichment score. (B) MitoTracker Green fluorescence of BRAF mutant (UACC-62 and UACC-257) or BRAF wild-type (MeWo) melanoma cell lines treated with PLX4720 (1µM, 72h) and subjected to analysis by flow cytometry. (C,D) MitoTracker Red fluorescence (C) or MitoSOX fluorescence (D) of UACC-62 cells treated with PLX4720. (E) Electron micrographs of UACC-62 cells treated with PLX4720 or vehicle control. Representative photographs of cells at 22000× (upper panel) or 44000× (lower panel) are shown. Arrows indicate mitochondria. (F) Lactate levels in media conditioned from the indicated cell lines treated with PLX4720 or vehicle control for 16 hours. (G) Cells in part (F) were concomitantly evaluated for ERK activity by Western blotting using phospho-ERK antibodies. Error bars represent SEM of at least three independent replicates. ***, p < 0.001; **, p < 0.01. See also Table S1 and Figure S1.
To directly evaluate the effect of BRAF inhibition on oxidative phosphorylation and mitochondrial number and function, we treated melanoma cell lines with PLX4720, a preclinical analog of vemurafenib (Tsai et al., 2008). PLX4720 increased the mitochondrial density of two BRAF mutant melanomas as detected by MitoTracker Green, which localizes to mitochondria independent of membrane potential. PLX4720 did not affect the mitochondrial density of MeWo cells that are BRAF wild-type (Figure 1b). PLX4720 also induced MitoTracker Red fluorescence, a measure of mitochondrial activity and mass (Figure 1c), and increased the production of mitochondrial oxidative stress measured by the MitoSOX fluorescence assay (Figure 1d). To confirm these findings, we also evaluated mitochondrial number by electron microscopy after treatment of the BRAF mutant melanoma cell line UACC62 with PLX4720. As seen in Figure 1e and Figure S1d, inhibition of BRAF led to a significant increase in the number of mitochondria per cell.
The conversion of glucose to lactate, as noted by Warburg, can be due to the shunting of pyruvate away from oxidative phosphorylation. In line with our findings above, PLX4720 reduced lactate levels in all BRAF mutant melanomas evaluated (Figure 1f). Lactate levels did not change upon treatment of a melanoma cell line that does not contain the BRAF mutation, consistent with the inability of PLX4720 to suppress ERK signaling in these cells (Figure 1g). Collectively our data suggest that BRAF suppresses oxidative phosphorylation gene expression and mitochondrial density in melanoma.
BRAF and MAPK activation suppresses PGC1α
Since oxidative phosphorylation depends on mitochondrial number and activity, their alteration may contribute to altered tumor metabolism. Candidate pathways, which physiologically regulate mitochondrial content and function include the transcription factors mitochondrial transcription factors A (TFAM) and B (TFB1M, TFB2M), nuclear respiratory factor 1 (NRF1), GA binding proteins (GABPA, GABPB2), peroxisome proliferator-activated receptors (PPARα, PPARβ), PPAR-γ coactivators (PGC1α, PGC1β), and PGC1-related coactivator 1 (PPRC1) (reviewed in Kelly, 2004).
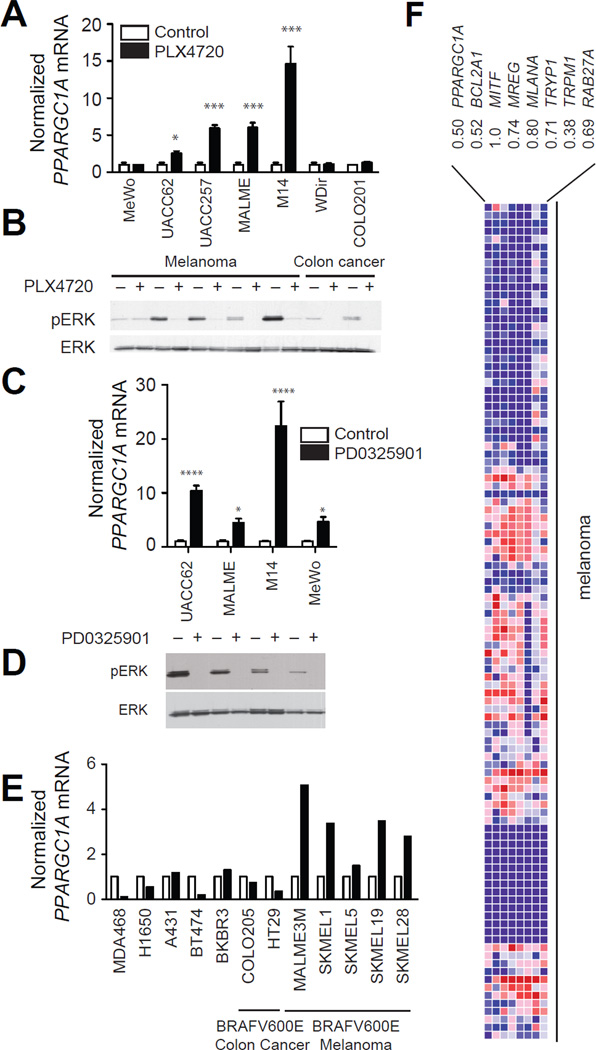
We observed that BRAF(V600E) expression suppressed PGC1α, a well-known regulator of mitochondrial metabolism in the microarray datasets above. To validate these observations, we treated a series of BRAF-mutant melanomas and non-melanoma cell lines with PLX4720 and evaluated the effect on PGC1α mRNA (Figure 2a). In all melanomas with BRAF mutations, PLX4720 induced 3–14 fold increases in PGC1α mRNA. We did not observe any changes in the expression of PGC1α in a BRAF wild-type MeWo cell line treated with PLX4720. Surprisingly, we did not observe any effects of PLX4720 on PGC1α expression in two BRAF mutant colon cancer cell lines, despite suppression of ERK phosphorylation similar to that seen in melanomas (Figure 2b). We did not observe any change in PGC1β mRNA upon treatment with PLX4720 or any effects in a BRAF-wild-type melanoma over 24 hours (Figure S2a,b). These data suggested that there might be lineage-specific differences in the regulation of PGC1α by BRAF. To validate our findings using a structurally unrelated small molecule, we treated several melanoma cell lines with the MEK inhibitor PD0325901. Induction of PGC1α mRNA (Figure 2c) and suppression of ERK phosphorylation (Figure 2d) were seen in all cell lines tested including the BRAF wild-type melanoma MeWo, suggesting that the BRAF/MEK/ERK pathway regulates PGC1α expression in melanoma cells. These results were also confirmed with additional NRAS-mutant melanoma cell lines treated with a MEK1/2 inhibitor (Figure S2c,d). Finally, we evaluated the expression of PGC1α in an independent dataset of A375 melanoma cells selected for resistance to BRAF inhibitors (Greger et al., 2012). We observed that PGC1α expression was 10-fold lower in cells that had acquired resistance to BRAF inhibitors (Figure S2e), likely reflecting their higher demonstrated basal MAPK activity.
Figure 2.
BRAF inhibitors induce PGC1α expression. PGC1α mRNA (A) and phospho-ERK levels (B) in melanoma or colon cancer cells treated with PLX4720 (1µM) for 24h. PGC1α mRNA (C) and ERK activity (D) in melanoma cells treated with the MEK inhibitor PD0325901 (10nM) for 24h. (E) Microarray analysis (GSE10086) of PGC1α mRNA in cell lines treated with 10nM PD0325901 for 24h. (F) Comparison of PGC1α mRNA with MITF, melanocytic markers, and MITF _targets in 105 melanoma cell cultures (Hoek et al., 2006). Pearson correlation coefficient is shown below each gene. Error bars represent SEM of at least three independent replicates. ****, p < 0.0001; ***, p < 0.001; *, p < 0.01. See also Figure S2.
We also interrogated a publically available microarray of 12 breast, lung, colon and melanoma cell lines treated with PD0325901 (Joseph et al., 2010). Suppression of MEK only affected PGC1α mRNA in melanoma cell lines (Figure 2e, p < 0.0001), suggesting that the regulation of PGC1α mRNA by the BRAF/MEK/ERK pathway is unique to the melanocytic lineage. Consistent with our results, we found that PGC1α expression was significantly correlated with melanocyte-specific antigen expression in a dataset comprising of 105 melanoma cell cultures (Figure 2f).
PGC1α mRNA is directly regulated by the MITF transcription factor
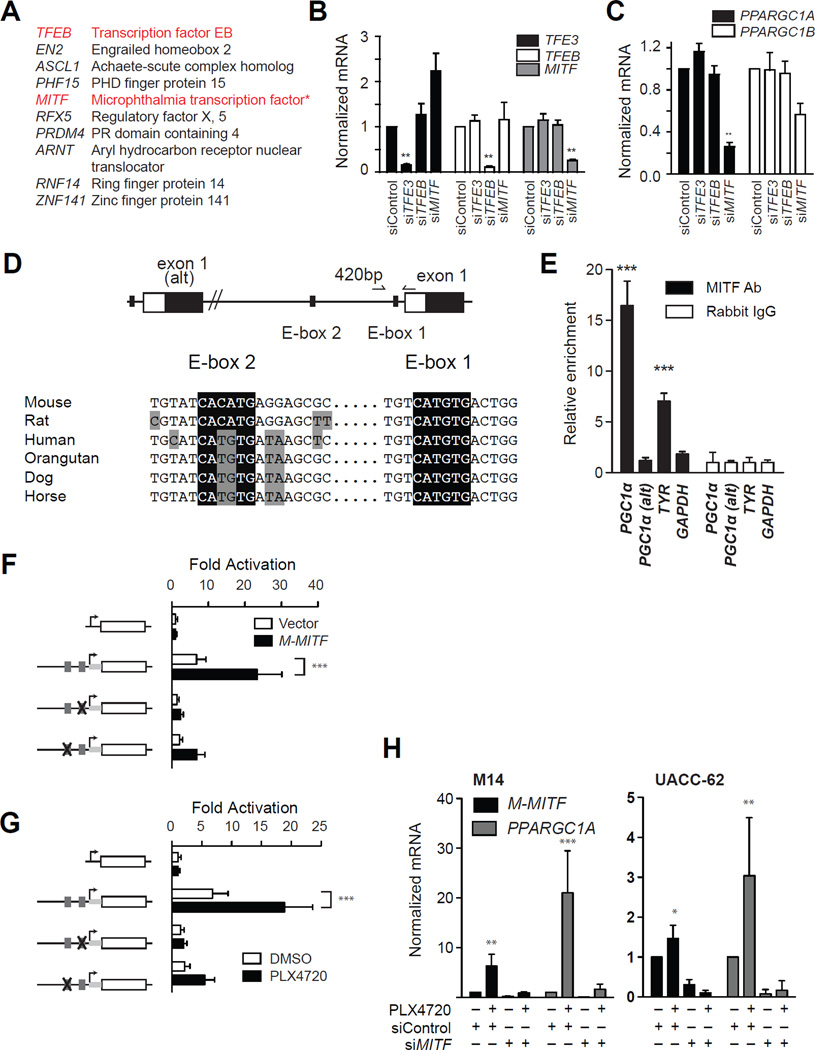
To elucidate how BRAF may regulate PGC1α, we compared the expression pattern of all human transcription factors to PGC1α in a large microarray dataset of short-term melanoma cultures (Lin et al., 2008). Among the transcription factors whose expression most closely paralleled PGC1α, we observed that both transcription factor EB (TFEB) and the micropthalmia-associated transcription factor (MITF) were significantly associated with PGC1α expression (q < 0.001) (Figure 3a). Both TFEB and MITF are members of a four-member family of distinctly encoded transcription factors (TFEB, TFEC, TFE3 and MITF) that share a common structure, binding recognition sequence, and function (Haq and Fisher, 2011). Whereas TFEB, TFEC, and TFE3 are ubiquitously expressed, MITF is largely restricted to the melanocytic lineage. The correlation of PGC1α to MITF was therefore interesting in light of our data suggesting BRAF regulation of PGC1α in melanoma but not in other lineages (see Figures 2a, 2e). MITF expression correlated to PGC1α as shown above (see Figure 2f).
Figure 3.
PGC1α is regulated by MITF in the melanocytic lineage. (A) Top 10 transcription factors correlated to PGC1α mRNA in (Lin et al., 2008). *, q < 0.05. (C) Requirement of MiT family members for PGC1α expression in M14 melanoma cells. Knockdown of each family member is shown in (B). ****, p < 0.0001; **, p < 0.01. (D) Structure of PGC1α promoter in mammalian species showing the location of alterative exon 1 and exon 1. Also depicted are the locations of E-box #1 and E-box #2 and primers used for chromatin-immunoprecipitation. (E) Chromatin immunopreciptation of indicated genomic region with anti-MITF, or rabbit IgG in primary melanocytes. Precipitated DNA was amplified using primers depicted in (D). ***, p < 0.001 compared to Rabbit IgG control. (F,G) Activity of PGC1α promoters upstream of the luciferase gene mutated as depicted in response to transfection of MITF (f) or treatment with PLX4720 (G). Grey boxes indicate the location of E-boxes. (H) Expression of PGC1α following knockdown of MITF (48h) and treatment with PLX4720 (1µM, 24h) in M14 cells (left) or UACC-62 cells (right). Error bars represent SEM of at least three independent replicates. ***, p < 0.001. See also Figure S3.
We therefore evaluated the requirement of TFE3, TFEB and MITF for the expression of PGC1α by siRNA. In both M14 melanoma cells and primary human melanocytes, suppression of MITF but not TFEB or TFE3 (Figure 3b,S3a) led to a significant suppression of PGC1α (Figure 3c, S3b) despite similar knockdown efficiency. We were unable to reliably detect TFEC expression in the cell lines tested (data not shown). MITF suppression in two other melanomas lines also significantly reduced PGC1α expression (Figure S3c), which was validated using two independent shRNAs (Figure S3d).
In sillico analysis of the PGC1α promoter identified three putative MITF recognition sequences (‘E-boxes’), which were conserved among mammalian species. One E-box was located approximately 420 bp upstream of the transcriptional start site, whereas a proximal Ebox was located within 20 bp of the start site (Figure 3d). Another E-box was located approximately 10 kilobases upstream of these sequences, in the promoter sequences of an alternative exon 1 (PGC1α-alt) (Miura et al., 2008; Chinsomboon et al., 2009). To evaluate if MITF could directly bind to the PGC1α promoter, we performed chromatin immunoprecipitation using primers located either near alternative exon 1 or exon 1 (see Figure 3d). As shown in Figure 3e, MITF was found to bind to the proximal PGC1α promoter but not to the PGC1α-alt promoter in primary melanocytes. Due to limits in the resolution of this assay, we were unable to distinguish if MITF binds to E-box #1, E-box #2 or both. We therefore utilized a PGC1α promoter cloned upstream of the luciferase reporter gene (Handschin et al., 2003) and mutated each E-box by site-directed mutagenesis (Figure 3f). MITF overexpression (Figure 3f) or treatment with PLX4720 (Figure 3g) led to the induction of the wild-type promoter, whereas mutation of either of the two E-boxes significantly inhibited this response. Collectively, these data indicate that MITF binds and directly regulates the PGC1α gene in the melanocyte lineage. To evaluate if BRAF regulates PGC1α via MITF, we suppressed MITF by siRNA, then treated with PLX4720. As shown in Figure 3h, treatment with PLX4720 strongly induced PGC1α mRNA in M14 cells and ~3 fold in UACC62 cells and this induction was absent in cells in which MITF was knocked down by siRNA. These data indicate that BRAF regulates PGC1α via MITF.
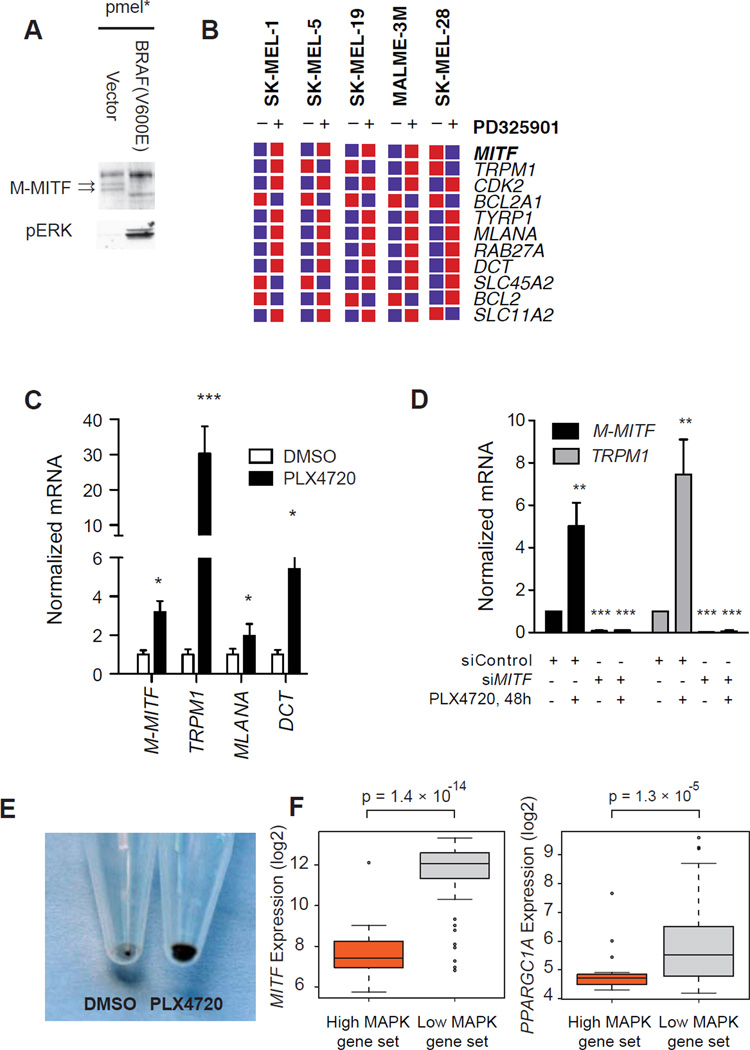
BRAF negatively regulates MITF activity
The relationship between BRAF and MITF is poorly understood since oncogenic BRAF and the ERK pathway promotes MITF activation but also leads to its degradation (Hemesath et al., 1998; Wu et al., 2000; Wellbrock, 2005; Wellbrock et al., 2008; Boni et al., 2010). We therefore examined the consequences of ectopic BRAF(V600E) expression in immortalized melanocytes. Introduction of oncogenic BRAF was associated with decreased levels of M-MITF protein (the melanocyte-specific isoform) but not other isoforms (Figure 4a). Conversely, the MEK inhibitor PD0325901 induced the expression of TRPM1 and other direct _targets of MITF in published microarrays (Figure 4b) and by qtPCR (Figure 4c). These effects on MITF _targets were dependent on MITF, since knockdown of MITF blocked induction of TRPM1 (Figure 4d). In line with these findings, we observed that treatment of UACC-257 cells with PLX4720 for 72h lead to increased pigmentation, reflecting MITF’s essential role in melanin synthesis and pigmentation (Figure 4e).
Figure 4.
BRAF suppresses MITF expression and activity. (A) Levels of M-MITF (arrows) and phosphorylated ERK in pmel* and pmel* BRAF (V600E). (B) Effects of MEK inhibitor PD0325901 on MITF mRNA and MITF _targets by published microarray (Pratilas et al., 2009). (C) Response of MITF and MITF _targets to PLX4720 in UACC-257 cells by quantitative PCR. *, p < 0.01; ***, p < 0.001 relative to DMSO control. (D) Effect of MITF suppression on induction of MITF-_target TRPM1 by PLX4720. **, p < 0.01; ***, p < 0.001 relative to siControl. Error bars represent SEM of at least three independent replicates. (E) Consequence of PLX4720 (72h) on UACC-257 pigmentation. Equal number of cells were pelleted by centrifugation. (F) Box-plots showing expression of MITF (left) and PGC1α in melanoma cells with high or low MAPK activation from 88 short-term melanoma cultures (Lin et al., 2008).
We were unable to detect a correlation between BRAF mutation status and PGC1α or MITF expression in short-term cultures. Given the prevalence of MAPK pathway mutations other than BRAF(V600E) in melanoma, we evaluated the expression of PGC1α and MITF in melanomas with either high or low expression of a MAPK activation gene signature. Both MITF (t-test, p = 1.4 × 10−14) and PGC1α (t-test, p = 1.34 × 10−5) expression inversely correlated with MAPK activity (Figure 4f). Collectively, these data suggest that BRAF/MAPK inhibition leads to activation of MITF mRNA and protein and that this induction leads to induction of MITF _targets including PGC1α.
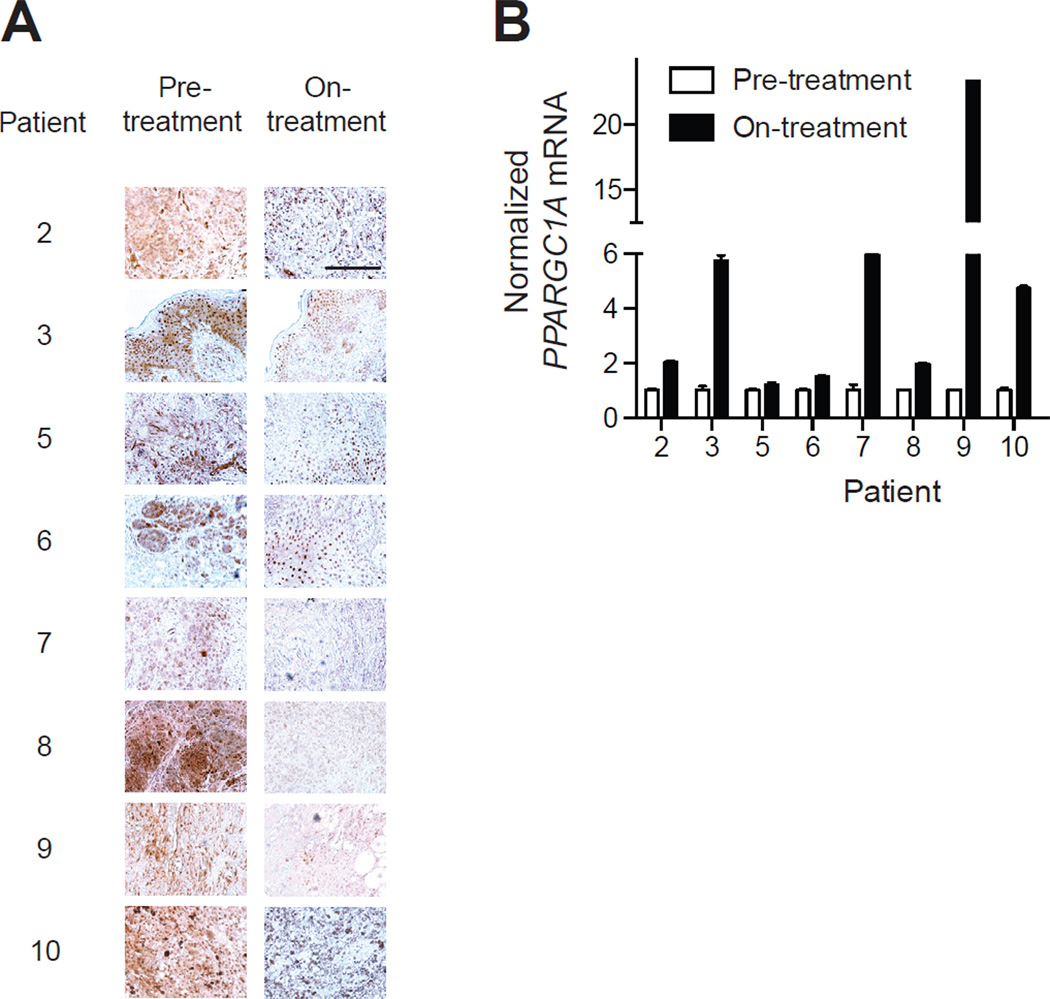
To evaluate if BRAF suppression alters the BRAF/MITF/PGC1α pathway described here in vivo, we obtained serial biopsies from 11 patients prior to treatment with BRAF/MEK inhibitors and 10–14 days after beginning treatment (patient characteristics described in Straussman et al., 2012). Of eight samples that had detectable phosphorylated ERK at baseline (Figure 5a), all had induction of PGC1α upon treatment (Figure 5b). Comparison to separately analyzed levels of M-MITF induction following BRAF _targeted therapy revealed a significant correlation to PGC1α induction within these patient-derived specimens (Wargo J et al., manuscript in preparation). Together these data indicate that the OXPHOS adaptive response described here exists in vivo.
Figure 5.
Validation of induction of PGC1α pathway in vivo following BRAF inhibition. (A) Expression of phospho-ERK in eight matched patient biopsies prior or during (10–14 days) of treatment with BRAF/MEK inhibitors. (B) Expression of PGC1α mRNA prior and during treatment with BRAF/MEK inhibitors. Error bars represent SEM of at least three technical replicates. Scale bar represents 100 µm.
MITF promotes expression of oxidative phosphorylation genes
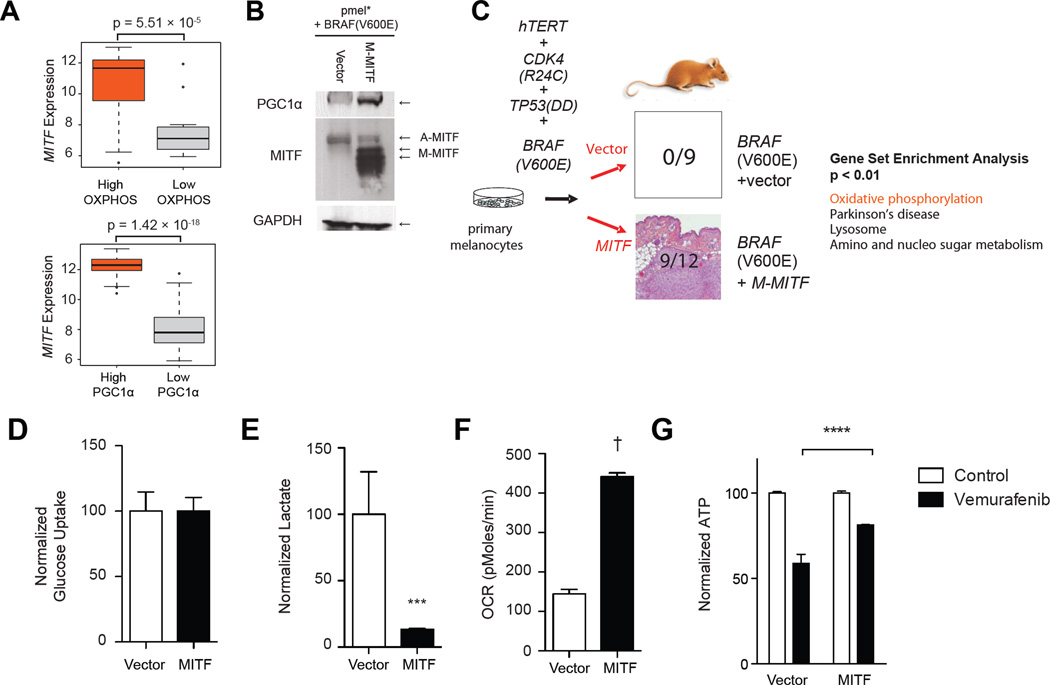
Up to 30% of melanomas harbor genomic amplifications of MITF (Garraway et al., 2005), and MITF is required for the survival of at least a subset of melanomas. Activating point mutations have also been identified in melanoma (Bertolotto et al., 2011; Yokoyama et al., 2011). These data have led to the designation of MITF as a lineage-specific melanoma oncogene (Garraway et al., 2005). Comprehensive expression profiling approaches have identified roles for MITF in promoting cell growth and survival, organelle biogenesis, oxidative stress response and miRNA regulation (Vachtenheim and Borovanský, 2010; Haq and Fisher, 2011). However a role for MITF in regulating metabolism has not been previously described. We therefore tested if MITF expression correlated with oxidative phosphorylation by classifying melanomas into two groups based on a previously defined oxidative phosphorylation signature. As seen in Figure 6a, melanomas with high MITF expression had significantly higher oxidative phosphorylation gene expression (p = 5.51 × 10−5). Similarly, MITF expression correlated with PGC1α -regulated gene expression (p = 1.42 × 10−15).
Figure 6.
MITF regulates oxidative phosphorylation. (A) Box plots depicting MITF expression in melanomas with high expression of a PGC1α-_target gene set (bottom) or oxidative phosphorylation gene set (top). (B) Western blotting showing expression of MITF and PGC1α in pmel* BRAF(V600E) with and without MITF overexpression. (C) Schema showing isogenic system evaluating the effect of MITF overexpression in BRAF(V600E) melanoma cells. The tumorigenicity of the paired cell lines was assessed in FoxNnu mice and the number of formed tumors per injection of each cell line is shown. Gene set enrichment analysis of the paired cell lines with the most highly induced gene sets is shown (right). Glucose uptake (D), lactate levels (E) and oxygen consumption (F) were measured as relative amounts in each cell line, normalized to cell number. (G) ATP levels, normalized to cell number in BRAF(V600E)+vector and BRAF(V600E)+MITF treated with PLX4032 (1µM) for 24h. ***, p < 0.001 compared to control cells. Error bars represent SEM of at least three independent replicates. See also Figure S4.
To evaluate if MITF was sufficient to drive oxidative phosphorylation in melanoma, we evaluated two matched human cell lines derived from primary melanocytes. These two cell lines (termed pmel*+BRAF(V600E) and pmel*+BRAF(V600E)+MITF) are derived from primary normal human melanocytes by immortalization using telomerase, with a constitutively active allele of cyclin-dependent kinase 4 and dominant negative p53 (Garraway et al., 2005). These cells are therefore isogenic with the exception of the expression of MITF. Consistent with published reports (Garraway et al., 2005) and our results above, expression of M-MITF protein was undetectable in pmel*+BRAF(V600E) cells, but strongly expressed in the derived MITFexpressing cells, which correlated with PGC1α expression (Figure 6b). BRAF(V600E) cells expressing MITF were able to form tumors in immunocompromised mice whereas control cells were not (Figure 6c), paralleling previous data from soft-agar growth (Garraway et al., 2005). Similar to our findings with BRAF inhibition, Gene Set Enrichment Analysis of microarray data identified a highly significant induction of oxidative phosphorylation gene set in MITF expressing cells compared to control cells (q ≃ 0.0, p < 5 × 10−4; see Figure 6c). We found a large majority of the oxidative phosphorylation gene set was induced by MITF overexpression (Figure S4a), consistent with the ability of PGC1α to strongly upregulate many oxidative phosphorylation genes (Mootha et al., 2003). We validated the expression of differentially expressed genes (chosen to represent different mitochondrial processes) by qtPCR (Figure S4b) and found significant increases in several oxidative phosphorylation genes. Similarly, expression of the oxidative phosphorylation genes were suppressed by oncogenic BRAF (Figure S4c).
To evaluate the metabolomic consequence of MITF overexpression directly, we evaluated glycolysis and oxidative phosphorylation in the isogenic cells. Overexpression of MITF did not increase glucose uptake (Figure 6d), but strikingly had decreased lactate production (Figure 6e) and oxygen consumption (Figure 6f), consistent with increased mitochondrial metabolism. Conversely, BRAF(V600E) cells had elevated sensitivity to knockdown of pyruvate kinase (muscle isoform), the final step in the glycolysis pathway, compared to isogenic cells expressing MITF (Figure S4d), despite similar degrees of knockdown efficiency in the two cell lines (Figure S4e).
To further validate these findings in patient-derived melanomas, we suppressed MITF by shRNA and performed gene expression profiling. We identified several genes involved in oxidative phosphorylation that were dependent on MITF (Figure S4f), consistent with the above gene expression data. With the exception of PGC1α, which was found to be directly bound by MITF, oxidative phosphorylation _targets were not bound by MITF but have been identified as PGC1α _targets in other cell lineages (Mootha et al., 2003). We conclude that MITF overexpression is sufficient and necessary to drive oxidative metabolism gene signature and metabolic reprogramming in the melanocyte lineage.
BRAF inhibition leads to bioenergic adaptation by induction of MITF and PGC1α
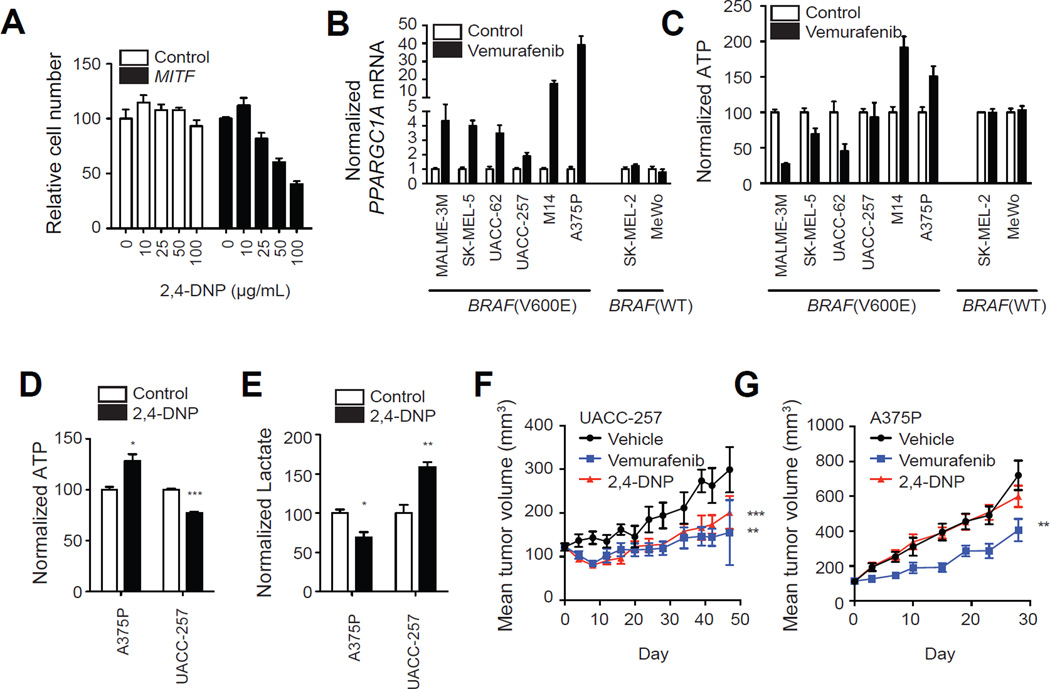
Collectively, the data above support the notion that BRAF inhibition endangers ATP production, which is rescued by concomitant induction of MITF, PGC1α and oxidative metabolism. To experimentally test this hypothesis, we evaluated the response of the isogenic cell lines expressing BRAF(V600E) with or without M-MITF overexpression to PLX4032. Inhibition of BRAF resulted in 42% drop in ATP in BRAF(V600E) parental cells whereas the magnitude of this drop was reduced in MITF overexpressing cells (Figure 6g), despite similar basal ATP concentrations. Consistent with MITF effects on oxidative metabolism, BRAF(V600E)+MITF cells were significantly more sensitive to the mitochondrial uncoupler 2,4-dinitrophenol (DNP; Figure 7a).
Figure 7.
Effects of 2,4-DNP on growth melanoma cells in vitro and in vivo. (A) Number of BRAF(V600E)+vector and BRAF(V600E)+MITF melanoma cells following treatment with 2,4- DNP with indicated dose for 72h. Levels of PGC1α mRNA (B) and ATP (C) in melanoma cell lines treated with vemurafenib (1µM) for 24h. Effects of 2,4-DNP (50 µg/mL, 24h) on ATP (D) and lactate levels (E) in indicated cell lines in vitro. (F,G) Effect of 2,4-DNP (20 mg/kg/day) or vemurafenib (75 mg/kg/day) on murine xenografts of indicated cell line (N = 7–8 per group). **, p < 0.01 compared to vehicle group; ***, p < 0.001 compared to vehicle group. Error bars represent SEM of at least three independent replicates. See also Figure S5.
We also evaluated the magnitude of bioenergetic compensation in 8 patient-derived melanoma cell lines. As shown in Figure 7b, in all cases there was induction of PGC1α following vemurafenib treatment, which varied in magnitude by cell line. We also measured ATP levels before and after treatment with vemurafenib, and we observed a significant correlation between induction of PGC1α and ATP levels in BRAF mutant cells (R = 0.72, p = 0.03; Figure 7c). Strikingly, the two lines with highest ATP levels following BRAF inhibition are the ones with the strongest induction of PGC1α. These data suggest that the metabolic switch to “normal” varies in magnitude among different melanoma cell lines and suggest that the inhibition of BRAF leads to a bioenergetic crisis that can be variably rescued by induction of the MITF/PGC1 α pathway.
We next asked whether oxidative metabolism could be exploited to inhibit the growth of melanoma cells. We compared the sensitivity of melanoma cells to primary melanocytes (Figure S5). All melanoma cells were more sensitive to DNP than primary melanocytes, except A375P. Treatment of UACC257 cells, but not A375P cells with DNP led to a decrease in ATP and increases in lactate, consistent with inhibition of OXPHOS (Figure 7d,e). To validate the effects of mitochondrial inhibitors on tumor growth in vivo, we treated animals bearing tumor xenografts of A375P and UACC257 melanoma cells with DNP. Consistent with the in vitro data, we found that longitudinal treatment of UACC257 xenografts profoundly inhibited tumor growth, similar to the effects of PLX4032 (Figure 7f), whereas A375P cells were insensitive to 2,4-DNP (Figure 7g).
Melanomas treated with BRAF inhibitors are addicted to oxidative metabolism
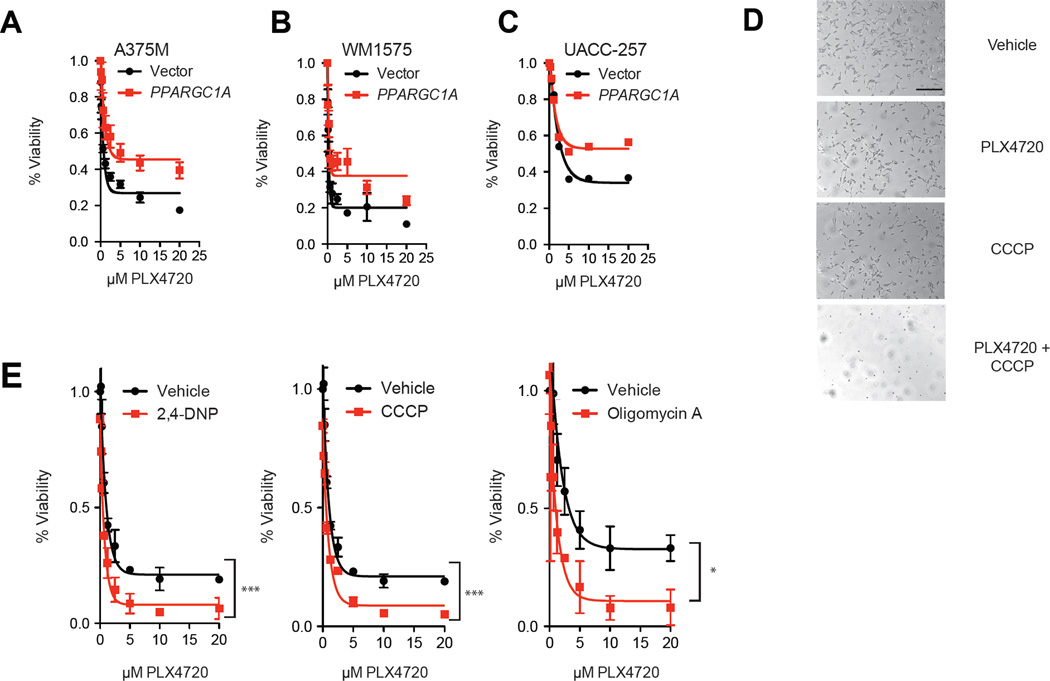
Our observations that BRAF and MITF regulate PGC1α expression prompted us to evaluate if oxidative metabolism affected response to BRAF inhibitors. We observed that high levels of PGC1α were associated with poorer prognosis of patients with Stage III melanoma (Figure S6a) (Bogunovic et al., 2009). To this end, forced expression of PGC1α protected cells from PLX4720, as demonstrated using three cell lines that expressed low levels of PGC1α (Figure 8a,b,c). As our data suggesting that BRAF regulates PGC1α and oxidative phosphorylation, we evaluated the effects of inhibitors of oxidative phosphorylation in combination with BRAF inhibitors. In addition, the cells were found relatively insensitive to the mitochondrial uncoupler CCCP, but addition of PLX4720 enhanced the cytotoxicity of this drug (Figure 8d). Melanoma cells treated with 2,4-Dinitrophenol (DNP), or oligomycin A, which inhibit oxidative phosphorylation through different mechanisms additively enhanced the efficacy of PLX4720 in vitro (Figure 8e). We also found similar data using rotenone and the complex II inhibitor, TTFA (Figure S6b,c). Together, the data indicate that adaptive induction of oxidative phosphorylation in response to BRAF inhibition limits their efficacy.
Figure 8.
BRAF inhibitors enhance dependence on mitochondrial metabolism. Sensitivity of A375M (A), WM1575 (B) and UACC-257 (C) melanoma cells overexpressing PGC1α to treatment with PLX4720 for 72 hours. (D) Photograph of M14 cells treated with PLX4720 (5µM), CCCP (20µM) or the combination for 72 hrs. Scale bar represents 100 µm. (E) Cell number following treatment with mitochondrial uncouplers oligomycin A (1µM), CCCP (5µM) or 2,4-DNP (200 µg/mL). Cell number was estimated after 72 hrs of treatment. *, p < 0.05 compared to control; **, p < 0.01 compared to control; ***, p < 0.001 compared to control. Error bars represent SEM of at least three independent replicates. See also Figure S6.
Discussion
Previous studies have suggested a role of BRAF signaling in the regulation of tumor metabolism. Clinically, patients with BRAF mutant melanomas have higher levels of serum lactate consistent with diminished oxidative phosphorylation (Board et al., 2009). In addition, BRAF mutant melanomas have an order of magnitude increased uptake of glucose compared to normal tissues in vivo as assessed by functional imaging (Bollag et al., 2010). However, the mechanism by which BRAF regulates metabolism in melanoma is poorly understood. This study identifies a pathway by which the oncogenic BRAF pathway regulates energy metabolism in melanoma. Our findings show that BRAF activation is associated with diminished oxidative enzymes, diminished mitochondrial number and function and increased production of lactate. This metabolic reprogramming triggered by BRAF(V600E) is accompanied by a suppression of MITF and PGC1α, a major regulator of mitochondrial biogenesis and function.
We identify the melanocyte master regulator MITF as a direct and essential mediator of BRAF-regulated PGC1α transcription. Consistent with the restricted expression of MITF to the melanocyte lineage, the ERK pathway does not appear to regulate PGC1α transcription in BRAF mutant cancers that lack MITF expression. The MITF-PGC1 connection thus explains the lineage-specific effects of BRAF activation and inhibition.
Although BRAF inhibitors can suppress glycolysis and induce oxidative phosphorylation, we found that MITF only regulates mitochondrial respiration. Consistent with this, MITF and PGC1α expression correlate with oxidative phosphorylation genes in a large series of melanoma short term cultures, but we have not observed an inverse correlation with glycolytic gene expression. We also found that MITF expression did not affect glucose uptake, but decreased lactate and increased oxygen consumption, consistent with oxidative metabolism. Thus, consistent with Warburg’s initial hypothesis, the activation of glycolysis and the suppression of oxidative metabolism as shown here to be initiated by oncogenic BRAF, are likely separate processes.
Overall, our data suggest that MITF is a major regulator of mitochondrial respiration in the melanocyte lineage by acting via PGC1α. Tumors likely generate ATP via both glycolysis and oxidative phosphorylation (Colombi et al., 2010; Weinberg et al., 2010) and tumor cells may require mitochondria for functions other than ATP generation, such as fatty acid synthesis and glutaminolysis in some cases (Wise and Thompson, 2010; Dang, 2012). However, we show here that MITF-expressing melanomas have higher level of oxidative gene expression. These data therefore suggest that MITF expression may serve as a biomarker for greater dependence on mitochondrial function. Since MITF has been difficult to drug directly (Haq and Fisher, 2011), the dependence of MITF dependent melanoma on oxidative phosphorylation thus presents a theoretical therapeutic approach. However, MITF paradoxically can promote tumorigenesis (e.g. Garraway et al., 2005; Yokoyama et al. 2011), whereas activation of MITF expression in normal melanocytes typically induces differentiation, which likely antagonizes tumorigenesis (D’Orazio et al., 2006; Carreira S et al., 2006; Haq and Fisher, 2011). A rheostat model has been proposed to explain the apparent paradox of MITF (Goding C, 2011) but definitive evidence of this model remains an area of active investigation. We show BRAF inhibitors induce some, but not all MITF _target genes, implying that the context in which MITF is regulated may also contribute to its physiologic effects. Finally, it is also highly likely that MITF has pro-tumorigenic effects outside of its induction of OXPHOS, so that any anti-tumorigenic effect of inducing OXPHOS may well be countered by other pro-tumorigenic effects. In the future it will be of great interest to examine how MITF can coordinate its numerous downstream effects.
Eight patients treated with vemurafenib had induction of PGC1α but a larger sample size will therefore be needed to evaluate the diagnostic and predictive role of PGC1α induction in responses to BRAF inhibitors. Interestingly, mutations in PGC1α have been detected in recent whole genome sequencing efforts (Prickett et al., 2009; Stark et al., 2012). Our data nonetheless suggest that dysregulation of PGC1α may have profound effects on metabolism of melanoma cells, and may contribute to oncogenesis in certain cases.
We found that BRAF-mutant melanomas treated with PLX4720 are dependent on ATP generation by mitochondria. Our data suggest that inhibition of mitochondrial metabolism may be most effective therapeutically as initial therapy, as most patients that have relapsed following BRAF inhibitors have reactivation of the MAPK pathway, which we have shown correlates with decreased level of MITF and PGC1α. While mitochondrial functions would likely be difficult to _target therapeutically in many cancer types, agents that exploit bioenergetic and metabolic alterations in mitochondria have been proposed (Fantin and Leder, 2006). We find that mitochondrial uncouplers enhanced the efficacy of PLX4720 in BRAF mutant melanomas, but demonstration of in vivo efficacy of this combination remains to be firmly established, which possibly will involve derivation of improved mitochondrial pharmacologic agents. Though available drugs have generally unfavorable pharmacologic properties, 2,4-dintirophenol (DNP) had been used extensively in diet pills (Cutting and Tainter, 1933) and over 100,000 people had been treated world-wide with the drug at the time of its discontinuation (Tainter et al., 1934). Cases of dangerous side effects such as fatal hyperthermia led to its official discontinuation by 1938. Given the toxicities of the drugs, further development of alternative oxidative phosphorylation inhibitors should be considered. Despite the recent successes of BRAF inhibition in the clinical arena, recurrence rates remain high, and survival is only extended several months. While further in vivo studies will be crucial, uncouplers such as DNP, or other inhibitors of oxidative phosphorylation may an alternative approach to enhance the effect of BRAF inhibitors in melanoma patients.
Experimental Procedures
Gene expression and bioinformatics
Immortal melanocytes (pmel*) and their transformed counterparts, pmel*BRAF(V600E)-vector and pmel*BRAF(V600E)+MITF were maintained as described (Garraway et al., 2005). Global gene expression analysis was carried out using HG-U133A microarrays (Affymetrix).
RNA isolation, chromatin immunoprecipitation and quantitative real-time PCR
Chromatin immunoprecipitation (ChIP) was performed in primary human melanocytes using previously described methods (Cui et al., 2007). Chromatin was immunoprecipitated using rabbit polyclonal anti-MITF, or normal rabbit IgG as a control. Results are normalized to input DNA.
Clinical samples
All patients gave informed consent for tissue acquisition as per an IRB-approved protocol (Office for Human Research Studies, Dana-Farber/Harvard Cancer Center). Tumors were biopsied before treatment (day 0), at 10–14 days during treatment.
siRNA delivery and analysis
siRNAs SMARTpools (Dharmacon) were delivered using the lipidoid delivery agent C12-133-B as described (Li et al., 2012).
Promoter assays and luciferase experiments
The murine PGC1α promoter was obtained from Addgene. Mutagenesis was performed using the QuickChange Mutagenesis Kit (Stratagene). Mutant promoters were verified by sequencing. UACC-62 cells were transfected with each promoter construct, pRL-CMV Renilla control and a M-MITF overexpression vector. PLX4720 treatment was for 48h. Results reported are averages of at least three independent experiments, normalized for transfection efficiency using Renilla luciferase.
Xenograft tumor studies
All mouse experiments were done in accordance with Institutional Animal Care and Use Committee (IACUC) approved animal protocols at Dana-Farber Cancer Institute as described in the Supplemental Experimental Procedures.
Electron microscopy
Electron microscopy of UACC-62 melanoma cells treated with vehicle or PLX4720 (3µM, 72hrs) was performed at the PMB Microscopy Core as described in Supplemental Experimental Procedures.
Microarray data
Expression array data are deposited under GEO accession GSE38007.
Supplementary Material
BRAF suppresses mitochondrial density and oxidative phosphorylation metabolism
MITF directly regulates the mitochondrial biogenesis factor, PGC1α
Melanomas with activation of the MAPK pathway have lower levels of MITF and PGC1α
Mitochondrial uncouplers may have therapeutic use in combination with BRAF inhibitors
Significance.
BRAF mutations are the most common genetic aberrations in melanoma but the mechanisms by which they promote oncogenesis are poorly understood. Here we show that activated BRAF promotes metabolic reprogramming by suppression of oxidative phosphorylation through the actions of the melanocyte lineage factor MITF and the mitochondrial master regulator PGC1α. BRAF inhibitors, which transiently suppress melanoma growth in vitro and in patients, induce PGC1α and oxidative phosphorylation. This addiction to oxidative phosphorylation in melanomas treated with BRAF _targeted therapy therefore suggests that mitochondrial inhibitors should be evaluated in combination with BRAF pathway inhibitors in vivo.
Acknowledgements
We thank A. Winant for assistance with manuscript writing, M. McKee for electron microscopy, A. Saur at the Luria Family Imaging Center (Dana-Farber Cancer Institute) for assistance with mouse xenograft experiments and J. Asara for metabolomic profiling. Funding: American Skin Association (R.H.), NIAMS/NIH (R01 AR043369-16), the Melanoma Research Alliance, and the Dr. Miriam and Sheldon Adelson Medical Research Foundation (D.E.F); The National Cancer Institute (J.S.S., R01CA163336) and H.R.W. (P50CA093683).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MNC, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d’Hayer B, Mohamdi H, Remenieras A, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- Bhalla K, Hwang BJ, Dewi RE, Ou L, Twaddel W, Fang HB, Vafai SB, Vazquez F, Puigserver P, Boros L, et al. PGC1 Promotes Tumor Growth by Inducing Gene Expression Programs Supporting Lipogenesis. Cancer Research. 2011;71:6888–6898. doi: 10.1158/0008-5472.CAN-11-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board RE, Ellison G, Orr MCM, Kemsley KR, McWalter G, Blockley LY, Dearden SP, Morris C, Ranson M, Cantarini MV, et al. Detection of BRAF mutations in the tumour and serum of patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma phase II study. Br J Cancer. 2009;101:1724–1730. doi: 10.1038/sj.bjc.6605371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic D, O'Neill DW, Belitskaya-Levy I, Vacic V, Yu Y-L, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proceedings of the National Academy of Sciences. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. Clinical efficacy of a RAF inhibitor needs broad _target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni A, Cogdill AP, Dang P, Udayakumar D, Njauw CNJ, Sloss CM, Ferrone CR, Flaherty KT, Lawrence DP, Fisher DE, et al. Selective BRAFV600E Inhibition Enhances T-Cell Recognition of Melanoma without Affecting Lymphocyte Function. Cancer Research. 2010;70:5213–5219. doi: 10.1158/0008-5472.CAN-10-0118. [DOI] [PubMed] [Google Scholar]
- Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsomboon J, Ruas J, Gupta RK, Thom R, Shoag J, Rowe GC, Sawada N, Raghuram S, Arany Z. The transcriptional coactivator PGC-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proceedings of the National Academy of Sciences. 2009;106:21401–21406. doi: 10.1073/pnas.0909131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombi M, Molle KD, Benjamin D, Rattenbacher-Kiser K, Schaefer C, Betz C, Thiemeyer A, Regenass U, Hall MN, Moroni C. Genome-wide shRNA screen reveals increased mitochondrial dependence upon mTORC2 addiction. Oncogene. 2010;30:1551–1565. doi: 10.1038/onc.2010.539. [DOI] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D'Orazio J, Fung CY, Schanbacher CF, Granter SR, et al. Central Role of p53 in the Suntan Response and Pathologic Hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho K-H, Aiba S, Bröcker E-B, LeBoit PE, et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Cutting WC, Tainter M. METABOLIC ACTIONS OF DINITROPHENOL. Journal of the American Medical Association. 1933;101:2099–2012. [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Research. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Links between metabolism and cancer. Genes & Development. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, You MJ, DePinho RA, McMahon M, Bosenberg M. BrafV600E cooperates with Pten loss to induce metastatic melanoma. Nature Genetics. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Eichner LJ, Perry M-C, Dufour CR, Bertos N, Park M, St-Pierre J, Giguère V. miR-378* Mediates Metabolic Shift in Breast Cancer Cells via the PGC-1β/ERRγ Transcriptional Pathway. Cell Metabolism. 2010;12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Fantin VR, Leder P. Mitochondriotoxic compounds for cancer therapy. Oncogene. 2006;25:4787–4797. doi: 10.1038/sj.onc.1209599. [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- Finck BN. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Girnun GD. The diverse role of the PPARγ coactivator 1 family of transcriptional coactivators in cancer. Seminars in Cell & Developmental Biology. 2012;23:381–388. doi: 10.1016/j.semcdb.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new _targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, Dickerson SH, Laquerre SG, Liu L, Gilmer TM. Combinations of BRAF, MEK, and PI3K/mTOR Inhibitors Overcome Acquired Resistance to the BRAF Inhibitor GSK2118436 Dabrafenib, Mediated by NRAS or MEK Mutations. Molecular Cancer Therapeutics. 2012;11:909–920. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- Handschin C. The biology of PGC-1α and its therapeutic potential. Trends in Pharmacological Sciences. 2009;30:322–329. doi: 10.1016/j.tips.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc. Natl. Acad. Sci. U.S.a. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Fisher DE. Biology and Clinical Relevance of the Micropthalmia Family of Transcription Factors in Human Cancer. Journal of Clinical Oncology. 2011;29:3474–3482. doi: 10.1200/JCO.2010.32.6223. [DOI] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-*1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Jacobetz MA, Robertson GP, Herlyn M, Tuveson DA. Suppression of BRAF(V599E) in human melanoma abrogates transformation. Cancer Research. 2003;63:5198–5202. [PubMed] [Google Scholar]
- Hoeflich KP. Oncogenic BRAF Is Required for Tumor Growth and Maintenance in Melanoma Models. Cancer Research. 2006;66:999–1006. doi: 10.1158/0008-5472.CAN-05-2720. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Research. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proceedings of the National Academy of Sciences. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, Friedlos F, Ogilvie L, Hedley D, Martin J, Marshall CJ, et al. B-RAF is a therapeutic _target in melanoma. Oncogene. 2004;23:6292–6298. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- Kelly DP. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes & Development. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- Klimcakova E, Chenard V, McGuirk S, Germain D, Avizonis D, Muller WJ, St-Pierre J. PGC-1 Promotes the Growth of ErbB2/Neu-Induced Mammary Tumors by Regulating Nutrient Supply. Cancer Research. 2012;72:1538–1546. doi: 10.1158/0008-5472.CAN-11-2967. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Kelly DP. Transcriptional Control of Cardiac Fuel Metabolism and Mitochondrial Function. Cold Spring Harb. Symp. Quant. Biol. 2011;LXXXVI:1–7. doi: 10.1101/sqb.2011.76.011965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Song JS, Bell RJA, Tran T-NT, Haq R, Liu H, Love KT, Langer R, Anderson DG, Larue L, et al. YY1 Regulates Melanocyte Development and Function by Cooperating with MITF. PLoS Genet. 2012;8:e1002688. doi: 10.1371/journal.pgen.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005a;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang C-Y, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin J, Yang R, Tarr PT, Wu P-H, Handschin C, Li S, Yang W, Pei L, Uldry M, Tontonoz P. Hyperlipidemic Effects of Dietary Saturated Fats Mediated through PGC- 1β Coactivation of SREBP. Cell. 2005b;120:261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, Laine E, Greulich H, Tseng H, Gates C, et al. Modeling Genomic Diversity and Tumor Dependency in Malignant Melanoma. Cancer Research. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S. p53 Regulates Mitochondrial Respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Miura S, Kai Y, Kamei Y, Ezaki O. Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to beta2-adrenergic receptor activation and exercise. Endocrinology. 2008;149:4527–4533. doi: 10.1210/en.2008-0466. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehár J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer Cell. 2011;19:11–15. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proceedings of the National Academy of Sciences. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nature Genetics. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MS, Woods SL, Gartside MG, Bonazzi VF, Dutton-Regester K, Aoude LG, Chow D, Sereduk C, Niemi NM, Tang N, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nature Genetics. 2012;44:165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainter ML, Cutting WC, Stockton AB. Use of Dinitrophenol in Nutritional Disorders : A Critical Survey of Clinical Results. Am J Public Health Nations Health. 1934;24:1045–1053. doi: 10.2105/ajph.24.10.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences. 2008;105:3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metabolism. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Vachtenheim J, Borovanský J. “Transcription physiology” of pigment formation in melanocytes: central role of MITF. Experimental Dermatology. 2010;19:617–627. doi: 10.1111/j.1600-0625.2009.01053.x. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan PTC, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Mechanism of activation of the RAFERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Moraes CT. Increases in mitochondrial biogenesis impair carcinogenesis at multiple levels. Molecular Oncology. 2011:1–11. doi: 10.1016/j.molonc.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GRS, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C. Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. The Journal of Cell Biology. 2005;170:703–708. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS ONE. 2008;3:e2734. doi: 10.1371/journal.pone.0002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic _target in cancer. Trends in Biochemical Sciences. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Stoffel M. Coactivation of Foxa2 through Pgc-1beta promotes liver fatty acid oxidation and triglyceride/VLDL secretion. Cell Metabolism. 2006;3:99–110. doi: 10.1016/j.cmet.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes & Development. 2000;14:301–312. [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JKV, Markowitz S, Zhou S, et al. Glucose Deprivation Contributes to the Development of KRAS Pathway Mutations in Tumor Cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.