Abstract
Defining the virus–host interactions responsible for HIV-1 transmission, including the phenotypic requirements of viruses capable of establishing de novo infections, could be important for AIDS vaccine development. Previous analyses have failed to identify phenotypic properties other than chemokine receptor 5 (CCR5) and CD4+ T-cell tropism that are preferentially associated with viral transmission. However, most of these studies were limited to examining envelope (Env) function in the context of pseudoviruses. Here, we generated infectious molecular clones of transmitted founder (TF; n = 27) and chronic control (CC; n = 14) viruses of subtypes B (n = 18) and C (n = 23) and compared their phenotypic properties in assays specifically designed to probe the earliest stages of HIV-1 infection. We found that TF virions were 1.7-fold more infectious (P = 0.049) and contained 1.9-fold more Env per particle (P = 0.048) compared with CC viruses. TF viruses were also captured by monocyte-derived dendritic cells 1.7-fold more efficiently (P = 0.035) and more readily transferred to CD4+ T cells (P = 0.025). In primary CD4+ T cells, TF and CC viruses replicated with comparable kinetics; however, when propagated in the presence of IFN-α, TF viruses replicated to higher titers than CC viruses. This difference was significant for subtype B (P = 0.000013) but not subtype C (P = 0.53) viruses, possibly reflecting demographic differences of the respective patient cohorts. Together, these data indicate that TF viruses are enriched for higher Env content, enhanced cell-free infectivity, improved dendritic cell interaction, and relative IFN-α resistance. These viral properties, which likely act in concert, should be considered in the development and testing of AIDS vaccines.
Keywords: mucosal HIV-1 transmission, acute HIV-1 infection, innate immunity, epidemic HIV-1 spread
Understanding the host and viral factors that influence the ability of HIV-1 to cross mucosal barriers may be critical for the development of an effective AIDS vaccine (1). HIV-1 virions or infected cells are believed to cross the epithelium shortly after sexual exposure, although the mechanisms by which this transfer occurs remain largely unknown. Within the mucosa, viruses are believed to interact with dendritic cells (DCs), such as Langerhans cells, but do not productively infect these cells (2). In the simian model of HIV-1, the first cells to become productively infected are resting intraepithelial CD4+ T cells, which represent the most abundant _target cell type in the lamina propria (3). Simultaneous with initial infection events, local innate immune responses are elicited, with plasmacytoid DCs accumulating rapidly at sites of virus entry. These cells secrete cytokines and chemokines, such as macrophage inflammatory protein (MIP)1-β and type I IFNs, and orchestrate an early local innate immune response (4). After infection in mucosal and submucosal tissues is established, HIV-1 spreads to regional and distant lymphoid tissues, including the gut-associated lymphoid tissue, where virus expands exponentially, triggering a systemic cytokine storm preceding peak viremia (5, 6).
Many host factors can influence whether virus exposure leads to productive infection, including the physical barrier of the mucosa and its associated mucous secretions (7, 8), _target cell availability (9, 10), immune activation (11), genital inflammation (12), and altered mucosal microbiota (13). Although chronic HIV-1 infection is characterized by high genotypic and phenotypic diversity, the extent to which this variation influences the transmission process remains unclear (14, 15). Transmission across intact mucosal barriers is inherently inefficient and invariably associated with a viral population bottleneck (1, 16). Indeed, in 60–80% of mucosal infections, a single transmitted founder (TF) virus is responsible for productive clinical infection (16). This finding has raised the question of whether the transmission process represents a stochastic event, in which every replication-competent virus has an equal chance of establishing a new infection, or whether the bottleneck selects for viruses that exhibit particular biological properties that predispose them to establish new infections more efficiently (14). In support of the latter, transmitted viruses have generally been found to exhibit chemokine receptor 5 (CCR5) tropism, infect CD4+ T cells but not macrophages, and share certain genetic features, including shorter variable loops, fewer potential N-linked glycosylation sites, and amino acid signatures, which may affect envelope glycoprotein (Env) surface expression and/or other viral properties (17–22). However, to date, no consistent phenotypic correlate of these genetic signatures has been identified.
Initial phenotypic studies of viruses obtained during acute and early infection were limited to analyses of Env functions, which were almost exclusively conducted in the context of pseudoviruses. These analyses led to a number of hypotheses as to how transmitted viruses might differ from their chronic counterparts, including that they engage their receptor and/or coreceptor more efficiently (23, 24), that they are more sensitive to neutralizing antibodies (17, 25), and that their Env glycoproteins interact preferentially with the integrin pair-α4β7 (26). However, most of these studies used Envs obtained weeks or months postinfection after substantial virus evolution and selection had already occurred. We, thus, used single-genome amplification and a model of random virus evolution to generate Env clones corresponding to actual TF genomes (16, 27). Although we confirmed that TF Envs use CCR5 as the coreceptor for entry, we failed to find evidence that TF Envs used CD4 and CCR5 particularly efficiently (28, 29), interacted specifically with α4β7 (29), or exhibited a preferential tropism for particular CD4+ T-cell subsets (28, 29). We also failed to identify an enhanced overall sensitivity of TF Envs to neutralization, although subtype B TF Envs were slightly more sensitive to CD4 binding site neutralizing antibodies than subtype C TF Envs (28). Most recently, we discovered that TF Envs are more completely inhibited by small-molecule CCR5 antagonists than control Env constructs, suggesting differences in CCR5 interaction (30); however, the biological relevance of this finding remains to be determined (30). Thus, except for preferential CCR5 coreceptor use and CD4+ T-cell tropism, no other reproducible phenotypic difference between TF and chronic viruses has been identified.
Env pseudotypes are generated by cotransfection of an env expression cassette with an env-minus proviral backbone. Because this backbone represents a standard HIV-1 genome, the contribution of gene products other than Env to the TF phenotype cannot be assessed. Moreover, overexpression of Env, which is inherent in the cotransfection assay, precludes a meaningful assessment of virus–host cell interactions and particle composition. Reasoning that these shortcomings may have obscured important biological differences, we developed methods to clone full-length TF genomes and showed that these produced replication-competent viruses (31–33). However, a systematic evaluation of the TF phenotype also required a sufficient number of molecularly cloned chronic control (CC) viruses for comparison. Here, we describe the generation and biological characterization of a comprehensive panel of TF (n = 27) and CC (n = 14) infectious molecular clones (IMCs), representing two major group M subtypes. Using these reagents, we compared biological properties that would be expected to influence viral fitness during the earliest stages of the transmission process. Our results reveal that TF viruses share common traits that likely enhance their fitness in crossing mucosal surfaces and promoting the establishment of a productive initial infection.
Results
IMCs of TF and CC Viruses.
Although a limited number of TF and CC IMCs has previously been reported (29, 31–33), available clones, especially from chronically infected individuals, were too few to conduct meaningful phenotypic comparisons (29). To create a more balanced panel, with respect to both the number of TF and CC IMCs and their subtype representation, we cloned additional viral genomes from individuals enrolled in acute and chronic HIV-1 infection cohorts. Using previously reported methods (31–33), we inferred 12 additional TF genomes (Fig. S1) representing mucosally transmitted viruses from single (n = 8) and multivariant (n = 4) infections. Together with existing constructs, these clones comprised a panel of 27 TF IMCs, with equal representation of subtypes B (n = 13) and C (n = 14) viruses (Table S1).
Chronically HIV-1–infected individuals harbor complex quasispecies of genetically diverse HIV-1 variants. Because it is impossible to predict based on sequence inspection alone which variants are biologically active and which variants are functionally impaired, we amplified between 20 and 40 env genes or 3′ half-genomes from chronic infection plasmas and searched for clusters of nearly identical sequences as indicators of recent clonal expansions (Fig. S2). We reasoned that the inferred common ancestors of these clusters must encode persistently replicating viruses and thus, represent relevant controls for biological comparisons with TF viruses. Consistent with this interpretation, we found that all chronic IMCs generated from such expansion rakes produced viruses that grew to high titers in CD4+ T cells. However, not all chronic infection plasmas were suitable for IMC construction. Analyzing specimens from over 60 individuals, we identified only 14, including 4 reported previously (29), that exhibited clonal expansion rakes in both 3′ and 5′ halves of their viral genomes (Fig. S2). These chronic plasma samples were used to construct CC IMCs representing both subtypes B (n = 5) and C (n = 9) infections (Table S1).
To determine the coreceptor use of 22 newly derived IMCs, we infected CCR5- and CXC chemokine receptor type 4 (CXCR4)-expressing reporter cells in both the presence and absence of their respective inhibitors (16). Consistent with previous analyses of Env pseudotypes (16, 28, 29, 34), the results indicated that all IMCs were CCR5 tropic, except for one TF and two CC viruses that were dual tropic for CCR and CXCR4 (Table S1).
Phenotypic Studies.
All biological experiments were performed using viral stocks that were CD4+ T cell-derived, sucrose-purified, and depleted of CD45+ microvesicles. Virus was quantified by measuring reverse transcriptase (RT) activity, viral RNA copy number, and Gag p24 antigen content. Comparing these values, we noticed that subtype C stocks seemed to contain about fivefold less p24 antigen per unit of RT activity than subtype B stocks (Fig. S3A). This discrepancy was also seen when p24 antigen was normalized using RNA copy numbers (Fig. S3B). However, no such difference was observed when virion RT activity was compared with RNA copy number (Fig. S3 C and D). Similar results were obtained when viral stocks were tested in two additional p24 detection assays (Fig. S3 E and F). Thus, three commonly used p24 capture assays bound subtype C core proteins considerably less efficiently than subtype B core proteins, leading to a systematic underestimation of the number of viral particles in subtype C viral stocks. We, therefore, used RT activity to normalize virus input and measure virus replication in all subsequent experiments.
To determine whether TF and CC viruses differed in their phenotypic properties, we used two complementary statistical approaches. First, we used a conservative nonparametric permutation test (perm test) to address the central question of whether TF and CC viruses exhibited reproducible phenotypic differences. Second, we used a generalized linear model (GLM) to test for interactions between different parameters, such as subtype and cell donors in replicate experiments, and test for the independence of viruses from the same subjects. Because both virus status (TF or CC) and subtype (B or C) influenced the biological outcome, the GLM test allowed us to interpret the data with these variables taken into account.
TF Viruses Exhibit Enhanced Infectivity.
Plasma virus collected during ramp-up stages of acute simian immunodeficiency virus (SIV) infection has been shown to be significantly more infectious than virus collected from later plasma samples (35). To examine whether this observation was also true for HIV-1, we used a single-round infection assay to determine whether TF virions were more infectious on a per-particle basis than CC virions. Serial dilutions of CD4+ T cell-derived viral stocks were used to infect TZM-bl cells (which express luciferase under the control of an HIV-1 promoter), and the resulting relative light units were expressed as a function of the input RT activity. The results showed that TF viruses as a group were 1.7-fold more infectious than CC viruses (Fig. 1A), although this difference was only marginally significant (P = 0.049 by perm test; P = 0.062 by GLM). Similar results were obtained when subtypes B and C viruses were considered separately (there was no subtype dependence of infectivity) (Fig. 1B). To enhance virus infectivity, we repeated these experiments in the presence of diethylaminoethyl (DEAE) dextran, which is known to increase virus attachment to _target cells (36). As expected, DEAE dextran increased the per-particle infectivity of all viruses by up to three orders of magnitude (Fig. 1C), with a significantly greater effect on subtype C than subtype B virions (P = 0.015 by GLM) (Fig. 1D). However, in the presence of DEAE dextran, the marginally significant difference between TF and CC viruses shifted to nonsignificance (P = 0.068 by perm test; 0.083 by GLM). Taken together, these data indicate that TF viruses are, on average, nearly two times as infectious as viruses that persist during chronic infection. However, when normal barriers to cell-free infection are mitigated by DEAE dextran, this difference is no longer significant (although a trend was still evident).
Fig. 1.
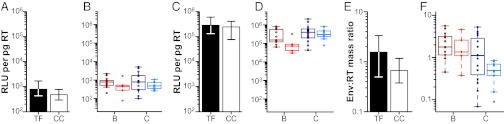
Virion infectivity and Env content. (A–D) Infectivity values for TF and CC viruses (x axis) are expressed as relative light units (RLUs) per picogram of viral RT activity (y axis). (A) Bars indicate the median infectivity of TF (filled) and CC (open) viruses, with interquartile ranges indicated. TF viruses were 1.7-fold more infectious than CC viruses (P = 0.049). (B) Infectivity values are shown for each virus. Subtypes B and C viruses are shown in red and blue, respectively, with TF viruses indicated in dark colors and CC viruses indicated in light colors, respectively. Values represent averages from four independent experiments. (C and D) Infectivity values are shown for TF and CC viruses as in A and B, except that infections were performed in the presence of DEAE dextran. Values represent averages from three independent experiments. (E and F) Env content of TF and CC virions (x axis) is expressed as the mass ratio of Env and RT content (y axis). (E) Bars indicate the median values of Env content for TF (filled) and CC (open) viruses, with interquartile ranges indicated. TF viruses contained 1.9 times more Env per unit of RT activity than CC viruses (P = 0.048). (F) Env content is shown for each virus and color-coded as in B and D. Values represent averages from two independent experiments.
TF Virions Have a Higher Env Content.
Previous computational analyses of TF and CC env gene sequences revealed genetic signatures in TF viruses that increased Env expression and particle incorporation when tested in the context of pseudoviruses (37). We, thus, examined whether TF virions packaged more Env per particle than CC virions. To detect both subtypes B and C Env proteins with comparable efficiency, we developed an ELISA that used antibodies previously shown to bind genetically highly diverse envelope (gp120) proteins. To capture Env, we used CD4-218.3-E51, a chimeric antibody that contains the first and second domains of human CD4 linked to the CD4-induced monoclonal E51 (38). To detect bound Env, we selected affinity-purified anti-gp120–specific polyclonal antibodies that were isolated from human plasma (Advanced Bioscience Laboratories, Inc.). Using this ELISA, we found that TF viruses contained 1.9 times more Env per unit of RT activity than CC viruses (P = 0.048 by perm test; P = 0.057 by GLM) (Fig. 1E). Subtype B viruses appeared to package 2.4-fold more Env per particle than subtype C viruses (Fig. 1F), but this result was largely caused by a binding preference of the capture antibody, which recognized subtype B Env glycoproteins two times more efficiently than subtype C Env glycoproteins. Nonetheless, this subtype bias did not affect the differential between TF and CC viruses, which was seen for both subtypes B and C, and it was controlled for in the statistical analyses (Fig. 1F). As expected, there also was a significant correlation between Env content and particle infectivity (r = 0.33; P = 0.036).
TF Viruses Bind DCs More Efficiently.
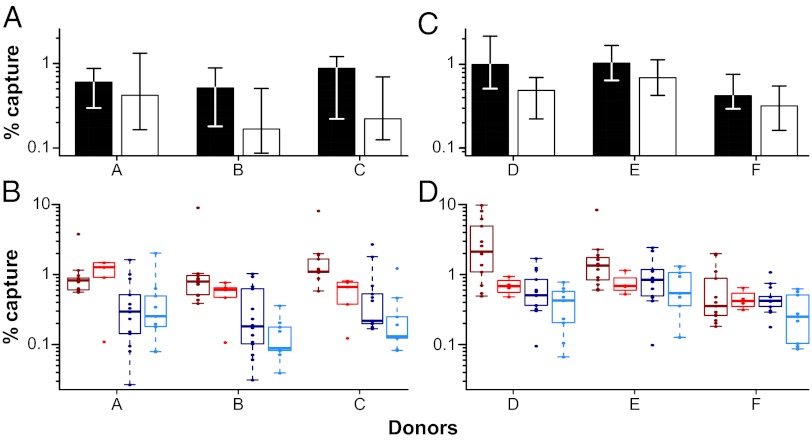
DCs have been proposed to play an important role in HIV-1 transmission, because they are located in the mucosa (39, 40), capture viruses using lectins (41, 42) and glycosphingolipid receptors (43), and efficiently transmit infectious particles to CD4+ T cells (44–46). To examine whether TF and CC viruses differ in their ability to bind DCs, we pulsed immature monocyte-derived DCs (moDCs) with equal amounts of virus (normalized by RT activity), washed the cells extensively to remove cell-free virions, and then lysed the cells to quantify the amount of cell-associated virus. Using cells from three different donors (Fig. 2A), we found that moDCs captured TF viruses 1.6 times more efficiently than CC viruses (P = 0.040 by perm test; P = 0.060 by GLM). This increase was observed for both subtypes B and C TF viruses (Fig. 2B), although subtype B viruses were captured 3.4 times more efficiently than subtype C viruses (P = 4.6 × 10−6 by GLM) (Fig. 2B).
Fig. 2.
Virus binding to moDCs. The percent of captured TF and CC virus is plotted (y axis) for moDC cell preparations from six different donors labeled A through F (x axis). (A and B) Virus input was normalized by RT activity. (C and D) Virus input was normalized by p24 content. (A and C) Bars indicate median values of moDC capture for TF (filled) and CC (open) viruses, with interquartile ranges indicated. TF viruses were captured 1.7-fold more efficiently than CC viruses (P = 0.035). (B and D) Values are plotted for each virus individually (color-coding for TF and CC viruses from subtypes B and C as in Fig. 1). Subtype B viruses were captured 3.4 times more efficiently than subtype C viruses (P = 4.6 × 10−6 by GLM).
Because in the above experiment, we had normalized virus input by RT activity but measured virion capture using the more sensitive p24 assay, we considered the possibility that the subtype-specific differences were an artifact of the p24 antigen capture assay. To explore this possibility, we repeated the binding experiment with cells from three additional donors, but this time, we normalized virus input by p24 content (Fig. 2 C and D). Despite adding an estimated fivefold excess of subtype C virions, TF viruses were again captured 1.8 times more efficiently than CC viruses (P = 0.030 by perm test; P = 0.014 by GLM). Moreover, there was still a subtype-specific difference, with subtype B viruses being captured 1.9 times more efficiently than subtype C viruses (P = 0.0029 by GLM). As a control, we treated an aliquot of the pulsed moDCs with 0.25% trypsin-EDTA and showed that this treatment removed all detectable cell-associated virus, confirming that most of the DC-associated virus was surface-exposed (47). We also cultured virus-exposed moDCs and showed that these cells were not productively infected. Finally, we asked whether the percentage of captured virus for each strain correlated between different donors. This association was indeed observed regardless of whether virus input was normalized by RT activity or p24 content, thus validating the DC binding assay (Fig. S4A).
Although the combined data from all six donors indicated that DCs captured TF viruses 1.7-fold more efficiently than CC viruses (P = 0.035 by perm test; P = 0.005 by GLM), there was considerable donor variability, with moDCs from two donors (A and F in Fig. 2) failing to yield significant binding differences (Fig. S4). To examine potential reasons, we compared surface expression levels of DC-specific intracellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN) and macrophage mannose receptor (MMR) by flow cytometry. Interestingly, moDCs of donors A and F expressed lower levels of these molecules (Fig. S4 B and C), suggesting that the number of Env-specific binding sites on their cells was limited. For the remaining four donors, moDC virus capture tended to correlate with particle Env content (Fig. S4D). These data suggest that virion Env content represents one important determinant in moDCs binding, although expression of lectins and other factors clearly also play a role (48).
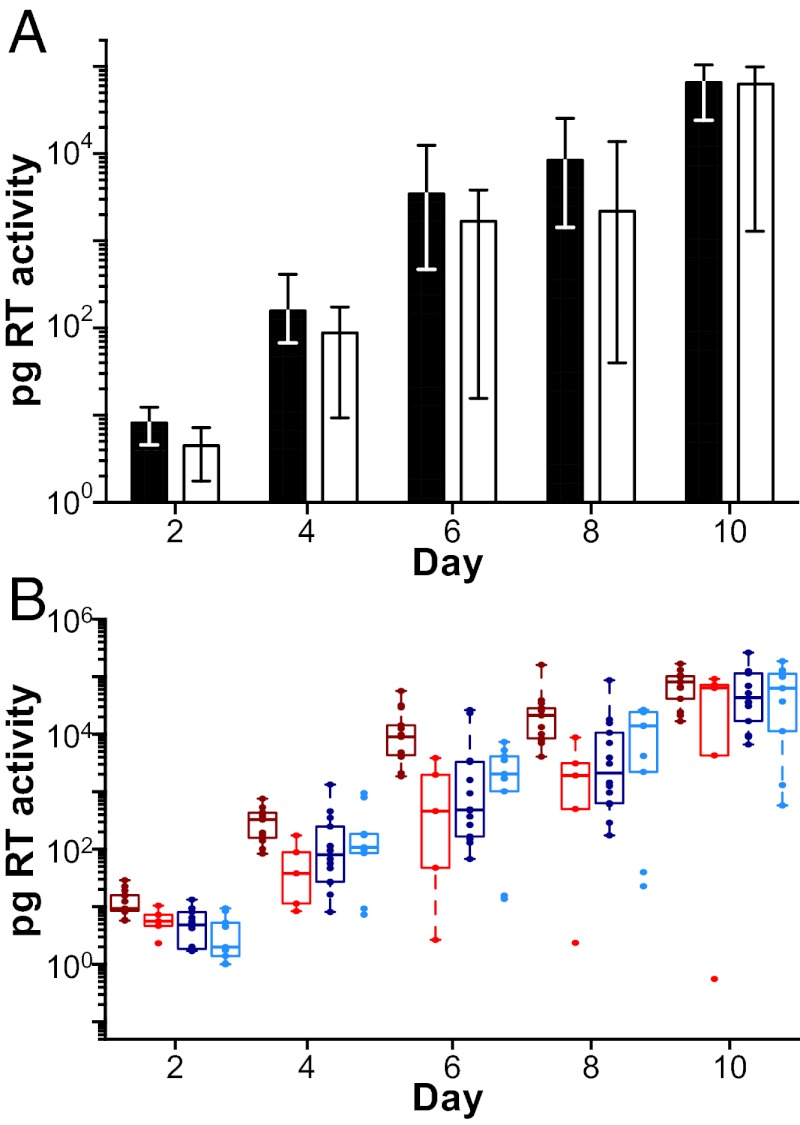
To examine the efficiency of virus transfer from moDC to CD4+ T cells, we pulsed moDCs with equivalent amounts of TF and CC viruses, cocultured these cells with autologous CD4+ T cells, and then measured RT activity in culture supernatants as an indicator of virus replication. Analysis of cells from two different donors showed that TF viruses replicated to slightly higher titers than CC viruses each day over the course of 10 d (one representative donor is shown in Fig. 3). Although these differences were not significant in the perm test (P = 0.104), significant differences in TF and CC virus titers were observed in the GLM analysis (P = 0.025). To explore the reason for this apparent discordance, we ran the perm test on subtypes B and C viruses separately (Fig. 3B), and we found that subtype B TF viruses (P = 0.004), but not subtype C TF viruses (P = 0.23), grew to significantly higher titers than their respective chronic controls. Averaging data from both donors and across all time points, this difference was 11.2-fold for subtype B viruses but not significant for subtype C viruses. Because subtype B virions bound moDCs 3.5-fold more efficiently than subtype C viruses (Fig. 2 B and D), we interpret these findings to indicate that virions that bind moDCs more efficiently are also transferred to CD4+ T cells more efficiently.
Fig. 3.
DC-mediated trans infection. (A) Virus replication expressed as picograms of RT activity per milliliter of culture supernatant (y axis) is shown after cocultivation of virus-pulsed moDC with CD4+ T cells for one representative donor (of two analyzed) over 10 d (x axis). Bars indicate median values of viral replication for TF (filled) and CC (open) viruses, with interquartile ranges indicated (there was no significant difference between TF and CC viruses). (B) Values are plotted for each virus individually (color-coding for TF and CC viruses from subtypes B and C as in Fig. 1). Averaging data from two different donors, subtype B TF viruses grew to 11.2-fold higher titers than subtype B CC viruses (P = 0.004), whereas no significant differences were observed for subtype C TF and CC viruses (P = 0.23).
TF Viruses Are Relatively More Resistant to IFN-α.
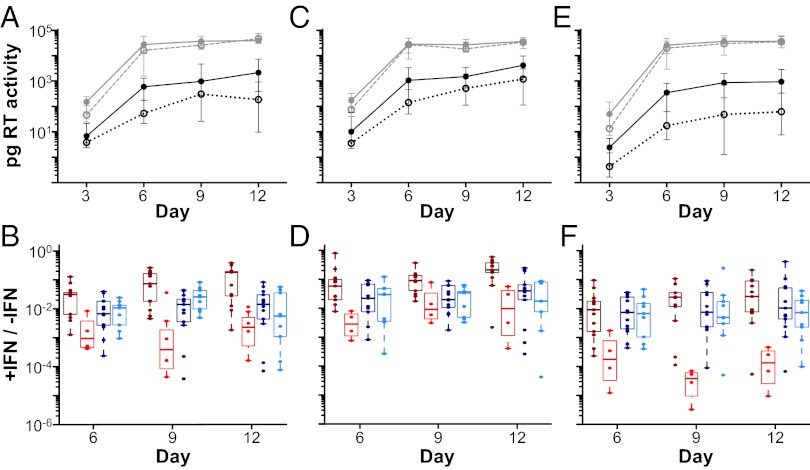
CD4+ T cells represent the predominant _target cells in gut-associated lymphoid tissues and the first cell type to become productively infected after transmission (49). To determine the growth potential of TF and CC viruses in this cell type, we infected activated CD4+ T cells from three healthy donors with equivalent amounts of virus and monitored viral growth by measuring RT activity in culture supernatants (Fig. 4 A, C, and E, gray lines). Although TF viruses replicated to slightly higher titers, especially at the earliest time point postinfection, this difference was not statistically significant (P = 0.16 by perm test; P = 0.186 by GLM). Thus, in activated CD4+ T cells, TF and CC viruses replicated with comparable efficiency.
Fig. 4.
Virus replication in CD4+ T cells in the presence and absence of IFN-α. (A, C, and E) The replication kinetics of TF (solid lines) and CC (broken lines) viruses are shown in CD4+ T cells from three donors in the absence (gray lines) and presence (black lines) of 500 U IFN-α. RT activity indicated as picograms per milliliter of culture supernatant (y axis) was measured every 3 d (x axis). Data points indicate median values of virus production, with interquartile ranges indicated. Averaging data from all donors and points, TF viruses grew to 24-fold higher titers than CC viruses in the presence of IFN-α (P = 0.012). (B, D, and F) The ratio of virus production in the presence and absence of IFN-α is plotted for each virus (y axis) at different time points (days) postinfection (x axis; color-coding for TF and CC viruses from subtypes B and C as in Fig. 1). Averaging data from all donors, subtype B TF viruses grew to 62-fold higher titers than subtype B CC viruses (P = 0.000013), whereas subtype C TF viruses grew only 1.7-fold more efficiently than subtype C CC viruses (P = 0.53). Note that these replication differentials represent cumulative totals for the 12-d culture period.
IFN-α is produced early in infection by plasmacytoid DCs (50), which are among the first cells to be recruited to the mucosal site of virus entry (4). This cytokine stimulates expression of hundreds of host genes [IFN-stimulated genes (ISGs)] (51), many of which have anti–HIV-1 activity (52). IFN-α has also been shown to limit Env incorporation into virus particles that are released from infected CD4+ T cells (53). To determine the sensitivity of TF and CC viruses to the antiviral activity of this cytokine, we analyzed their replication kinetics in CD4+ T cells in the presence of 500 U/mL IFN-α. Determining RT activity in culture supernatants as a measure of viral replication, we found significant differences in the growth rate of TF and CC viruses (Fig. 4 A, C, and E, black lines). However, this effect was highly subtype-specific (Fig. S5). Averaging replication data from all donors and across all time points, we found that subtype B TF viruses grew to 62-fold higher titers than subtype B CC viruses (P = 0.000013 by GLM). In contrast, subtype C TF viruses grew only 1.7-fold more efficiently than their respective CC viruses, which was not significant (P = 0.53 by GLM). Importantly, this difference was not because of a lack of IFN-α resistance of the subtype C TF viruses but an increased IFN-α resistance of the respective chronic controls (Fig. 4 B, D, and F). Inclusion of sex and risk group information into the statistical analyses indicated that neither was significantly associated with IFN-α resistance; however, the study was not sufficiently powered to examine the impact of sex and risk group, and a contribution of these factors cannot, therefore, be excluded (Fig. S5).
As a different way to analyze these data, we also calculated the ratio of virus production in the presence and absence of IFN-α for each virus strain (Fig. 4 B, D, and F). Comparing across all donors, we again found that TF viruses were significantly more resistant to inhibition by IFN-α than chronic viruses (P = 0.027 by perm test). However, when considering the two clades separately, this finding was significant only for subtype B (P = 0.0004 by one-sided Wilcoxon test) and not for subtype C (P = 0.84). Viruses that were highly resistant in one donor were also highly resistant in the other donors (Fig. S6), indicating that their IFN-α phenotype was consistent between donors.
To determine whether the higher titers of TF viruses were because of increased particle release or differences in viral spread, we used flow cytometry to measure the percent of infected cells in IFN-α–treated cultures from two donors (Fig. S7). In the presence of IFN-α, TF viruses infected a larger number of cells than CC viruses, although the overall effect was only marginally significant (P = 0.049 by perm test). However, GLM analysis again detected a significant interaction between subtype and TF/CC status (P = 0.041), indicating that the differences were primarily caused by subtype B viruses. Because the ratio of virus production in the presence and absence of IFN-α correlated well with the percent of Gag-positive cells (P < 0.0001) (Fig. S7 B and C), it is likely that IFN-α resistance represents an enhanced ability to spread between CD4+ T cells. Interestingly, this phenotype correlated only weakly with particle infectivity and Env content, indicating that the higher levels of particle-associated Env in TF viruses are not the main drivers of their relative IFN-α resistance.
Discussion
A primary goal of AIDS vaccine development is to prevent acquisition of HIV-1 at mucosal surfaces. In this context, it is critical to determine whether newly transmitted HIV-1 strains share traits that provide novel _targets for effective immunization. For such an analysis, the use of full-length IMCs is essential, because they contain the complete genetic information of viruses that successfully transmit infection. Moreover, genetic linkage is maintained to preserve potentially important regulatory and structural protein interactions (16, 31–33). Characterizing a large set of TF and CC IMCs from two major HIV-1 group M subtypes, we found that TF viruses were slightly more infectious than CC viruses on a per particle basis, packaged slightly more Env, bound to moDCs more efficiently, and were relatively more resistant to the antiviral effects of IFN-α. These biological differences could act in concert to enhance cell-free infection and virus replication in the face of an early innate immune response.
Analyzing particle composition and infectivity, we found that TF virions were, on average, two times as infectious and contained two times as much Env as CC virions. These phenotypic differences are consistent with data from a recent genetic study (22) that identified sequence signatures in the signal peptide of TF Env sequences. When tested in the context of pseudoviruses, these signatures increased steady-state Env expression and particle incorporation (37). Assuming 35 enzymatically active RT dimers per virion, we estimate that the median TF virion contains 18 Env trimer spikes, whereas the median CC virion contains 7 spikes, a value essentially identical to previous estimates (54). Because only one or a few functional Env trimers are believed to be necessary for cell fusion and infection (55), it would seem that TF viruses carry an excess of envelope glycoprotein. Such excess is likely of advantage during the earliest stages of HIV-1 infection, because viruses with a higher Env content may also contain more functional trimers. Particle-associated Env may also play a role beyond the mere fusion and cell entry function. For example, excess Env could ensure more stable attachment of virions to _target cells and/or be involved in receptor and/or coreceptor mediated signaling (56). In addition, excess Env may compensate for shedding or inactivation by semen (57) and cervicovaginal mucous (58). Thus, it seems likely that the increased Env content and associated enhanced infectivity of TF viruses serve to increase their transmission fitness. Conversely, the lower Env content of CC viruses may be more advantageous after infection is established and may even be selected for by Env-specific neutralizing antibodies.
We also found that TF viruses bind more efficiently to moDCs than their chronic counterparts. Although moDCs are only a proxy for mucosal DC subsets, previous studies have shown that tissue- and in vitro-derived DCs have similar virus capture capabilities (59, 60). Thus, it is likely that the improved interaction of TF viruses with tissue culture-derived moDCs is also a reflection of their in vivo biological properties. The differential binding of TF viruses was statistically significant across different donors and across both clades, and thus, it seems to represent a general feature of TF viruses. There are at least two steps in the transmission process where this phenotype may confer an advantage. First, more efficient binding to tissue resident DCs may increase the likelihood of a successful transfer to CD4+ T cells. Second, more efficient binding to emigrating DCs may promote the seeding of regional lymph nodes (61). In this context, it is of interest that adaptive changes that conferred mucosal transmissibility to a commonly used simian-human immunodeficiency virus (SHIV-SF162P3) increased the binding capacity of its Env glycoprotein to DC-SIGN (62). Our data are, thus, consistent with the view that DCs are among the first cells to interact with HIV-1 after exposure and that this interaction is important for transmission, although DCs themselves are not productively infected.
One unexpected finding was that TF viruses replicated and spread in CD4+ T cells in the presence of IFN-α more efficiently than CC viruses. Although this effect was most pronounced for subtype B TF viruses, a trend was also observed for subtype C TF viruses. This finding raises the question of whether IFN-α responses in the mucosa are protective against HIV-1 infection. On first glance, a review of the literature would suggest that the answer to this question is no. Previous studies in macaques concluded that the initial innate immune response is ineffective at containing SIV infection and may even enhance early viral replication (4). Moreover, mucosal pretreatment with toll-like receptor agonists induced IFN-α but did not protect animals from SIV challenge (63). However, these studies were not only assessing the effects of IFN-α but also causing immune activation by inducing proinflammatory cytokines and chemokines. In fact, it has been shown more recently that blockade of type 1 IFN during acute SIV infection resulted in accelerated progression to AIDS and death (64). It, thus, seems that early IFN responses can be protective against HIV-1/SIV infection. The fact that IFN-α levels (65) as well as IFN regulatory factor polymorphisms (66) have been associated with protection in highly exposed but uninfected individuals is consistent with this hypothesis. Thus, the induction of an effective early antiviral state seems to contribute to the population bottleneck associated with mucosal HIV-1 transmission.
We hypothesized that virions containing more Env would be more resistant to IFN-α, because in previous studies, this cytokine was reported to decrease cell-free virus transmission (67), particle release (68), and particle infectivity (53). However, we failed to see a significant correlation between IFN-α resistance and Env content or virion infectivity. Thus, higher baseline Env incorporation cannot explain the IFN-α–resistant phenotype of TF viruses. Nonetheless, the ability of TF viruses to replicate in the presence of IFN-α indicates that resistance-conferring determinants must exist. There are several candidates, such as the accessory proteins Vpu and Vif, that are known to counteract the IFN-stimulated genes tetherin and apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G), respectively (69, 70), as well as the Tat protein, which has been reported to modulate the IFN-induced RNase L antiviral pathway (71) and down-regulate the ISG protein kinase R (72). Structural proteins, such as Gag, may also contribute to IFN-α resistance by exhibiting differential sensitivity to the tripartite motif family of proteins (73–75). Finally, IFN-α resistance may not only be protein-mediated, because variation in the number of transcription factor binding sites in the long terminal repeat (LTR) region could also influence IFN-α sensitivity (76, 77). Notably, there is a relationship between IFN-α resistance and efficiency of transmission in other viruses, including Venezuelan Equine Encephalitis virus (78) and Bovine Viral Diarrhea Virus (79), suggesting that increased IFN-α resistance may be a general property of transmitted founder viruses.
If IFN-α resistance was a prerequisite for efficient HIV-1 transmission, one would expect this viral property to be conserved across all clades. The lack of significance between subtype C TF and CC viruses seems to argue against this conclusion; however, Fig. 4 shows that the lack of significance is not caused by a loss of IFN-α resistance of subtype C TF viruses. Instead, the 62-fold cumulative replication differential observed for subtype B viruses reflects a much greater IFN-α sensitivity of the respective chronic controls. Although our panel of TF and CC IMCs is much larger than any previously tested, there are epidemiological differences between the patient cohorts from which they were derived. For example, all but one of the chronic subtype B IMCs were derived from men who had sex with men, whereas seven of eight chronic subtype C IMCs were derived from heterosexual women (Table S1). This sex bias may have contributed to the increased IFN-α resistance of chronic subtype C viruses, because plasmacytoid dendritic cells (pDCs) of women have been reported to generate more IFN-α in response to HIV-1 than pDCs of men (80). In addition, all subtype C viruses were derived from individuals living in Africa, whereas all subtype B viruses were derived from patients residing in the United States (Table S1). Thus, environmental factors, such as concurrent infections that increase general inflammation levels, may have selected for the much higher IFN-α resistance of the respective chronic viruses. Finally, if loss of IFN-α resistance is the result of pressure from host adaptive responses, then duration of infection may have been an important variable. We have recently found that subtype B viral isolates obtained from subjects later in infection are more sensitive to IFN-α than the viruses isolated shortly after transmission. Thus, it seems clear that, to decipher the reasons for the IFN-α resistance differences observed here, future studies will need to consider patient demographics, route of transmission, and duration of infection in addition to TF and CC virus status. Such studies may also shed light on factors that fuel the HIV-1 epidemic in Africa.
One of the motivations to characterize the phenotypic properties of TF viruses is the possibility that virus traits might be uncovered that could represent useful vaccine _targets. In this context, the increased Env content of TF viruses may increase their sensitivity to neutralization by allowing antibodies to bind bivalently and potentially cross-link more densely packed Env spikes (81). Similarly, the identification of key ISGs that contribute significantly to viral control during the most vulnerable stages of HIV-1 infection may provide a new vaccination strategy, by either vaccine vector-mediated type I IFN production and/or induction of T cells that express the antiviral ISGs.
The molecular identification and biological analysis of TF viral genomes are powerful enabling tools for characterizing the transmission requirements and subsequent evolution of HIV-1, SIV, and other viruses (16, 31–33, 82). In the present study, we used TF analyses to characterize the biological properties of full-length HIV-1 genomes captured at the moment of transmission. Although we identified significant phenotypic differences, none of these differences completely differentiated TF from CC viruses, and the magnitude of the observed differences was generally modest. This finding is not surprising, because TF viruses, by necessity, represent a subset of a much larger and more diverse set of viruses that replicates persistently throughout chronic infection. We speculate that the viral population bottleneck associated with mucosal transmission is largely caused by inherent physical barriers but also specific virus–host cell interactions necessary for early virus infection and replication, each of which may pose hurdles to virus acquisition. Thus, even slight advantages of TF viruses at one or more of these biophysical checkpoints could significantly increase their transmission fitness. Defining the viral determinants that underlie this fitness will provide additional insights into the biology of mucosal HIV-1 transmission and may also inform vaccine design.
Materials and Methods
IMC Construction.
The sequences of full-length TF and CC viruses were inferred and molecularly cloned as described (27, 29, 31–33). GenBank accession numbers are listed in Table S2.
Viral Stocks.
CD4+ T cells pooled from nine donors were infected with 293T cell-derived transfection supernatants. Viral stocks were harvested after 11 d and depleted of CD45+ microvesicles. Viral p24 core protein content was measured using the PerkinElmer High-Sensitivity AlphaLISA, the PerkinElmer Alliance ELISA, and the method described by Biancotto et al. (83). RT activity was measured using the Roche Reverse Transcriptase Assay. Viral RNA was quantified using the Roche COBAS AmpliPrep/COBAS Taqman HIV-1 v. 2.0 Test.
Phenotypic Analyses.
Coreceptor use was determined as described (33). Virion infectivity was measured using the TZM-bl reporter cell line. Particle-associated Env content was measured using a newly developed ELISA (SI Materials and Methods). To assess DC capture and CD4+ T-cell transfer, peripheral blood monocytes were differentiated to moDCs using 100 IU/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) and 200 IU/mL IL-4. For moDC capture experiments, 1 × 105 cells were incubated with viral stocks containing either 30 pg RT activity or 600 pg p24 protein. For CD4+ T-cell transfer experiments, viral stocks containing 50 pg RT activity were added, and moDCs were cultured at a 1:5 ratio with autologous CD4+ T cells. For CD4+ T-cell infection studies, 2 × 106 negatively selected CD4+ T cells were infected overnight with viral stocks containing 30 pg RT activity. Cells were then propagated in media containing 30 U/mL IL-2 in the presence and absence of 500 U/mL IFN-α. Infected cells were identified by flow cytometry (SI Materials and Methods).
Statistical Analyses.
Data were analyzed using a nonparametric perm test that controlled for virus subtype, donor cells, and time of sampling as well as a GLM that tested for interactions between and the relative importance of these variables (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank John Moore, Frank Kirchhoff, Paul Sharp, and Stuart Shapiro for helpful discussions; the University of Pennsylvania’s Center for AIDS Research (CFAR) Human Immunology, Flow Cytometry, and Viral and Molecular Core facilities for reagents and protocols; and Patricia Crystal for artwork and manuscript preparation. This work was supported by National Institutes of Health (NIH) Grants R01 AI45378, R01 AI04088, P30 AI45008, and P30 AI27767, Center for HIV/AIDS Vaccine Immunology Grant U19 AI067854, Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Grant UM1 AI100645, and Bill and Melinda Gates Foundation Grant 37874. N.F.P., S.S.I., and C.B.W. were supported by NIH Training Grant T32 AI07632.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC149260–KC149299, KC149138–KC149185, KC149442–KC149450, KC156222–KC156499, KC312330–KC312539, KC312540–KC312600, JX972238–JX972249, JX972390–JX972410, JX972455–JX972487, JX972838–JX972845, JX972931–JX972964, JX972986–JX972998, JX973075–JX973101, JX973171–JX973180, JX973234–JX973270, JX973388–JX973412, JX973482–JX973502, JX973549–JX973555, JX973846–JX973854, JX973930–JX973938, JX973955–JX973969, JX974014–JX974023, JX974082–JX974094, and JX974138–JX974164).
See Profile on page 6613.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304288110/-/DCSupplemental.
References
- 1.Shaw GM, Hunter E. HIV transmission. Cold Spring Harb Perspect Med. 2012;2(11):pii:a006965. doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hladik F, McElrath MJ. Setting the stage: Host invasion by HIV. Nat Rev Immunol. 2008;8(6):447–457. doi: 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286(5443):1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458(7241):1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83(8):3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schacker T, et al. Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J Infect Dis. 2001;183(4):555–562. doi: 10.1086/318524. [DOI] [PubMed] [Google Scholar]
- 7.Moss GB, et al. Association of cervical ectopy with heterosexual transmission of human immunodeficiency virus: Results of a study of couples in Nairobi, Kenya. J Infect Dis. 1991;164(3):588–591. doi: 10.1093/infdis/164.3.588. [DOI] [PubMed] [Google Scholar]
- 8.Gray RH, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357(9263):1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 9.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86(3):367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, et al. Persistence of HIV-1 receptor-positive cells after HSV-2 reactivation is a potential mechanism for increased HIV-1 acquisition. Nat Med. 2009;15(8):886–892. doi: 10.1038/nm.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaren PJ, et al. HIV—exposed seronegative commercial sex workers show a quiescent phenotype in the CD4+ T cell compartment and reduced expression of HIV—dependent host factors. J Infect Dis. 2010;202(Suppl 3):S339–S344. doi: 10.1086/655968. [DOI] [PubMed] [Google Scholar]
- 12.Laga M, et al. Non-ulcerative sexually transmitted diseases as risk factors for HIV-1 transmission in women: Results from a cohort study. AIDS. 1993;7(1):95–102. doi: 10.1097/00002030-199301000-00015. [DOI] [PubMed] [Google Scholar]
- 13.Atashili J, Poole C, Ndumbe PM, Adimora AA, Smith JS. Bacterial vaginosis and HIV acquisition: A meta-analysis of published studies. AIDS. 2008;22(12):1493–1501. doi: 10.1097/QAD.0b013e3283021a37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeras DI, et al. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proc Natl Acad Sci USA. 2011;108(46):E1156–E1163. doi: 10.1073/pnas.1103764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bull ME, et al. Human immunodeficiency viruses appear compartmentalized to the female genital tract in cross-sectional analyses but genital lineages do not persist over time. J Infect Dis. 2013 doi: 10.1093/infdis/jit016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derdeyn CA, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303(5666):2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 18.Chohan B, et al. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79(10):6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, et al. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol. 2006;80(11):5211–5218. doi: 10.1128/JVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. Env length and N-linked glycosylation following transmission of human immunodeficiency virus Type 1 subtype B viruses. Virology. 2008;374(2):229–233. doi: 10.1016/j.virol.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curlin ME, et al. HIV-1 envelope subregion length variation during disease progression. PLoS Pathog. 2010;6(12):e1001228. doi: 10.1371/journal.ppat.1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnanakaran S, et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 2011;7(9):e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander M, et al. Donor and recipient envs from heterosexual human immunodeficiency virus subtype C transmission pairs require high receptor levels for entry. J Virol. 2010;84(8):4100–4104. doi: 10.1128/JVI.02068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaacman-Beck J, et al. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol. 2009;83(16):8208–8220. doi: 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Functional properties of the HIV-1 subtype C envelope glycoprotein associated with mother-to-child transmission. Virology. 2010;400(2):164–174. doi: 10.1016/j.virol.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nawaz F, et al. The genotype of early-transmitting HIV gp120s promotes α (4) β(7)-reactivity, revealing α (4) β(7) +/CD4+ T cells as key _targets in mucosal transmission. PLoS Pathog. 2011;7(2):e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar-Gonzalez JF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82(8):3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilen CB, et al. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol. 2011;85(17):8514–8527. doi: 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrish NF, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog. 2012;8(5):e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker ZF, et al. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol. 2013;87(5):2401–2411. doi: 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salazar-Gonzalez JF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206(6):1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6(5):e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochsenbauer C, et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol. 2012;86(5):2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang C, et al. Primary infection by a human immunodeficiency virus with atypical coreceptor tropism. J Virol. 2011;85(20):10669–10681. doi: 10.1128/JVI.05249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma ZM, et al. High specific infectivity of plasma virus from the pre-ramp-up and ramp-up stages of acute simian immunodeficiency virus infection. J Virol. 2009;83(7):3288–3297. doi: 10.1128/JVI.02423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt EJ, Kozak SL, Durnin JP, Hope TJ, Kabat D. Rapid dissociation of HIV-1 from cultured cells severely limits infectivity assays, causes the inactivation ascribed to entry inhibitors, and masks the inherently high level of infectivity of virions. J Virol. 2010;84(6):3106–3110. doi: 10.1128/JVI.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asmal M, et al. A signature in HIV-1 envelope leader peptide associated with transition from acute to chronic infection impacts envelope processing and infectivity. PLoS One. 2011;6(8):e23673. doi: 10.1371/journal.pone.0023673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West AP, Jr, et al. Evaluation of CD4-CD4i antibody architectures yields potent, broadly cross-reactive anti-human immunodeficiency virus reagents. J Virol. 2010;84(1):261–269. doi: 10.1128/JVI.01528-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hladik F, et al. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity. 2007;26(2):257–270. doi: 10.1016/j.immuni.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fahrbach KM, Barry SM, Anderson MR, Hope TJ. Enhanced cellular responses and environmental sampling within inner foreskin explants: Implications for the foreskin’s role in HIV transmission. Mucosal Immunol. 2010;3(4):410–418. doi: 10.1038/mi.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geijtenbeek TB, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100(5):587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 42.Turville SG, et al. Diversity of receptors binding HIV on dendritic cell subsets. Nat Immunol. 2002;3(10):975–983. doi: 10.1038/ni841. [DOI] [PubMed] [Google Scholar]
- 43.Puryear WB, Yu X, Ramirez NP, Reinhard BM, Gummuluru S. HIV-1 incorporation of host-cell-derived glycosphingolipid GM3 allows for capture by mature dendritic cells. Proc Natl Acad Sci USA. 2012;109(19):7475–7480. doi: 10.1073/pnas.1201104109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cameron PU, et al. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science. 1992;257(5068):383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 45.Fahrbach KM, et al. Activated CD34-derived Langerhans cells mediate transinfection with human immunodeficiency virus. J Virol. 2007;81(13):6858–6868. doi: 10.1128/JVI.02472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ballweber L, et al. Vaginal langerhans cells nonproductively transporting HIV-1 mediate infection of T cells. J Virol. 2011;85(24):13443–13447. doi: 10.1128/JVI.05615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pöhlmann S, et al. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J Virol. 2001;75(10):4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gummuluru S, Rogel M, Stamatatos L, Emerman M. Binding of human immunodeficiency virus type 1 to immature dendritic cells can occur independently of DC-SIGN and mannose binding C-type lectin receptors via a cholesterol-dependent pathway. J Virol. 2003;77(23):12865–12874. doi: 10.1128/JVI.77.23.12865-12874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehandru S, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200(6):761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegal FP, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284(5421):1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 51.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen BD, et al. Loss of infectivity by progeny virus from alpha interferon-treated human immunodeficiency virus type 1-infected T cells is associated with defective assembly of envelope gp120. J Virol. 1992;66(12):7543–7548. doi: 10.1128/jvi.66.12.7543-7548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chertova E, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 2002;76(11):5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol. 2005;79(19):12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bachrach E, et al. Effects of virion surface gp120 density on infection by HIV-1 and viral production by infected cells. Virology. 2005;332(1):418–429. doi: 10.1016/j.virol.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 57.Martellini JA, et al. Cationic polypeptides contribute to the anti-HIV-1 activity of human seminal plasma. FASEB J. 2009;23(10):3609–3618. doi: 10.1096/fj.09-131961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ongradi J, Ceccherini-Nelli L, Pistello M, Specter S, Bendinelli M. Acid sensitivity of cell-free and cell-associated HIV-1: Clinical implications. AIDS Res Hum Retroviruses. 1990;6(12):1433–1436. doi: 10.1089/aid.1990.6.1433. [DOI] [PubMed] [Google Scholar]
- 59.Izquierdo-Useros N, et al. Maturation of blood-derived dendritic cells enhances human immunodeficiency virus type 1 capture and transmission. J Virol. 2007;81(14):7559–7570. doi: 10.1128/JVI.02572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Granelli-Piperno A, et al. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J Exp Med. 1996;184(6):2433–2438. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spira AI, et al. Cellular _targets of infection and route of viral dissemination after an intravaginal inoculation of simian immunodeficiency virus into rhesus macaques. J Exp Med. 1996;183(1):215–225. doi: 10.1084/jem.183.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsu M, et al. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIV(SF162P3) maps to envelope gp120. J Virol. 2003;77(2):989–998. doi: 10.1128/JVI.77.2.989-998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, et al. The Toll-like receptor 7 (TLR7) agonist, imiquimod, and the TLR9 agonist, CpG ODN, induce antiviral cytokines and chemokines but do not prevent vaginal transmission of simian immunodeficiency virus when applied intravaginally to rhesus macaques. J Virol. 2005;79(22):14355–14370. doi: 10.1128/JVI.79.22.14355-14370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sandler N, et al. (2013) Blockade of Type I Interferon During Acute SIV Infection Results in Accelerated Progression to AIDS and Death. The Conference on Retroviruses and Opportunistic Infections: March 3–6, 2013 (Atlanta, Georgia). Available at http://www.retroconference.org/2013b/Abstracts/46260.htm. Accessed March 17, 2013.
- 65.Hirbod T, et al. Upregulation of interferon-alpha and RANTES in the cervix of HIV-1-seronegative women with high-risk behavior. J Acquir Immune Defic Syndr. 2006;43(2):137–143. doi: 10.1097/01.qai.0000229016.85192.60. [DOI] [PubMed] [Google Scholar]
- 66.Ball TB, et al. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS. 2007;21(9):1091–1101. doi: 10.1097/QAD.0b013e3280ef6ae1. [DOI] [PubMed] [Google Scholar]
- 67.Vendrame D, Sourisseau M, Perrin V, Schwartz O, Mammano F. Partial inhibition of human immunodeficiency virus replication by type I interferons: Impact of cell-to-cell viral transfer. J Virol. 2009;83(20):10527–10537. doi: 10.1128/JVI.01235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yasuda Y, et al. Interferon-alpha treatment leads to accumulation of virus particles on the surface of cells persistently infected with the human immunodeficiency virus type 1. J Acquir Immune Defic Syndr. 1990;3(11):1046–1051. [PubMed] [Google Scholar]
- 69.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 70.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 71.Schröder HC, et al. Binding of Tat protein to TAR region of human immunodeficiency virus type 1 blocks TAR-mediated activation of (2′-5′)oligoadenylate synthetase. AIDS Res Hum Retroviruses. 1990;6(5):659–672. doi: 10.1089/aid.1990.6.659. [DOI] [PubMed] [Google Scholar]
- 72.Roy S, et al. Control of the interferon-induced 68-kilodalton protein kinase by the HIV-1 tat gene product. Science. 1990;247(4947):1216–1219. doi: 10.1126/science.2180064. [DOI] [PubMed] [Google Scholar]
- 73.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4(2):e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reynolds MR, et al. The TRIM5α genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J Virol. 2011;85(18):9637–9640. doi: 10.1128/JVI.05074-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeh WW, et al. The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J Virol. 2011;85(19):10389–10398. doi: 10.1128/JVI.00854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeeninga RE, et al. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol. 2000;74(8):3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bachu M, et al. Multiple NF-κB sites in HIV-1 subtype C long terminal repeat confer superior magnitude of transcription and thereby the enhanced viral predominance. J Biol Chem. 2012;287(53):44714–44735. doi: 10.1074/jbc.M112.397158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spotts DR, Reich RM, Kalkhan MA, Kinney RM, Roehrig JT. Resistance to alpha/beta interferons correlates with the epizootic and virulence potential of Venezuelan equine encephalitis viruses and is determined by the 5′ noncoding region and glycoproteins. J Virol. 1998;72(12):10286–10291. doi: 10.1128/jvi.72.12.10286-10291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Charleston B, Fray MD, Baigent S, Carr BV, Morrison WI. Establishment of persistent infection with non-cytopathic bovine viral diarrhoea virus in cattle is associated with a failure to induce type I interferon. J Gen Virol. 2001;82(Pt 8):1893–1897. doi: 10.1099/0022-1317-82-8-1893. [DOI] [PubMed] [Google Scholar]
- 80.Meier A, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med. 2009;15(8):955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klein JS, Bjorkman PJ. Few and far between: How HIV may be evading antibody avidity. PLoS Pathog. 2010;6(5):e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, et al. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS Pathog. 2012;8(8):e1002880. doi: 10.1371/journal.ppat.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biancotto A, et al. A highly sensitive and dynamic immunofluorescent cytometric bead assay for the detection of HIV-1 p24. J Virol Methods. 2009;157(1):98–101. doi: 10.1016/j.jviromet.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.