Background: The role of the lysine acetyltransferase GCN5 in cancer development remains largely unknown.
Results: GCN5 expression correlates with lung cancer tumor size, directly enhances the expression of E2F1, cyclin E1, and cyclin D1, and potentiates lung cancer growth.
Conclusion: GCN5 potentiates lung cancer growth in an E2F1-dependent manner.
Significance: GCN5 is critical for lung cancer growth and represents a potential _target for the treatment of lung cancer.
Keywords: Cancer biology, Cancer Prevention, Cell Cycle, Gene Regulation, Histone Acetylase, E2F1, GCN5, Lung Cancer, Lysine Acetyltransferase (KAT)
Abstract
The lysine acetyltransferases play crucial but complex roles in cancer development. GCN5 is a lysine acetyltransferase that generally regulates gene expression, but its role in cancer development remains largely unknown. In this study, we report that GCN5 is highly expressed in non-small cell lung cancer tissues and that its expression correlates with tumor size. We found that the expression of GCN5 promotes cell growth and the G1/S phase transition in multiple lung cancer cell lines. Further study revealed that GCN5 regulates the expression of E2F1, cyclin D1, and cyclin E1. Our reporter assays indicated that the expression of GCN5 enhances the activities of the E2F1, cyclin D1, and cyclin E1 promoters. ChIP experiments suggested that GCN5 binds directly to these promoters and increases the extent of histone acetylation within these regions. Mechanistic studies suggested that GCN5 interacts with E2F1 and is recruited by E2F1 to the E2F1, cyclin D1, and cyclin E1 promoters. The function of GCN5 in lung cancer cells is abrogated by the knockdown of E2F1. Finally, we confirmed that GCN5 regulates the expression of E2F1, cyclin D1, and cyclin E1 and potentiates lung cancer cell growth in a mouse tumor model. Taken together, our results demonstrate that GCN5 specifically potentiates lung cancer growth by directly promoting the expression of E2F1, cyclin D1, and cyclin E1 in an E2F1-dependent manner. Our study identifies a specific and novel function of GCN5 in lung cancer development and suggests that the GCN5-E2F1 interaction represents a potential _target for lung cancer treatment.
Introduction
Histone acetylation plays a central role in establishing an active chromatin environment for transcriptional regulation. The balance between histone acetylation and deacetylation is maintained in cells by histone lysine acetyltransferases (KATs)3 and histone deacetylases (HDACs) (1, 2). Hypoacetylation of a specific set of genes results from an imbalance between KATs and HDACs and is associated with the transformed state of cancer cells (3–6). HDAC inhibitors, which can suppress HDAC enzymatic activity and reactivate epigenetically silenced antitumor genes, represent useful tools for cancer prevention and therapy (7). Currently, clinical trials employing HDAC inhibitors have been hampered by their lack of specificity. Most HDAC inhibitors _target a panel of HDAC superfamily members, and individual HDAC members regulate diverse cellular pathways (7, 8). Therefore, the inhibition of HDAC enzymatic activity using HDAC inhibitors often results in unintentional gene activation and the development of drug resistance (9, 10). Together with HDACs, KATs balance the control of histone acetylation, increasing histone acetylation in a more specific manner than that elicited by HDAC inhibitors (11, 12). Hence, the function of KATs in cancer warrants investigation.
Previous studies have shown that KATs play crucial and complicated roles in cancer development (5, 13, 14). The KAT p300 has been recognized as a potential anti-cancer drug _target because the p300 gene is mutated in most colon cancer cell lines exhibiting microsatellite instability and in some primary tumors (15, 16). Other studies have indicated that p300 activity is important for tumor growth and that the down-regulation or inhibition of this activity abrogates the growth of tumor cells. Aggressive cases of prostate cancers have been found to exhibit overexpression of the KAT Tip60, the extent of which correlates with disease progression (17, 18). In contrast, another report has suggested that Tip60 is required for the expression of the tumor metastasis suppressor KAI1 in prostate cancer cell lines, suggesting that Tip60 is a tumor suppressor (19). PCAF is a KAT that is essential for central nervous system tumors (20). GCN5, the first identified transcription-related KAT, is a crucial catalytic component of a transcriptional regulatory complex. GCN5 exhibits a broad range of functions with regard to cell proliferation, differentiation, cell cycle, and DNA damage repair (11, 12, 21). There are only two reports that have previously investigated the potential involvement of GCN5 in cancer development (22, 23). Whether GCN5 is directly and specifically involved in cancer growth is unknown.
Notably, the functional contribution of HDACs to transcription requires their association with transcription factor (TF)-containing complexes. For example, HDAC2 participates in several TF complexes, including the Sin3, NuRD, CoREST, and NcoR-SMRT complexes (24), which are required for transcriptional suppression. HDACs associate with sequence-specific DNA-binding proteins or with transcription factors to regulate the transcription of specific sets of genes (24). The study of HDAC inhibitors has suggested that, in terms of anticancer therapy, disrupting the interaction of HDAC/TF represents a more optimal approach than the chemical inhibition of their enzymatic activities. Similarly, the KATs also exert their function and specificity through their interaction with TFs. Identifying a cancer type-specific KAT and its cognate specificity-determining transcription factor is important for the cancer treatment.
Lung cancer is one of the major causes of mortality worldwide (25). The identification of biomarkers and the dissection of the mechanisms underlying the development of lung cancer are urgently needed for the development of therapeutic strategies (26, 27). In an analysis of human non-small cell lung cancer (NSCLC) tissue arrays, we found that GCN5 is highly expressed in lung cancer tissues, and its levels significantly correlate with tumor size. We further investigated the role of GCN5 in lung cancer growth and found that GCN5 potentiates lung cancer cell growth in an E2F1-dependent manner.
EXPERIMENTAL PROCEDURES
Tissue Microarray and Evaluation of Immunostaining
Lung cancer tissue arrays were purchased from the National Engineering Center for BioChips in Shanghai, China. The expression of GCN5 or E2F1 in the tissues was evaluated by immunohistochemical staining with either a GCN5-specific or E2F1-specific antibody. The staining was scored according to the staining intensity and the percentage of cells stained. The final staining scores were calculated as the product of staining intensity multiplied by the percentage of stained cells.
Cell Culture
The A549, H460, and H358 human lung cancer cell lines were purchased from the Cell Bank at the Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences and were maintained as instructed by the provider. Human embryonic kidney (HEK) 293T cells were purchased from the American Type Culture Collection (ATCC). All of the cells were maintained at 37 °C with 5% CO2.
Plasmids, Transfections, and Virus Production
For the overexpression of the GCN5 and E2F1, we used the lentiviral vector FUGW, in which the GFP-coding region was replaced with the GCN5 or E2F1 coding sequences. The lentiviral vector pLKO.1 was used for gene knockdown. The following two siRNAs were selected for human GCN5: sense 5′-GCUCUACACAACCCUCAAATT-3′ and antisense 5′-UUUGAGGGUUGUGUAGAGCTT-3′; sense 5′-GCAUGCCUAAGGAGUAUAUTT-3′ and antisense 5′-AUAUACUCCUUAGGCAUGCTT-3′. The following two siRNAs were selected for human E2F1: sense 5′-GCGCAUCUAUGACAUCACCTT-3′ and antisense 5′-GGUGAUGUCAUAGAUGCGCTT-3′; sense 5′-CCUCUUCGACUGUGACUUUTT-3′ and antisense 5′-AAAGUCACAGUCGAAGAGGTT-3. DNA oligonucleotides for these siRNAs were synthesized, annealed, and cloned into pLKO.1 according to the protocol recommended by Addgene. HEK293T cells were transfected with the ExcellGene transfection reagent. Lentiviruses were generated from the HEK293T cells as described previously (28), and lung cancer cells were infected with the virus in the presence of 8 μg/ml Polybrene.
Cell Proliferation Assays, Cell Cycle Analysis, and Annexin V-PI Staining
Cell proliferation and apoptosis were evaluated using the Cell Counting Kit-8 (CCK8, DOJINDO, Japan) and the annexin V-PI staining kit (Roche Applied Science) according to the manufacturer's protocols. Cellular proliferation was measured at 12, 24, 48, and 72 h post-seeding.
Western Blotting, Immunoprecipitation, Immunofluorescence Staining, and ChIP Assays
Western blotting, immunoprecipitation, and immunofluorescence were performed as described previously (29, 30). The following antibodies were used: GCN5, sc-36532, and sc-20698; cyclin D1 (M-20), sc-718; cyclin E (M-20), sc-481; PCAF (E-8), sc-13124; E2F1, sc-251; p-Rb, BS4164; Rb, BS1310; GAPDH (0411), sc-47724; p53 (BP53.122), sc-73566; c-Myc (A-14), sc-789; Bcl-2, CST 2876; Bax, CST 2772; p27 (N-20), sc-527; p21, Abcam ab7960; acetyl histone H3 (Millipore 17–615); acetyl histone H4 (Millipore 17–630). ChIP assays were performed with EZ ChIP kit (Millipore). Ten million cells were used per immunoprecipitation reaction. Immunoprecipitated DNA and whole cell-extracted DNA were used as templates for RT-qPCR with primers that _targeted the flanking sequences of the E2F1-binding site in the promoters of cyclin D1, cyclin E1, and E2F1.
Tumor Xenografts in Nude Mice
Our animal experiments were approved by the Institutional Animal Care and Use Committee of Tongji University and were in compliance with all regulatory guidelines. Five-week-old nude mice were purchased from the National Resource Center for Rodent Laboratory Animals of China. Six mice were injected subcutaneously with 5.0 × 106 of GCN5-overexpressing (FUW-GCN5) or vector control (FUW) cells. The tumors were monitored, and the volumes were calculated at 3-day intervals.
RESULTS
GCN5 Expression Correlates Significantly with the Size of Lung Tumors
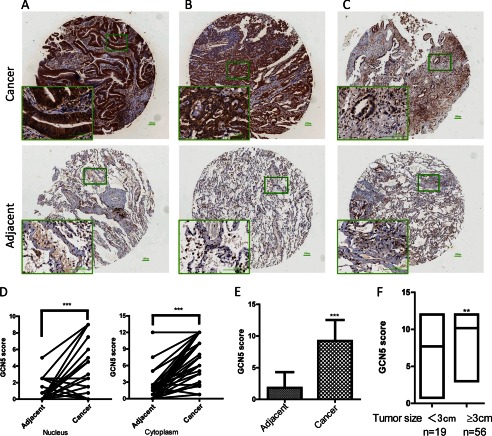
To examine the physiological relevance of GCN5 in human lung cancer development, we analyzed the GCN5 protein levels and its distribution in an NSCLC tissue array by immunohistochemical staining using a GCN5-specific antibody. The array included the cancer tissue samples and corresponding adjacent normal tissue samples from 75 lung cancer patients. The patients consisted of 41 males and 34 females, with a median age of 61 years (range, 37–84 years). Among the patients, 24 (32%) died of tumor-related causes, and 51 (68%) were still alive as of the last follow-up (Table 1). The GCN5 levels in the tissue array specimens were assessed by measurement of the staining density scores. We found that in the cancer tissues, 42 cases exhibited strong immunopositivity (56%) (Fig. 1A), 30 cases exhibited moderate-to-strong immunopositivity (40%) (Fig. 1B), and three cases exhibited weak immunopositivity (4%) (Fig. 1C). Statistical analysis revealed that nuclear, cytoplasmic (Fig. 1D), and total levels (Fig. 1E) of GCN5 were significantly increased in the cancer tissues compared with the normal tissues. Remarkably, the GCN5 expression levels were significantly correlated with tumor size (Fig. 1F) (p = 0.0024), progressively advanced regional lymph node metastasis (p = 0.016), and TNM stage III (Table 1). Our lung cancer tissue array analysis revealed that GCN5 is overexpressed in NSCLC, and its expression is tightly linked to tumor proliferation and development.
TABLE 1.
Clinical and pathological characteristics of patients with lung cancer (n = 75)
| Characteristics | n | GCN5 score | p value |
|---|---|---|---|
| Sex | |||
| Male | 41 | 9.323 | p = 0.7761 |
| Female | 34 | 9.102 | |
| Age (years) | |||
| <60 | 29 (37–59) | 9.617 | p = 0.8578 |
| ≥60 | 46 (60–84) | 9.168 | |
| Tumor size | |||
| <3 cm | 19 | 7.710 | p = 0.0024 |
| ≥3 cm | 56 | 9.791 | |
| T stage | |||
| 1–2 | 49 | 9.041 | p = 0.2423 |
| 3–4 | 26 | 10.13 | |
| Lymph node metastasis | |||
| Negative | 35 | 8.414 | p = 0.0167 |
| Positive | 40 | 10.06 | |
| TNM stage | |||
| I | 11 | 9.250 | p = 0.0333 |
| II | 37 | 8.513 | |
| III | 27 | 10.18 | |
FIGURE 1.
Increased expression of GCN5 and its correlation with tumor size in human lung cancers. A–C, representative images of GCN5 expression in lung cancer tissue samples (upper row) and corresponding adjacent normal tissue samples (lower row), showing high (A), medium (B), and low (C) levels of GCN5 expression in cancer tissues. D, plot representation of scores based on the nuclear and cytoplasmic expression of GCN5 in 75 lung cancer patients compared with matched normal tissues, p < 0.001. E, total GCN5 score in adjacent normal and tumor tissues. F, GCN5 score correlates with tumor size. **, p < 0.01; ***, p < 0.0001. Error bars, S.D.
GCN5 Promotes Lung Cancer Cell Growth by Facilitating the G1/S Transition of the Cell Cycle
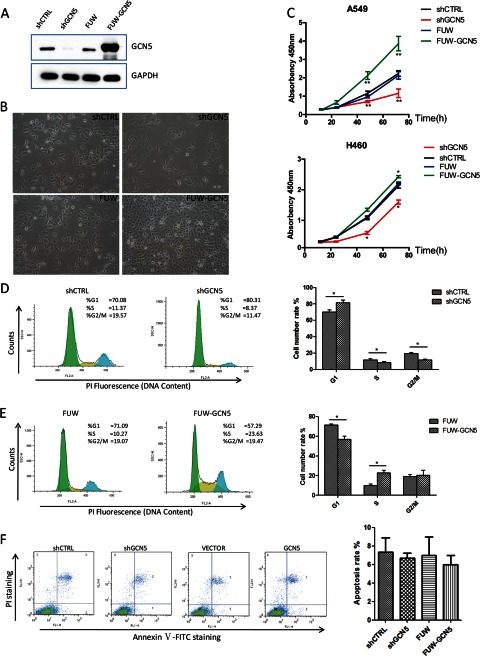
The increased expression of GCN5 in the NSCLC tissues suggests a role for GCN5 in the regulation of lung cancer cell growth. To address whether GCN5 affects NSCLC cell growth, we were able to stably knock down GCN5 (shGCN5) as well as to overexpress GCN5 (FUW-GCN5) in the A549 lung cancer cell line. shRNA oligonucleotides for _targeting two different published regions of GCN5 were cloned into the lentiviral vector pLKO.1. The shRNA exhibiting the highest knockdown efficiency was used in the remainder of this study. GCN5 was efficiently knocked down (shCTRL versus shGCN5) or overexpressed (FUW versus FUW-GCN5) in A549 cells (Fig. 2A). We observed apparent changes in growth rate when we passaged these stable cell lines. The GCN5-knockdown cell line exhibited the most significant growth impairment (Fig. 2B, upper row), and the GCN5-overexpressing cell line grew significantly faster than the control cell line (Fig. 2B, lower row). We further quantified the cell growth rate of these cell lines using a CCK8 assay and confirmed that the overexpression of GCN5 promotes cell growth, whereas its knockdown inhibits the cell growth (Fig. 2C, upper panel). Importantly, the knockdown or overexpression of GCN5 in the other two lung cancer cell lines H460 and H358 (Fig. 2C, lower panel and data not shown) resulted in similar phenotypes.
FIGURE 2.
GCN5 affects cell cycle progression and cell proliferation. A, Western blotting verification of the knockdown and overexpression of GCN5 in stable A549 cell lines. GAPDH was used as a loading control. B, morphology of the stable cell lines at 48 h after plating the same number of cells in the plates. C, proliferation of stable cell lines derived from A549 (upper panel) and H460 (lower panel) cells was examined using a CCK8 assay. D and E, representative profiles of the cell cycle distribution of the A549 stable cell lines (left panels) and statistical analysis of three independent assays. F, apoptotic analysis of the A549 stable cell lines by annexin V and PI staining followed by FACS; representative profiles of cell population distribution (left panels) and statistical analysis of three independent assays. *, p < 0.05; **, p < 0.01. Error bars, S.D.
To determine how the growth of A549 cell lines is regulated by GCN5, we analyzed their cell cycle dynamics by PI staining and fluorescence-activated cell sorting (FACS). We found that compared with the control cell line (shCTRL), the G1 population was significantly increased in the GCN5-knockdown cell line (shGCN5), with a corresponding decrease in S and G2/M phases of more than 10% (Fig. 2D). Consistently, in the GCN5-overexpressing cell line (FUW-GCN5), the S phase population was significantly increased compared with the control cell line (FUW), with a corresponding decrease in the G1 population of ∼13% (Fig. 2E). The data suggest that GCN5 is required for the G1-to-S phase transition, and its overexpression promotes the growth of A549 cells by facilitating the G1/S phase transition of the cell cycle. In contrast to the significant changes in cell cycle dynamics caused by the overexpression or knockdown of GCN5, we observed no significant effect on apoptosis, as indicated by annexin V and PI co-staining (Fig. 2F). Neither the overexpression nor the knockdown of GCN5 induced other types of cell death, as measured by trypan blue staining (supplemental Fig. S1). Importantly, the knockdown of PCAF, a KAT with 73% homology to GCN5, in A549 cells does not significantly affect cell proliferation (supplemental Fig. S2).
GCN5 Promotes the Expression of Cyclin D1, Cyclin E1, and E2F1
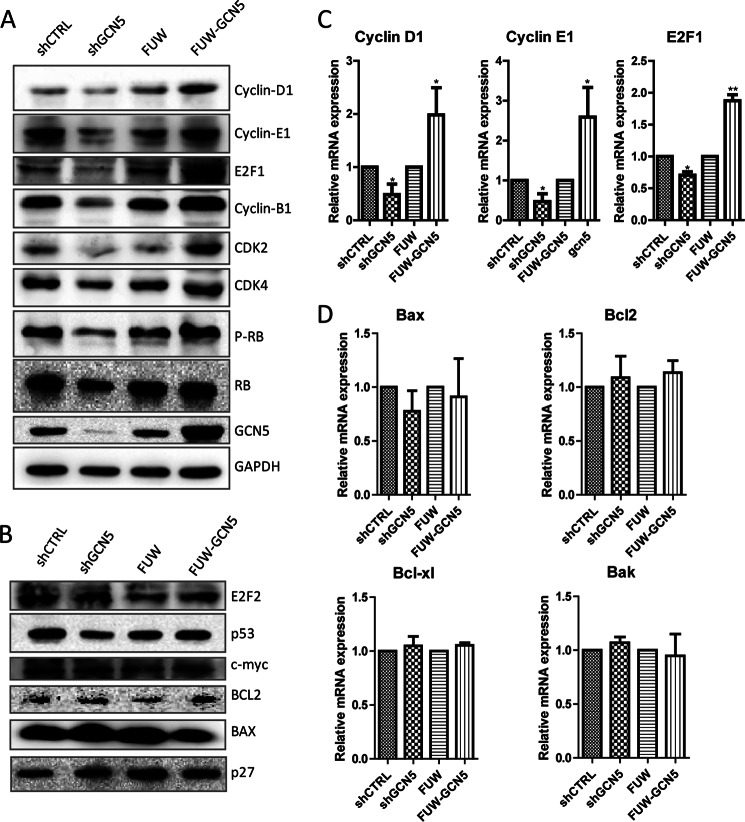
Because the knockdown or overexpression of GCN5 significantly altered cell cycle phase distribution, we further measured the protein levels of the cell cycle and apoptotic regulators (Fig. 3, A and B) to determine the potential _targets that are attributable to the altered cell cycle dynamics. The knockdown or overexpression of GCN5 elicited pleiotropic effects on a variety of genes (Fig. 3A) and marginal effect on E2F2, p53, c-myc, BCL2, BAX, and p27 (Fig. 3B). Particularly, we noticed consistent changes in cyclin D1, cyclin E1, E2F1, and CDK2 in GCN5 knockdown and overexpressing cells. These _targets were down-regulated upon GCN5 knockdown and up-regulated upon GCN5 overexpression.
FIGURE 3.
GCN5 increases both mRNA and protein levels of cyclin D1, cyclin E1, and E2F1. A and B, Western blotting analysis of the protein levels of cell cycle regulators in GCN5-knockdown A549 cells, or GCN5-overexpressing A549 cells and control cell lines. GAPDH was included to show that equivalent amounts of protein were loaded in each lane. A, proteins that are regulated by GCN5. B, proteins that are not regulated by GCN5. C, RT-qPCR analysis of cyclin D1, cyclin E1, and E2F1 mRNA in the GCN5-knockdown cells, GCN5-overexpressing A549 cells, and control cell lines. D, RT-qPCR analysis of apoptosis genes in GCN5-knockdown cells, GCN5-overexpressing cells, and control cell lines. *, p < 0.05; **, p < 0.01. Error bars, S.D.
We further measured the mRNA levels of E2F1, cyclin D1, cyclin E1 (Fig. 3C), Bax, Bcl2, Bcl-xl, and Bak (Fig. 3D) in these cell lines using real time quantitative-PCR (RT-qPCR). The mRNA measurements were consistent with the protein level measurements. cyclin D1, cyclin E1, and E2F1 were regulated by GCN5, whereas the apoptosis-related genes were not. The mRNA measurements not only confirmed the protein level measurements but also suggested that GCN5 transcriptionally regulates key cell cycle-related genes.
GCN5 Directly Binds to and Increases the Histone H3 and H4 Acetylation of the Cyclin E1, Cyclin D1, and E2F1 Promoters
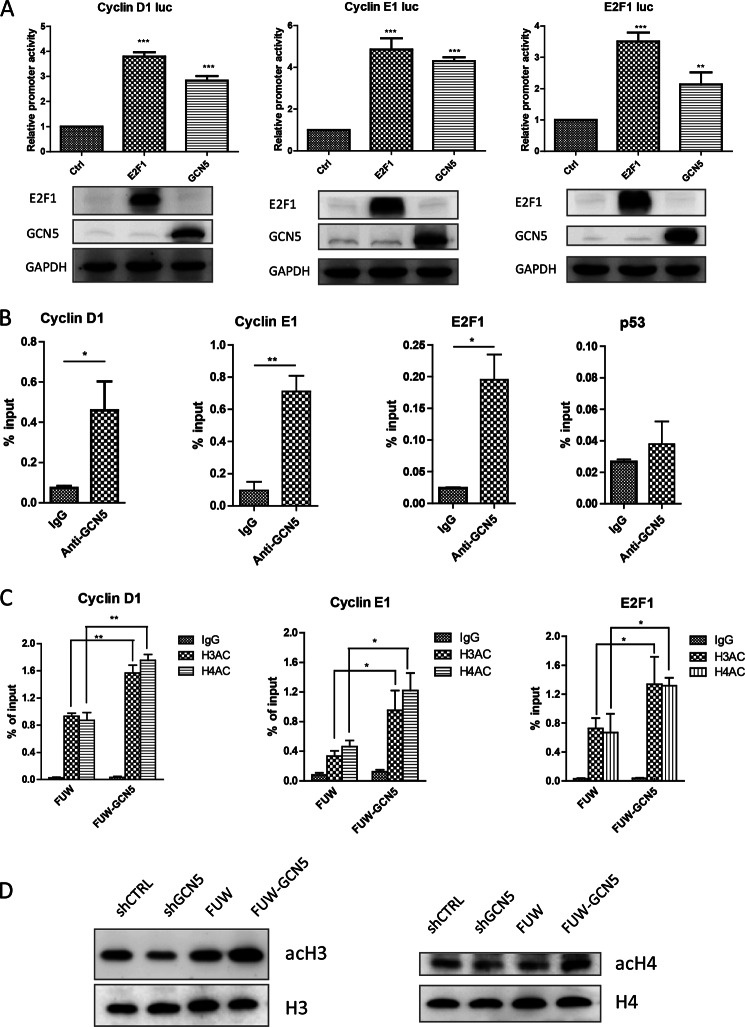
We found that the expression levels of cyclin D1, cyclin E1, and E2F1 were markedly regulated by GCN5 at both the protein and mRNA levels. We hypothesized that GCN5 might directly regulate the expression of these genes. To test this possibility, luciferase reporters driven by the promoter regions from cyclin D1 (Addgene 32726), cyclin E1 (Addgene 8458), or E2F1 (Addgene 20950) (31–33) were used to evaluate the effect of GCN5 on the promoter activity of these genes. These promoter regions contain E2F1-binding sites, and we observed that the overexpression of E2F1 significantly increased the activities of these reporters, reflecting the robust E2F1 responsiveness of these reporters. The overexpression of GCN5 enhanced the activity of these reporters to similar levels as those induced by the overexpression of E2F1 (Fig. 4A). Chromatin immunoprecipitation (ChIP) showed that the promoter regions of these genes were significantly immunoprecipitated using the GCN5 antibody, indicating that endogenous GCN5 binds directly to the cyclin D1, cyclin E1, or E2F1 promoters (Fig. 4B). The control p53 promoter exhibited no enrichment of GCN5 (Fig. 4B, far right panel). Additional ChIP assays using the acetyl histone antibody revealed that the overexpression of GCN5 resulted in increased histone H3 and H4 acetylation at the cyclin D1, cyclin E1, and E2F1 promoters (Fig. 4C). We verified the lysine acetylation levels of histone H3 and H4 in the four cell lines that we created. Both histone H3 and H4 acetylation were increased upon GCN5 overexpression and decreased upon GCN5 knockdown (Fig. 4D). Taken together, these experiments demonstrated that GCN5 binds directly to the cyclin D1, cyclin E1, and E2F1 promoters, increasing the histone acetylation at these promoters.
FIGURE 4.
GCN5 directly binds to the cyclin E1, cyclin D1, and E2F1 promoters, increasing histone H3 and H4 acetylation levels within these regions. A, cyclin E1, cyclin D1, and E2F1 luciferase reporter assay performed in HEK293T cells. Empty control vector and E2F1- or GCN5-overexpression plasmids were co-transfected with cyclin E1, cyclin D1, or E2F1 luciferase reporter plasmids and were then assayed for luciferase activity at 48 h post-transfection. Protein bolts were used to confirm the overexpression of GCN5 and E2F1. B, GCN5 binding within the promoter region of cyclin E1, cyclin D1, and E2F1 in A549 cells. ChIP was performed with control IgG and GCN5 antibodies. Shown are the results of three independent experiments. C, comparison of histone H3 and H4 acetylation of the cyclin E1, cyclin D1, or E2F1 promoters between control and GCN5-overexpressing cell lines. Shown are the results of three independent experiments. *, p < 0.05; **, p < 0.01. Error bars, S.D. D, acetylation levels of histones H3 and H4 (acH3 and acH4) in GCN5-overexpressing or knockdown cells were detected with acetyl histone antibodies. Histone H3 and H4 levels (H3 and H4) were detected with histone antibodies.
E2F1 Is Required for the GCN5-mediated Regulation of Lung Cancer Cell Growth and for the G1/S Transition
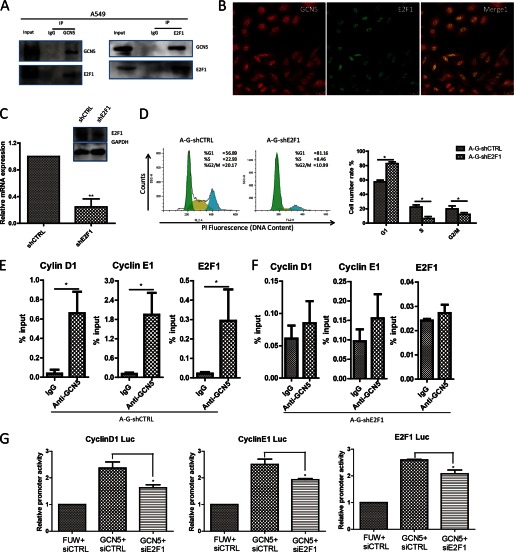
Unlike a transcription factor, the GCN5 protein does not contain a DNA-binding domain. Rather than binding to DNA directly, GCN5 is recruited by transcription factors to specific regions to regulate gene transcription. Previous studies have found that E2F1 associates with and recruits GCN5 to sites of DNA damage (34). We also noted that cyclin D1 and cyclin E1 are downstream _targets of E2F1 (35) and, furthermore, that GCN5 is enriched at the E2F1-binding site of the cyclin D1, cyclin E1, or E2F1 promoters in our ChIP assays. We hypothesized that E2F1 might be the transcription factor mediating the transcriptional function of GCN5 in lung cancer cell growth. To examine this hypothesis, we first performed co-immunoprecipitation. We found that endogenous GCN5 co-immunoprecipitated with E2F1 (Fig. 5A). Confocal microscopy analysis revealed that in A549 cells, both GCN5 and E2F1 localize within the nucleus, and both GCN5 and E2F1 signals exhibited significant degrees of overlap (Fig. 5B). The data suggest that GCN5 interacts with E2F1 in cells. We also observed significant cytoplasmic GCN5, consistent with the cytoplasmic GCN5 observed in patient cancer tissue (Fig. 1).
FIGURE 5.
E2F1 is required for GCN5 to promote the proliferation of A549 cells. A, GCN5 interacts with endogenous E2F1. Immunoprecipitation (IP) was performed using control IgG or GCN5 antibody, followed by Western blot analysis for E2F1 and GCN5. B, intracellular localization of GCN5 and E2F1 in A549 cells. Immunostaining was performed using GCN5 and E2F1 antibodies. C, RT-qPCR and Western blotting analysis of E2F1 knockdown by shRNA in the GCN5-overexpressing stable cell line. Error bars, S.D. (n = 3 biological replicates). **, p < 0.01.D, altered cell cycle distribution after E2F1 knockdown in A549 GCN5-overexpressing cell lines. E and F, comparison of GCN5-binding at cyclin E1, cyclin D1, and E2F1 promoters in the GCN5-overexpressing cell line after E2F1 knockdown. G, effect of E2F1 knockdown on the GCN5-mediated regulation of cyclin E1, cyclin D1, and E2F1 promoter activity in 293T cells. The GCN5 overexpression plasmid was co-transfected with control (siCTRL) or E2F1 siRNA, and the luciferase reporter activity was assayed 48 h post-transfection. *, p < 0.05. Error bars, S.D.
To further evaluate the role of E2F1 in GCN5-mediated lung cancer cell growth, we knocked down E2F1 in the GCN5-overexpressing cell line (Fig. 5C). We found that the knockdown of E2F1 abrogated the growth-promoting effects of GCN5, as evidenced by the significant accumulation of the GCN5-overexpressing E2F1 shRNA (A-G-shE2F1)-treated A549 cells in G1 phase (Fig. 5D). The binding of GCN5 to the cyclin D1, cyclin E1, or E2F1 promoters in GCN5-overexpressing cells was significantly reduced when E2F1 was knocked down (Fig. 5, E versus F). The effect of GCN5 overexpression on cyclin D1, cyclin E1, and E2F1 promoter-driven luciferase reporters was also impaired by treatment with E2F1 siRNA. Collectively, our results suggest that GCN5 is recruited by E2F1 to the cyclin D1, cyclin E1, and E2F1 promoters to regulate lung cancer cell growth in an E2F1-dependent manner.
Overexpression of GCN5 Accelerates Tumorigenesis in Vivo
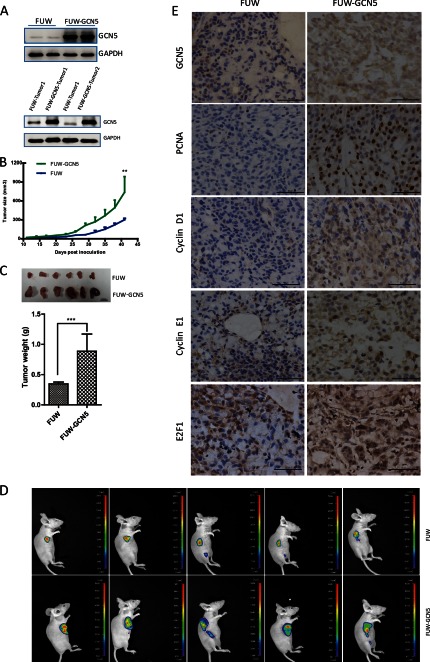
We examined whether GCN5 promotes tumor growth in vivo. Two GCN5-overexpressing cell lines (FUW-GCN5) and two control cell lines expressing an empty vector (FUW) were independently generated. The overexpression of GCN5 was assessed prior to injection into mice (Fig. 6A, upper panel). Lentiviruses encoding firefly luciferase were also introduced into these cell lines to allow for the in vivo imaging of tumor size and tumor metastasis. The ectopic overexpression of GCN5 was detectable in the isolated tumors (Fig. 6A, lower panel). We found that the time of tumor onset (defined as the day when the tumors become palpable after injection) was reduced to 7 days when GCN5 was overexpressed. The growth of tumors initiated by GCN5-overexpressing cells was significantly faster than those initiated by the control cells (Fig. 6B). At 6 weeks after injection, the tumors were isolated and weighed. The GCN5-overexpressing cells produced significantly larger tumors than the control cells (Fig. 6C). In vivo imaging also confirmed that the GCN5-overexpressing cells produced larger tumors than control cells (Fig. 6D). We further compared the protein levels of cyclin D1, cyclin E1, and E2F1 in the tumor tissues isolated from GCN5-overexpressing and control groups by immunostaining (Fig. 6E). The protein levels of all three genes in the tumors initiated by the GCN5-overexpressing cell line were increased compared with those of the tumors derived from the control cell line and correlated with the PCAN level, a growth potential marker. Our animal experiment confirmed that GCN5 potentiates lung cancer growth and promotes the expression of cyclin D1, cyclin E1, and E2F1 in vivo.
FIGURE 6.
Overexpression of GCN5 accelerates tumorigenesis in vivo. A, upper panel, the overexpression of GCN5 in two stable cell lines was confirmed by Western blotting prior to injection. Lower panel, sustained GCN5 expression in tumors isolated from mice at 6 weeks post-injection. Two tumors isolated from mice injected with control (FUW) and GCN5-overexpressing cells (FUW-GCN5) were analyzed. B, tumor growth of control and GCN5-overexpressing cell lines in mice. The tumor size was monitored every 3 days. Error bars, S.D. C, tumors derived from control and GCN5-overexpressing cell lines (upper panel) and statistical analysis of tumor weight at 6 weeks post-injection (lower panel). D, in vivo imaging of tumors derived from the control and GCN5-overexpressing cell lines. E, immunochemical staining for GCN5, cyclin D1, Cyclin E1, E2F1, and proliferating cell nuclear antigen (PCNA) in tumors derived from the control and GCN5-overexpressing cell lines. **, p < 0.01; ***, p < 0.001. Error bars, S.D.
DISCUSSION
Histone acetylation has significant effects on the proliferation and metastasis of lung cancer cells (36, 37), suggesting roles for KATs and HDACs in this cancer (38, 39). The major obstacle for the clinical application of HDAC inhibitors as a treatment is their lack of specificity. To design more selective inhibitors, it is essential to identify specific isoforms of HDACs or KATs that are present in cancers (40). In this study, we found that GCN5 is highly expressed in lung cancer, and its expression levels are tightly linked to tumor size. Our in vitro assays using lung cancer cell lines demonstrated that the overexpression of GCN5 promotes the growth of multiple lung cancer cell lines and facilitates the G1/S transition, which could be the major biological process underlying the increased cell growth. More importantly, we found that the overexpression of GCN5 potentiates lung cancer growth in an in vivo mouse model. Notably, GCN5 has a unique function in promoting the growth of lung cancer cells but not their apoptosis or death, thereby conferring a net growth advantage on the cancer cells. Another KAT, PCAF, which is closely related to GCN5 and exhibits ∼73% sequence homology to GCN5, has no effects on the growth of lung cancer cells, demonstrating the specificity of GCN5 for lung cancer growth. Our study reveals a unique and specific tumorigenic function for GCN5 in lung cancer growth. These findings provide evidence that GCN5 has prognostic significance and might represent a potential therapeutic _target for the control of lung cancer growth.
In the cases described thus far, KATs do not function independently but, rather, act as a component within multiprotein complexes. Previous data have indicated that GCN5 facilitates gene transcription by virtue of being a general acetyltransferase component within the transcription complexes. The formation of such complexes helps GCN5 to exert its catalytic activity more effectively and/or specifically by providing a substrate-recognition context. Consistent with this notion, we found that GCN5 potentiates lung cancer cell growth in conjunction with the transcription factor E2F1. Our co-immunoprecipitation and immunofluorescence assays confirmed that there is an interaction between GCN5 and E2F1. The ChIP results demonstrated that GCN5 is recruited by E2F1 to E2F1-binding sites within the promoter regions of E2F1 _target genes. GCN5 catalyzes the histone acetylation at these promoter regions, enhancing the transcription of these genes. Interestingly, our data suggest that GCN5 plays a profound role in the E2F1 pathway. It not only participates in regulating the E2F1 downstream _targets cyclin E1 and cyclin D1, but it is also required for the positive feed-forward regulation of E2F1 expression. The transcription factor E2F1 controls the transcription of cyclin E1 and cyclin D1 (41, 42) and mediates the regulation of G1/S progression during tumor growth. E2F1 also mediates the DNA damage response and promotes apoptosis, thereby functioning as a tumor suppressor (43–45). This study reveals that E2F1 is required for the growth of lung cancer. Our lung cancer tissue array analysis using an E2F1-specific antibody revealed that E2F1 expression is significantly increased in lung cancer than in normal tissues (supplemental Fig. S3). There are reports on the interplay between GCN5 and the transcription factors c-Myc and p53 (46). Our findings underscore that, in lung cancer cell growth control, the function of GCN5 is predominantly determined by the transcription factor E2F1 and that GCN5 is a critical KAT that regulates the E2F1 pathway in lung cancer growth.
In contrast to the antitumor effect elicited by increasing acetylation using HDAC inhibitors, in this study the increased expression of the lysine acetyltransferase GCN5 promoted tumor development, although the acetylation of both histone and non-histone proteins might be involved. This finding suggests that acetylation is bifunctional, either suppressing or promoting tumors, depending on what genes are expressed and what proteins are regulated. In terms of cancer treatment, both the acetylation of histone and non-histone proteins should be tightly regulated rather than globally increased. Our findings provide new insights into the previously under-appreciated roles of histone hyper-acetylation during cancer development. Recently, small molecule KAT inhibitors have demonstrated relevance in oncology (17, 47), thereby providing an additional strategy for combating cancer through the manipulation of histone acetylation. Given the diverse cellular roles of KATs and their working mechanism within transcription factor-containing complexes, KAT inhibitors face the same challenge of a lack of specificity that hampers their therapeutic application in cancer cases. To achieve more specific therapeutic effects, it is essential to disrupt the tumorigenic interaction between the transcription factor and the histone-modifying enzyme (either HDAC or KAT). Tumor-promoting protein interactions, such as the MDM2-p53, mediate cancer development, and small molecules that inhibit such interactions represent attractive candidates for the treatment of cancer (48–50). Disrupting the GCN5 and E2F1 interaction that we have discovered in lung cancer provides a potential strategy for blocking lung cancer growth.
This work was supported by Ministry of Science and Technology Grants 2011CB965100, 2010CB944902, 2011CBA01106, 2011DFA30480, 2010CB945000, and 2012CB966603; National Natural Science Foundation of China Grants 91219305, 31171432, 31101061, 31210103905, 30971451, 31071306, 31000378, and 81170499; Science and Technology Commission of Shanghai Municipality Grant 11ZR1438500 and 11XD1405300; Ministry of Education Grants IRT1168 and 20110072110039 from “Chen Guang” Project from Shanghai Municipal Education Commission; Shanghai Education Development Foundation Grant 12CG19, and the Fundamental Research Funds for the Central Universities.

This article contains supplemental Figs. S1–S3.
- KAT
- lysine acetyltransferase
- TF
- transcription factor
- PI
- propidium iodide
- NSCLC
- non-small cell lung cancer
- qPCR
- quantitative PCR.
REFERENCES
- 1. Yuan H., Reddy M. A., Sun G., Lanting L., Wang M., Kato M., Natarajan R. (2013) Involvement of p300/CBP and epigenetic histone acetylation in TGF-β1-mediated gene transcription in mesangial cells. Am. J. Physiol. Renal Physiol. 304, F601–F613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Icardi L., De Bosscher K., Tavernier J. (2012) The HAT/HDAC interplay: multilevel control of STAT signaling. Cytokine Growth Factor Rev. 23, 283–291 [DOI] [PubMed] [Google Scholar]
- 3. Ahmad M., Hamid A., Hussain A., Majeed R., Qurishi Y., Bhat J. A., Najar R. A., Qazi A. K., Zargar M. A., Singh S. K., Saxena A. K. (2012) Understanding histone deacetylases in the cancer development and treatment: an epigenetic perspective of cancer chemotherapy. DNA Cell Biol. 31, S62–S71 [DOI] [PubMed] [Google Scholar]
- 4. Ma J., Wang J. D., Zhang W. J., Zou B., Chen W. J., Lam C. S., Chen M. H., Pang R., Tan V. P., Hung I. F., Lan H. Y., Wang Q. Y., Wong B. C. (2010) Promoter hypermethylation and histone hypoacetylation contribute to pancreatic-duodenal homeobox 1 silencing in gastric cancer. Carcinogenesis 31, 1552–1560 [DOI] [PubMed] [Google Scholar]
- 5. Suzuki J., Chen Y. Y., Scott G. K., Devries S., Chin K., Benz C. C., Waldman F. M., Hwang E. S. (2009) Protein acetylation and histone deacetylase expression associated with malignant breast cancer progression. Clin. Cancer Res. 15, 3163–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armas-Pineda C., Arenas-Huertero F., Pérezpeñia-Diazconti M., Chico-Ponce de León F., Sosa-Sáinz G., Lezama P., Recillas-Targa F. (2007) Expression of PCAF, p300, and Gcn5 and more highly acetylated histone H4 in pediatric tumors. J. Exp. Clin. Cancer Res. 26, 269–276 [PubMed] [Google Scholar]
- 7. Giannini G., Cabri W., Fattorusso C., Rodriquez M. (2012) Histone deacetylase inhibitors in the treatment of cancer: overview and perspectives. Future Med. Chem. 4, 1439–1460 [DOI] [PubMed] [Google Scholar]
- 8. Ropero S., Esteller M. (2007) The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 1, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee J. H., Choy M. L., Marks P. A. (2012) Mechanisms of resistance to histone deacetylase inhibitors. Adv. Cancer Res. 116, 39–86 [DOI] [PubMed] [Google Scholar]
- 10. Fantin V. R., Richon V. M. (2007) Mechanisms of resistance to histone deacetylase inhibitors and their therapeutic implications. Clin. Cancer Res. 13, 7237–7242 [DOI] [PubMed] [Google Scholar]
- 11. Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y., Ge K. (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nagy Z., Tora L. (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357 [DOI] [PubMed] [Google Scholar]
- 13. Akil A., Ezzikouri S., El Feydi A. E., Benazzouz M., Afifi R., Diagne A. G., Benjouad A., Dejean A., Pineau P., Benjelloun S. (2012) Associations of genetic variants in the transcriptional coactivators EP300 and PCAF with hepatocellular carcinoma. Cancer Epidemiol. 36, e300–305 [DOI] [PubMed] [Google Scholar]
- 14. Yang X. J. (2004) The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 32, 959–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang F., Marshall C. B., Ikura M. (2013) Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in _target recognition. Cell. Mol. Life Sci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shiota M., Yokomizo A., Tada Y., Uchiumi T., Inokuchi J., Tatsugami K., Kuroiwa K., Yamamoto K., Seki N., Naito S. (2010) P300/CBP-associated factor regulates Y-box binding protein-1 expression and promotes cancer cell growth, cancer invasion, and drug resistance. Cancer Sci. 101, 1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coffey K., Blackburn T. J., Cook S., Golding B. T., Griffin R. J., Hardcastle I. R., Hewitt L., Huberman K., McNeill H. V., Newell D. R., Roche C., Ryan-Munden C. A., Watson A., Robson C. N. (2012) Characterisation of a Tip60-specific inhibitor, NU9056, in prostate cancer. PloS One 7, e45539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Halkidou K., Gnanapragasam V. J., Mehta P. B., Logan I. R., Brady M. E., Cook S., Leung H. Y., Neal D. E., Robson C. N. (2003) Expression of Tip60, an androgen receptor coactivator, and its role in prostate cancer development. Oncogene 22, 2466–2477 [DOI] [PubMed] [Google Scholar]
- 19. Kim J. H., Kim B., Cai L., Choi H. J., Ohgi K. A., Tran C., Chen C., Chung C. H., Huber O., Rose D. W., Sawyers C. L., Rosenfeld M. G., Baek S. H. (2005) Transcriptional regulation of a metastasis suppressor gene by Tip60 and β-catenin complexes. Nature 434, 921–926 [DOI] [PubMed] [Google Scholar]
- 20. Okumura K., Mendoza M., Bachoo R. M., DePinho R. A., Cavenee W. K., Furnari F. B. (2006) PCAF modulates PTEN activity. J. Biol. Chem. 281, 26562–26568 [DOI] [PubMed] [Google Scholar]
- 21. Li S., Shogren-Knaak M. A. (2009) The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J. Biol. Chem. 284, 9411–9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Germaniuk-Kurowska A., Nag A., Zhao X., Dimri M., Band H., Band V. (2007) Ada3 requirement for HAT recruitment to estrogen receptors and estrogen-dependent breast cancer cell proliferation. Cancer Res. 67, 11789–11797 [DOI] [PubMed] [Google Scholar]
- 23. Chen J., Luo Q., Yuan Y., Huang X., Cai W., Li C., Wei T., Zhang L., Yang M., Liu Q., Ye G., Dai X., Li B. (2010) Pygo2 associates with MLL2 histone methyltransferase and GCN5 histone acetyltransferase complexes to augment Wnt _target gene expression and breast cancer stem-like cell expansion. Mol. Cell. Biol. 30, 5621–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayakawa T., Nakayama J. (2011) Physiological roles of class I HDAC complex and histone demethylase. J. Biomed. Biotechnol. 2011, 129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Son J. W. (2012) Year-in-review of lung cancer. Tuberc. Respir. Dis. 73, 137–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cagle P. T., Allen T. C. (2012) Lung cancer genotype-based therapy and predictive biomarkers: present and future. Arch. Pathol. Lab. Med. 136, 1482–1491 [DOI] [PubMed] [Google Scholar]
- 27. Al Dayel F. (2012) EGFR mutation testing in non-small cell lung cancer (NSCLC). J. Infect. Public Health 5, S31–S34 [DOI] [PubMed] [Google Scholar]
- 28. Yan H., Zhu S., Song C., Liu N., Kang J. (2012) Bone morphogenetic protein (BMP) signaling regulates mitotic checkpoint protein levels in human breast cancer cells. Cell. Signal. 24, 961–968 [DOI] [PubMed] [Google Scholar]
- 29. Zhu S., Wang W., Clarke D. C., Liu X. (2007) Activation of Mps1 promotes transforming growth factor-β-independent Smad signaling. J. Biol. Chem. 282, 18327–18338 [DOI] [PubMed] [Google Scholar]
- 30. Xu Q., Zhu S., Wang W., Zhang X., Old W., Ahn N., Liu X. (2009) Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by Mps1 phosphorylation. Mol. Biol. Cell 20, 10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tetsu O., McCormick F. (1999) β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 32. Geng Y., Eaton E. N., Picón M., Roberts J. M., Lundberg A. S., Gifford A., Sardet C., Weinberg R. A. (1996) Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 12, 1173–1180 [PubMed] [Google Scholar]
- 33. Neuman E., Flemington E. K., Sellers W. R., Kaelin W. G., Jr. (1994) Transcription of the E2F-1 gene is rendered cell cycle-dependent by E2F DNA-binding sites within its promoter. Mol. Cell. Biol. 14, 6607–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo R., Chen J., Mitchell D. L., Johnson D. G. (2011) GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 39, 1390–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cartier J., Berthelet J., Marivin A., Gemble S., Edmond V., Plenchette S., Lagrange B., Hammann A., Dupoux A., Delva L., Eymin B., Solary E., Dubrez L. (2011) Cellular inhibitor of apoptosis protein-1 (cIAP1) can regulate E2F1 transcription factor-mediated control of cyclin transcription. J. Biol. Chem. 286, 26406–26417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Komatsu N., Kawamata N., Takeuchi S., Yin D., Chien W., Miller C. W., Koeffler H. P. (2006) SAHA, an HDAC inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncol. Rep. 15, 187–191 [PubMed] [Google Scholar]
- 37. Zhong S., Fields C. R., Su N., Pan Y. X., Robertson K. D. (2007) Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel _targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene 26, 2621–2634 [DOI] [PubMed] [Google Scholar]
- 38. Liu Y., Mayo M. W., Nagji A. S., Hall E. H., Shock L. S., Xiao A., Stelow E. B., Jones D. R. (2013) BRMS1 suppresses lung cancer metastases through an E3 ligase function on histone acetyltransferase p300. Cancer Res. 73, 1308–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kishimoto M., Kohno T., Okudela K., Otsuka A., Sasaki H., Tanabe C., Sakiyama T., Hirama C., Kitabayashi I., Minna J. D., Takenoshita S., Yokota J. (2005) Mutations and deletions of the CBP gene in human lung cancer. Clin. Cancer Res. 11, 512–519 [PubMed] [Google Scholar]
- 40. Ramsey M. R., He L., Forster N., Ory B., Ellisen L. W. (2011) Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. 71, 4373–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alla V., Engelmann D., Niemetz A., Pahnke J., Schmidt A., Kunz M., Emmrich S., Steder M., Koczan D., Pützer B. M. (2010) E2F1 in melanoma progression and metastasis. J. Natl. Cancer Inst. 102, 127–133 [DOI] [PubMed] [Google Scholar]
- 42. Inoshita S., Terada Y., Nakashima O., Kuwahara M., Sasaki S., Marumo F. (1999) Regulation of the G1/S transition phase in mesangial cells by E2F1. Kidney Int. 56, 1238–1241 [DOI] [PubMed] [Google Scholar]
- 43. Iwamoto M., Banerjee D., Menon L. G., Jurkiewicz A., Rao P. H., Kemeny N. E., Fong Y., Jhanwar S. C., Gorlick R., Bertino J. R. (2004) Overexpression of E2F-1 in lung and liver metastases of human colon cancer is associated with gene amplification. Cancer Biol. Ther. 3, 395–399 [PubMed] [Google Scholar]
- 44. Banerjee D., Gorlick R., Liefshitz A., Danenberg K., Danenberg P. C., Danenberg P. V., Klimstra D., Jhanwar S., Cordon-Cardo C., Fong Y., Kemeny N., Bertino J. R. (2000) Levels of E2F-1 expression are higher in lung metastasis of colon cancer as compared with hepatic metastasis and correlate with levels of thymidylate synthase. Cancer Res. 60, 2365–2367 [PubMed] [Google Scholar]
- 45. Engelmann D., Pützer B. M. (2012) The dark side of E2F1: in transit beyond apoptosis. Cancer Res. 72, 571–575 [DOI] [PubMed] [Google Scholar]
- 46. Udayakumar T., Shareef M. M., Diaz D. A., Ahmed M. M., Pollack A. (2010) The E2F1/Rb and p53/MDM2 pathways in DNA repair and apoptosis: understanding the cross-talk to develop novel strategies for prostate cancer radiotherapy. Semin. Radiat. Oncol. 20, 258–266 [DOI] [PubMed] [Google Scholar]
- 47. Park W. J., Ma E. (2012) Inhibition of PCAF histone acetyltransferase, cytotoxicity, and cell permeability of 2-acylamino-1-(3- or 4-carboxyphenyl)benzamides. Molecules 17, 13116–13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watanabe N., Osada H. (2012) Phosphorylation-dependent protein-protein interaction modules as potential molecular _targets for cancer therapy. Curr. Drug _targets 13, 1654–1658 [DOI] [PubMed] [Google Scholar]
- 49. Li J., Zhang S., Gao L., Chen Y., Xie X. (2011) A cell-based high-throughput assay for the screening of small-molecule inhibitors of p53-MDM2 interaction. J. Biomol. Screen. 16, 450–456 [DOI] [PubMed] [Google Scholar]
- 50. Vu B. T., Vassilev L. (2011) Small-molecule inhibitors of the p53-MDM2 interaction. Curr. Top. Microbiol. Immunol. 348, 151–172 [DOI] [PubMed] [Google Scholar]