Abstract
Despite advances in sequencing, the goal of obtaining a comprehensive view of genetic variation in populations is still far from reached. We sequenced 180 lines of A. thaliana from Sweden to obtain as complete a picture as possible of variation in a single region. Whereas simple polymorphisms in the unique portion of the genome are readily identified, other polymorphisms are not. The massive variation in genome size identified by flow cytometry seems largely to be due to 45S rDNA copy number variation, with lines from northern Sweden having particularly large numbers of copies. Strong selection is evident in the form of long-range linkage disequilibrium (LD), as well as in LD between nearby compensatory mutations. Many footprints of selective sweeps were found in lines from northern Sweden, and a massive global sweep was shown to have involved a 700-kb transposition.
The common weed A. thaliana is highly selfing and naturally exists as inbred lines that can be grown in replicate under controlled conditions. The species is widely distributed throughout the northern hemisphere and shows strong evidence of local adaptation1,2. The pattern of genetic polymorphism is compatible with isolation by distance on every scale3. Taken together, these features make A. thaliana an excellent model for studying the genetics of natural variation, and, indeed, shared inbred lines have been a resource for the Arabidopsis community since its inception4. More recently, over 1,300 lines have been genotyped for 250,000 SNPs using a custom Affymetrix SNP tiling array (AtSNPtile1) to facilitate genome-wide association studies (GWAS)5,6, and efforts are underway to sequence over 1,000 lines7–11.
Here we report the sequencing of 180 lines from Sweden. We contribute the largest sample by far from a single geographic region, which allows us to look for evidence of selection and to carry out GWAS in local populations for the first time. Our analysis emphasizes structural variation, which we show to be a major component of genetic variation.
RESULTS
Sequencing and polymorphism detection
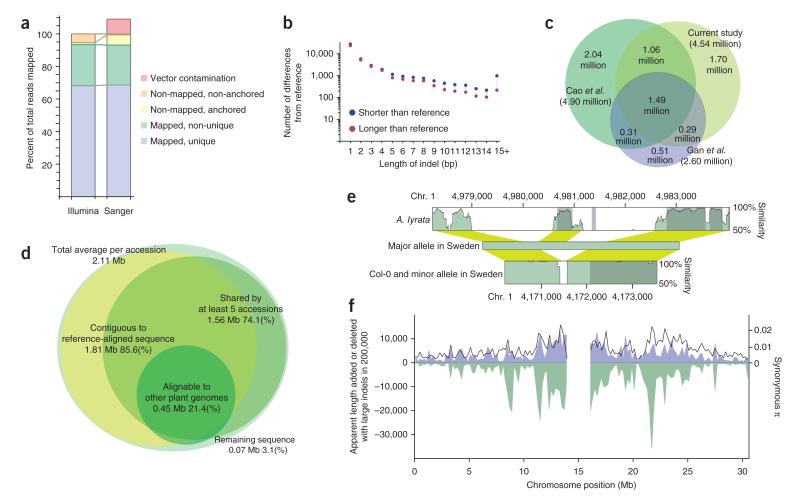
The analyzed lines were selected on the basis of low-density SNP data3 to obtain samples with distinct genotypes from both northern and southern Sweden (52 versus 128 lines, respectively; Supplementary Fig. 1). Using 76- or 100-bp Illumina paired-end reads and fragments of roughly 300 bp in size, we obtained an average of 39-fold coverage per line. We identified differences from the A. thaliana reference genome, including short insertion-deletion polymorphisms (indels) and other structural variants, using an ad hoc pipeline (Online Methods). This approach generated 4.5 million SNPs and almost 0.6 million structural variants, over 90% of which are indels shorter than 10 bp in length. The data had low error rates overall (Supplementary Table 1), but it is important to realize that the genome sequences are far from complete. Several important biases exist. First, we are only able to detect polymorphisms reliably in the roughly 85% of the genome that can be uniquely aligned to the reference genome (Fig. 1a). Second, some kinds of variants are easier to detect than others. For example, we estimated that false positive and false negative rates when detecting short indels (shorter than 15 bp) were roughly twice as high as when detecting SNPs (Supplementary Table 1), and these rates rose markedly with increasing length of the indel. Third, there are biases with respect to the reference genome. For example, we found more indels with the variant allele shorter than the reference genome than with the variant allele longer than the reference genome. These differences are unlikely to be real (as there is no evidence that the reference genome is unusually large) but can readily be explained by noting that alignment algorithms handle gaps better than inserted sequence. Consistent with this interpretation, such discrepancies were almost absent for polymorphisms of ≤4 bp but increased with the length of the indel (Fig. 1b).
Figure 1.
Polymorphism detection. (a) Comparison of Illumina reads and longer, dideoxy-sequenced, randomly cloned fragments (Sanger) with respect to how well they align to the reference genome. The distributions are very similar, except that longer reads that cannot be aligned are more likely to be anchored by a short stretch of presumably homologous sequence. (b) Average number of indels between the sequenced lines and the reference genome, divided into variants that are shorter and longer than the reference genome and shown as a function of the length of the variant. (c) Overlap between SNPs generated by this study and two previous resequencing studies9,10. (d) Characterization of new sequence identified by de novo assembly. (e) An example of a region containing new sequence. The graphs show sequence similarity (coding sequence in dark green, noncoding sequence in light green; yellow shows alignment) to the majority haplotype in Sweden, which contains a ~1-kb fragment of new sequence not found in the reference genome. The new fragment is also found in A. lyrata, indicating that it is ancestral; however, the region has been subject to several more rearrangements since the species diverged. The polymorphism may have functional consequences, as it affects putative coding sequence. (f) Distribution of large variants increasing length (blue; identified using de novo assembly), large variants decreasing length (green; inferred from sequencing coverage) and SNPs (synonymous nucleotide diversity, π black line) along chromosome 1. Chromosomes 2–5 show an analogous pattern (Supplementary Fig. 2).
The overlap between the SNPs identified here and by two previous resequencing efforts9,10 is shown in Figure 1c. Whereas the majority of SNPs were present in all data sets, there was also a very large number of new SNPs, as expected given that previous studies were smaller and did not include lines from Sweden. The total number identified was smaller than the number previously identified in 80 lines10 (4.54 versus 4.90 million, respectively), reflecting a combination of differences in SNP calling and real differences between the samples (mostly in population structure, as the average number of pairwise differences per site between individuals did not differ greatly: 0.49% for the 180 lines sequenced here, 0.53% for the 80 previously sequenced lines; based on regions with high alignment scores in our data). A rigorous analysis of the nature of these differences will require reprocessing the raw sequence data using a common pipeline.
Detection and characterization of new sequence
The biases that arise from aligning to a reference genome apply to all resequencing studies, but there is reason to believe them to be more serious for A. thaliana, which has a genome half the size of its nearest relative, Arabidopsis lyrata, apparently owing to deletions in the A. thaliana lineage12. If this reduction in genome size is still ongoing, individual A. thaliana genomes will harbor many ancestral chromosomal segments not present in the reference genome (and will lack equally many that are). With this in mind, we assembled all our lines individually, de novo, identifying 1.3–3.3 Mb of new sequence per line (compared to 181 kb for Col-0, the line corresponding to the reference genome), largely in segments shorter than 10 kb. Most of this new sequence seemed to be genuine A. thaliana genomic sequence: 96.5% of the new sequence was either anchored by a sequence that aligned well with the reference genome or was shared by at least five of the Swedish lines (Fig. 1d). Furthermore, 21% of the sequence showed similarity to sequence from other plant genomes, usually A. lyrata (Supplementary Note), and thus likely represents retained ancestral fragments; however, closer examination often identified complex polymorphisms, making the precise mutational events difficult to infer (Fig. 1e). The genomic distribution of the new sequence is similar to that of regions of missing coverage, as would be expected if the latter reflect segregating longer deletions of ancestral sequence (that we largely did not detect). Both distributions resembled that of SNPs, suggesting that all three types of polymorphism are influenced by similar evolutionary forces (Fig. 1f and Supplementary Fig. 2). On the basis of available annotation and preliminary mRNA sequencing data, the identified new sequence seems to contain around 200–300 genes or gene fragments per line, in agreement with previous estimates9. One might expect rapidly evolving gene families, such as F-box and NB-LRR genes12, to be overrepresented, but no evidence for this was found.
Massive variation in genome size
The above analyses suggest that, despite the recent marked decrease in the size of the A. thaliana genome, variation between lines is only on the order of 1%. Yet, flow cytometry analysis has suggested that there is up to 10% variation worldwide13. Using the same technique, we found that our lines varied by well over 10%, ranging from 161 Mb to 184 Mb in length. The estimate for the reference line, Col-0, was 166 Mb, making it one of the smallest, whereas the largest values were found exclusively in lines from northern Sweden. Extending the study by including 36 lines selected from the worldwide distribution of the species confirmed this impression: the variation in lines from southern Sweden was similar to that found worldwide, whereas the estimates in lines from northern Sweden were substantially greater (Supplementary Fig. 3).
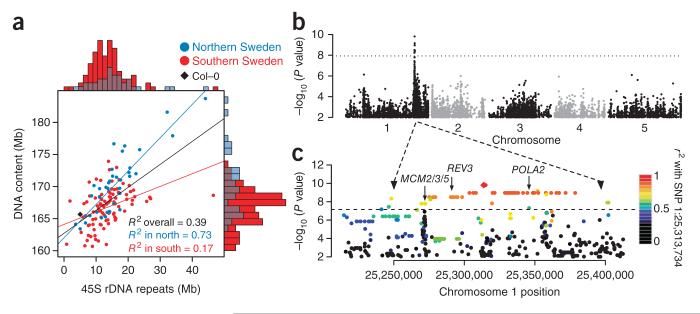
Given the analyses above, it seemed unlikely that the cause of this variation would lie in the unique portion of the genome. To investigate the role of repetitive sequence, we used sequence coverage to estimate copy number variation for 45S rDNA, 5S rDNA and centromeric repeats, as well as for transposable elements, and used the results to predict the flow cytometry–based estimates of genome size using linear regression. In a multiple regression, all four classes of repeats were significantly positively correlated with the flow cytometry–based estimates; however, 45S rDNA made the largest contribution by far (Table 1). Notably, both the flow cytometry–based estimates and the 45S rDNA copy number estimates showed a strong geographic pattern, with larger estimates being more prevalent and the correlation between the estimates being much stronger (R2 = 0.73) in lines from northern Sweden (Fig. 2a).
Table 1.
Multiple regression of flow cytometry–based estimates
| Feature | DF | SS | MS | F | P value | R 2 |
|---|---|---|---|---|---|---|
| 45S rDNA | 1 | 739 | 739 | 94 | 7.7 × 10−17 | 0.39 |
| 5S rDNA | 1 | 42 | 42 | 5 | 0.023 | 0.022 |
| Centromeres | 1 | 114 | 114 | 14 | 2.3 × 10−4 | 0.059 |
| TEs | 1 | 56 | 56 | 7 | 8.8 × 10−3 | 0.029 |
| Error | 123 | 968 | 8 | |||
| Total | 127 | 1,918 |
DF, degrees of freedom; SS, sum of squares; MS, mean square; TEs, novel transposable element insertions. Total R2 = 0.50; adjusted R2 = 0.48.
Figure 2.
Genome size variation. (a) Joint distribution of nuclear DNA content (estimated using flow cytometry) and total amount of 45S rDNA (estimated using sequencing coverage). Marginal distributions are shown along the axes. (b) Manhattan plot of genome-wide association results for the flow cytometry–based estimates of genome size. The dotted horizontal line marks a significance level of 0.05 after Bonferroni correction for 4 million tests. The two known 45S rDNA clusters are close to the left ends of chromosomes 2 and 4 (ref. 15). (c) Magnified view of the chromosome 1 peak in b including a roughly 100-kb region of extensive LD. Colors indicate the extent of LD with the most significant SNP at position 25,313,734. The positions of three replication-related candidate genes are shown: POLA2 (At1g67630), which encodes the B subunit of DNA polymerase α; REV3 (At1g67500), which encodes recovery protein 3, the catalytic subunit of DNA polymerase ζ; and MCM2/3/5 (At1g67460), which is related to the minichromosome maintenance family of proteins. Sequence analysis of these candidates identified no obvious candidate polymorphisms (multiple alignments are available on the project download site).
These results confirm that there is considerable natural variation in nuclear DNA content and demonstrate that this variation is mainly due to 45S rDNA, in agreement with findings from previous studies14. Because the flow cytometry and genome sequencing experiments used different plants as well as tissues (leaves and roots, respectively), it is clear that the variation is heritable. To investigate the genetics of this variation, we carried out a GWAS for the flow cytometry–based estimates of genome size. Unexpectedly, this analysis identified neither of the two known 45S rDNA clusters15. Instead, the scan identified a major locus in a euchromatic region of chromosome 1 that apparently explained 26% of the variation in genome size (Fig. 2b). Neither sequence analysis nor FISH found any evidence for new 45S rDNA clusters (Supplementary Fig. 4).
It would thus seem that the identified locus regulates DNA content in trans rather than in cis, and this, in turn, implies that the presumed ‘genome size variation’ should, at least partially, be regarded as a phenotype rather than a genotype. There is evidence of regulation of rDNA copy number in several organisms16–18, including A. thaliana19. Notably, mapping of variation in cytosine methylation of 45S rDNA repeat arrays, which is strongly correlated with copy number20, in a cross between two inbred lines has previously identified both cis and trans quantitative trait loci (QTLs)21. The two strongest QTLs corresponded to the 45S rDNA clusters, but the third strongest contained the GWAS peak reported here. These results are consistent with ours if the repeat number changes too rapidly to be mapped using GWAS but is inherited stably enough to be mapped in crosses. The trans-acting loci might modify the replication process, with different alleles effectively predisposing lines to large or small numbers of repeats. The peak of association contained at least three candidates that might affect replication (Fig. 2c)22–25.
However, it must be emphasized that the association may simply be spurious. GWAS on subsets of the lines showed that the chromosome 1 association was due to a relatively small number of lines from northern Sweden with very large genome size estimates (Supplementary Fig. 5). Although our analysis takes confounding from genome-wide population structure into account, it does not necessarily handle confounding caused by a small number of genes of large effect26,27. A spurious correlation could arise due to LD with the true causal loci, for example, the 45S rDNA clusters themselves, which we think we are unable to map owing to allelic heterogeneity. In other words, the peak on chromosome 1 could be a so-called synthetic association26,28. To resolve this, multigeneration experiments will be required.
Selection and LD
We searched for evidence that some of this genomic variation is adaptively important. With regard to the variation in nuclear DNA content, its marked geographic distribution (Fig. 2a) was suggestive of local adaptation, as the overall genetic divergence was much smaller. Less than 0.6% of SNPs showed a stronger correlation with location in northern versus southern Sweden than the flow cytometry–based estimates of genome size. However, if the variation is due to a very small number of genetic loci, then it might have been possible for genetic drift to cause the observed divergence in size. Resolving this will require further studies.
Given the apparent recent shrinkage of the A. thaliana genome, it is also natural to consider selection at indels. Previous work, using a small number of indels, has suggested that deletions are selectively favored relative to insertions, perhaps because of selection for a more compact genome12. Unfortunately, this kind of analysis is very sensitive to the kinds of biases we saw in our data and, even worse, depends on accurate inference of the ancestral state (that is, whether an indel is the result of a deletion or an insertion). Indels are often complex (see Fig. 1e for an example). For the 18% of indels we were able to classify unambiguously, there was no evidence of selection favoring deletions, in contradiction to previous results (Supplementary Fig. 6). However, it is dangerous to extrapolate from a biased minority of events, and our conclusion is that the divergence between A. thaliana and A. lyrata is probably too great for analyses that rely on the determination of the ancestral state of indels to be reliable.
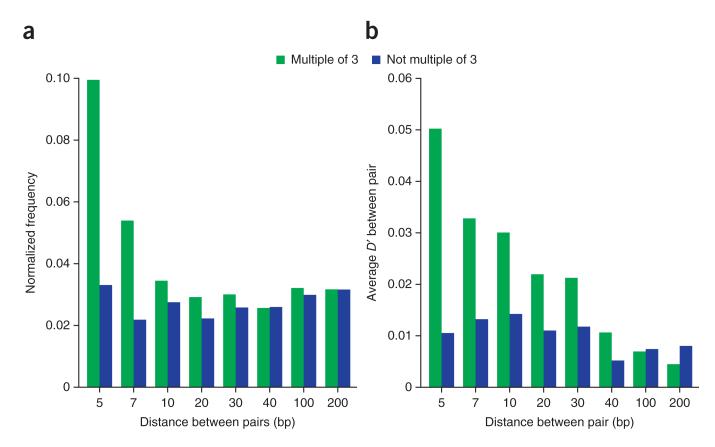
However, we found several other clear signals of strong selection. Recent resequencing efforts have notably identified many new protein-coding alleles involving apparently disruptive frameshift mutations and closely linked compensatory changes8–10. With our larger sample size, we were able to show that selection has a role in creating this diversity. Closely linked alleles that restored the reading frame were greatly overrepresented compared to those that did not, and positive LD between such alleles ensures that aberrant proteins occur at a lower frequency than expected from the marginal allele frequencies (Fig. 3). How these kinds of variant haplotypes arise is far from clear, as the evolution of compensatory changes involves crossing an adaptive valley29. One possibility is that the population structure of A. thaliana leads to local fixation of weakly deleterious mutations during colonization of new patches, which is followed by compensatory evolution as the local population size increases.
Figure 3.
Compensatory indels. (a) Over-representation of compensatory pairs of indels compared to their genome-wide frequency, plotted as a function of the distance between the indels. Compensatory pairs of indels are those whose sum length is a multiple of 3, thus restoring the reading frame. (b) LD (D’) between compensatory pairs of indel alleles as a function of the distance between the indels. Positive LD indicates an excess of non-reference alleles.
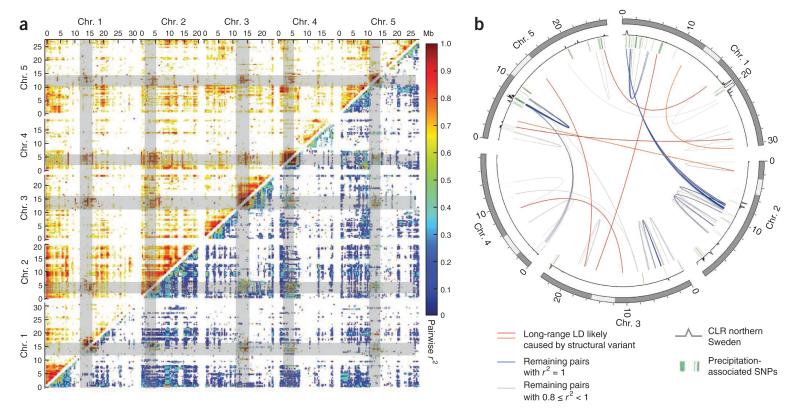
Strong selection can also cause LD between unlinked loci, especially in conjunction with local adaptation (in which case, there is no requirement for epistatic interactions between the loci). In agreement with previous results6,10,30–32, average LD in our sample decayed relatively quickly (on roughly the same scale as in humans) to high background levels that were largely determined by population structure (Supplementary Fig. 7). However, even after taking this structure into account, considerable long-range LD remained, including over 300,000 pairs of loci for which r2 was >0.8, even though the loci were separated by more than 1 Mb (Fig. 4a). Especially notable was the prevalent LD between all centromeres. Because it is difficult to imagine selection maintaining LD between all centromeres, it seemed likely that most of these patterns must be artifactual, perhaps because the SNP loci, in fact, map to multiple regions. Indeed, strict filtering for uniqueness resulted in the elimination of all but around 70,000 pairs with long-range LD (corresponding to 7,973 loci). From these, we selected 4 centromeric and 2 non-centromeric sets of SNPs for genotyping in informative crosses (Supplementary Table 2). Of the centromeric pairs, one showed complete linkage, despite the SNPs supposedly being located on different chromosomes, and the other three failed PCR, perhaps because they are associated with repetitive regions. These results illustrate the danger inherent in assuming that SNPs are located where they are supposed to be located and show that population genetics analysis may assist in identifying unreliable ones. However, both non-centromeric pairs segregated independently in crosses, showing that at least some of the long-range LD we observed must be due to normal population genetics forces, whether chance or natural selection. In support of the latter explanation, there was a significant enrichment of the remaining loci among SNPs exhibiting signs of having been involved in local adaptation (Fig. 4b and Supplementary Table 3).
Figure 4.
Long-range LD. (a) Genome-wide pairwise LD. Values before correcting for population structure are shown above the diagonal; for clarity, only values above 0.6 are shown. Values after applying a transformation to reduce the effects of population structure (related to the correction used in genome-wide association mapping; Supplementary Note) are shown below the diagonal. (b) Remaining long-range LD after extensive filtering, combined with positions of putatively selected loci. Green bars show the position of loci significantly associated with minimum precipitation and relative humidity in a global sample (Supplementary Table 3), and the gray curve indicates the signatures of local adaptation in the northern Swedish population (Fig. 5). Gray bars indicate centromeric regions.
Global and local selective sweeps
Population structure in A. thaliana is generally characterized by varying degrees of isolation by distance3. In previous studies, samples from southern Sweden have seemed to be part of a European continuum, whereas those from northern Sweden were quite distinct6,31. Our data confirmed this distribution (Supplementary Figs. 7 and 8)33 and further suggest that the divergence is due to changing allele frequencies rather than the accumulation of mutations (as would accompany ancient separation with little gene flow), as we found fewer private alleles in lines from northern Sweden than in lines from southern Sweden (18% versus 67%) and because pairwise sequence divergence was commensurate with the distance between the regions (Supplementary Fig. 9). However, within each region, the divergence increased more rapidly in lines from northern Sweden, consistent with previously reported greater population structure there3,6,31, as well as with field observations: whereas A. thaliana is a common weed in southern Sweden, its distribution in northern Sweden is often restricted to eroded south-facing slopes and is much more patchy (M.N., unpublished observation). Whereas most of the divergence between lines from northern and southern Sweden is likely due to genetic drift, there are clear differences in many traits that are likely to be adaptive, such as seed dormancy and flowering time (M.N., unpublished observation), and we thus decided to search our data for evidence of selective sweeps.
The results for lines from northern and southern Sweden were markedly different. SweepFinder34, an algorithm that uses the distribution of SNP allele frequencies to detect sweeps close to fixation, returned 22 strong signals in lines from northern Sweden and only a single signal in lines from southern Sweden (Fig. 5a, Supplementary Figs. 10–14 and Supplementary Note). The signals were extremely strong: the SweepFinder composite likelihood ratio for the strongest selective sweep was 178 times that corresponding to background, and those for the other sweeps were 30 times stronger on average. Most selective sweeps exhibited strong population subdivision, quantifed using FST (Fig. 5a), and were found by the FST-like cross-population statistic XP-CLR35, in agreement with the notion that they are due to local adaptation. The identified regions were also over-represented among SNPs that showed long-range LD as well as among SNPs that have previously been associated with environmental variables (Fig. 4b and Supplementary Note).
Figure 5.
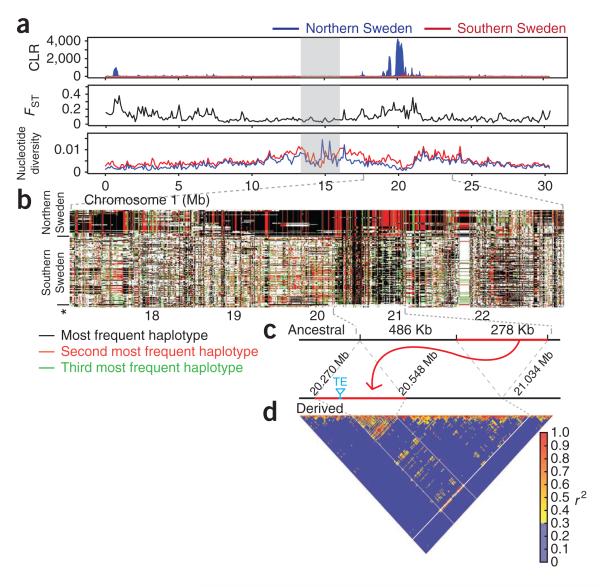
Characterization of selective sweeps on chromosome 1. (a) Values of three different statistics sensitive to selective sweeps plotted along the chromosome. Statistics were calculated separately for the lines from northern and southern Sweden. The CLR statistic clearly marks a strong sweep in the northern lines, and the same region also shows increased FST as well as decreased nucleotide diversity. The gray bar indicates the centromeric region. (b) Pattern of haplotype sharing underlying the major signal around 20 Mb. Shown are haplotypes derived from lines in northern and southern Sweden, as are the six presumed ancestral haplotypes (asterisk). Haplotype sharing is much more extensive in the lines from northern Sweden than in those from southern Sweden. (c) Schematic of the transposition event most likely responsible for the observed pattern. (d) Pattern of LD across the swept region (red bar in c).
The reason for the much greater number of sweep signals in lines from northern Sweden is not clear. Distinguishing between real signals of selection and artifactual ones due to demography is, as always, very difficult. However, at least one of the identified selective sweeps is almost certainly real. The single signal in lines from southern Sweden corresponded to the strongest signal in lines from northern Sweden (Fig. 5a), and the pattern of haplotype sharing showed that the selective sweep in lines from northern Sweden was simply more extensive (Fig. 5b). If the sweep signals in lines from northern Sweden were simply due to complicated demographics (for example, colonization bottlenecks and concomitant differences in local effective population size (Supplementary Figs. 7 and 15)), there would be no reason to expect them to overlap with sweep signals in lines from southern Sweden. Furthermore, the signal corresponded to a presumed global selective sweep, previously identified in a chip-based resequencing study of 20 lines, in which 18 of the 20 lines were found to share a haplotype of several hundred kilobases in length, with the remaining 2 lines hailing from Cape Verde and northern Sweden, at the southern and northern edges of the species range, respectively36. The simplest explanation for the observed pattern is thus that this is an ongoing global selective sweep and that the sweep is more recent in lines from northern Sweden than in those from southern Sweden. And, if this is true, then it seems likely that some of the other strong signals in lines from northern Sweden also represent genuine selective sweeps rather than artifacts due to demographic factors.
A curious feature of the previously reported selective sweep was that the shared haplotype appeared identical in all carriers36, which is inconsistent with the random action of recombination. Furthermore, the extent of haplotype sharing seemed far too great given the average decay of LD in global samples of A. thaliana. An obvious explanation was that the selective sweep was associated with some kind of large-scale structural variant that suppressed recombination locally. With this in mind, we examined the region more closely and discovered that the swept haplotype was associated with an intrachromosomal conservative transposition of 278 kb containing 72 genes to a new position 486 kb away (Fig. 5c and Supplementary Note). The A. thaliana reference line Col-0 carried the swept haplotype, as did most members of the species: using genome-wide SNP data6, we estimated that only 45 of 1,306 lines (3.4%) had escaped the selective sweep (Supplementary Note). Contrary to previous results, the ancestral haplotype was not just found at the extremes of the range but was also found at low frequency worldwide. Recombination in heterozygotes is likely to be effectively suppressed by selection against recombinants, given that crossing over within the region would lead to either duplication or deletion of the 72 transposed genes. The pattern of LD across the region was suggestive of the suppression of recombination (Fig. 5d). It should be noted that the strong signal of selection was not simply due to lack of recombination: it remained present even if we treated the entire transposed region as a single locus (SweepFinder scores based solely on SNPs outside the rearrangement decreases from 178 to 165 times the background).
The breakpoints of the identified transposition are consistent with the action of non-homologous end joining. Resealing at the donor site seems to have been facilitated by 5 bp of microhomology, leading to a 5-bp deletion, whereas a 9-bp _target site deletion occurred at the receptor site (Supplementary Fig. 16). Although we do not know the selective agent, the transposition seems to contain a relatively small number of derived variants that tag the sweep globally, including roughly 30 SNPs and 2 helitron insertions. Attempts to date the selective sweep on the basis of polymorphism among the swept haplotypes yielded estimates of 43,000 years for lines from southern Sweden and 17,000 years for lines from northern Sweden (Supplementary Note), which predate the end of the last glaciation in Sweden and are consistent with the lack of geographic structuring of the sweep3.
DISCUSSION
We have used next-generation sequencing to generate a high-quality polymorphism data set for a Swedish sample of A. thaliana. We provide a reasonable estimate of variation for SNPs and very short indels in the fraction of the genome that is accessible using these methods10, and, although biases complicate many kinds of evolutionary analyses, the data comprise an important resource, in particular for GWAS. At the same time, our findings highlight how much we may be missing by simply employing standard pipelines for polymorphism detection. Perhaps most notably, we discovered massive variation in nuclear DNA content and showed that it may be possible to map genes regulating this variation, suggesting that what we had assumed to be part of the genotype should partly be viewed as a phenotype. It is also clear that we have very little idea of how many large structural variants (especially inversions and transpositions) exist. By combining population genetics analysis with manual searches for putative breakpoints in the sequencing data, we uncovered a very large structural variant that seems to have undergone extremely strong selection. Our attempt to search for such variants systematically, using a novel method based on de novo assembly, identified several other noteworthy examples, including the 1.17-Mb inversion that gave rise to a heterochromatic knob on chromosome 4 (Supplementary Fig. 17) (ref. 37). However, there is every reason to believe that there is more to be found. Of the roughly 13 million SNPs that distinguish the A. thaliana and A. lyrata reference genomes, roughly 4.4% are polymorphic in our sample of A. thaliana genomes. The corresponding percentage for short indels is of the same magnitude. If similar selection pressures affect large structural variants, a similar proportion of the very large number of structural rearrangements between these two genomes11 should still be segregating. Many of these polymorphisms may be complex and very difficult to resolve using short-read sequencing data. Finally, our analyses found signs of selection at every level, from compensatory changes within single genes to local adaptation (giving rise to long-range LD and footprints of selective sweeps) and global selective sweeps. Even in an organism as well studied as A. thaliana, the genome is full of surprises.
ONLINE METHODS
Sequencing and polymorphism detection
Genomic DNA was fragmented, size selected to between 450 and 800 bp and subjected to paired-end Illumina sequencing with read length of 76 or 100 bp. Reads were mapped with Burrows-Wheeler aligner (BWA)38 to the TAIR 10 reference genome, allowing 4% mismatch and one indel. SNPs and short indels were called with SAMtools39 and the Genome Analysis Toolkit (GATK)40. Larger structural variants were called using a variety of tools. For further details, see the Supplementary Note. Tables summarizing the results are available on the project download site.
Error estimates and quality control
Considerable effort was devoted to quality control. Notably, we were not simply trying to ensure that identified SNPs were called correctly but also tried to estimate the underlying sequence, paying as much attention to what was missed as to what was found. We compared our data to 4 different kinds of data to estimate error rates for SNPs and short indels: (i) the reference line was resequenced using our pipeline, and all variants called were assumed to be false positives; (ii) our results were compared with previously published SNP chip data6 to provide estimates of the false negative rate (the rate at which we did not discover SNPs) and the genotyping error rate (the rate at which we made the wrong call for the ones we did detect); (iii) our results were directly compared with an old data set of close to 1,500 manually curated multiple alignments of PCR amplicons from Sanger sequencing of 95 lines31; and (iv) our results were directly compared with ~250 kb of sequence from a single accession that we generated by Sanger sequencing random shotgun clones. The results are summarized in Supplementary Table 1 (for details, see the Supplementary Note). In general, error rates were higher close to centromeres and decreased markedly as the quality of the mapping (alignment Q value) increased (Supplementary Figs. 18–20).
Detection and characterization of new sequence
Each line was assembled de novo using SOAPdenovo to identify fragments longer than 100 bp that were absent from the reference genome (see the Supplementary Note for details). The majority of such fragments could either be anchored to the reference genome by flanking sequence or were shared by more than five lines (Fig. 1d and Supplementary Fig. 21). Summaries are available on the project download site.
Variation in genome size
Flow cytometry was carried out on 128 of the Swedish lines, the reference line (Col-0) and 36 randomly chosen world-wide lines using 2-week-old leaves. Copy number variation for 45S rDNA and 5S rDNA and centromeric repeat number were estimated via normalized read coverage across the appropriate region of the reference genome. In simple single-factor analysis, only 45S and 5S rDNA contributed significantly to the flow cytometry–based estimates (Fig. 2a, Supplementary Fig. 22 and Supplementary Table 4). Estimates for 45S rDNA were validated via quantitative PCR (Supplementary Fig. 23). Further details are given in the Supplementary Note.
GWAS on genome size estimates were carried out with imputed SNP data from this study, accounting for population structure using a mixed model. The genome was scanned for new rDNA clusters bioinformatically (by using an algorithm that searches for read pairs with one read matching the relevant repeat and the other read anchored in unique sequence41), as well as by using FISH (Supplementary Note).
Selection on indels
Where alignment was possible, the ancestral state of each SNP and indel was defined using the A. lyrata genome as the outgroup. The criteria used are detailed in the Supplementary Note.
To test for selection on compensatory mutations (Fig. 3), the proportion of high-confidence indel pairs (no missing data, confirmed by GATK local realignment and less than three SNPs within 20 bp) within coding regions was normalized to the genome-wide count of indel pairs and binned for distance between the events. The count and LD for events that disrupted an ORF were compared with those for events that did not.
LD in structured populations
LD can be thought of as having three different sources: ‘true’ LD, population structure and chance or error42. The first source encompasses both short-range LD due to cosegregation of alleles (linkage) and LD (at any range) due to locus-specific deterministic forces (for example, selection). The other two sources act across the genome and are typically of no direct interest. However, if the sample is heavily structured, the second source will have a massive influence, making it difficult to draw conclusions about the first source. Two related methods for correcting LD estimates for population structure have been proposed43,44; however, the approach adopted in ref. 44 has the advantage that it results in symmetric r2 values. Our approach (Supplementary Note) is similar to the one in ref. 44 (it is also superficially related to the mixed-model correction we used in GWAS), although the underlying assumptions necessary for the derivation are different. In particular, we make no assumptions about the existence of discrete subpopulations, something that would be inappropriate for an organism in which the pattern of variation is characterized by isolation by distance3. The transformed LD estimates were generally lower than the original ones, as the inflation caused by population structure had been removed (Supplementary Fig. 24); however, large numbers of pairs with long-range LD remained (Fig. 4a). Indeed, the presence of strong long-range LD, within as well as between chromosomes, between about 8,000 loci was robust to (i) correction for population structure; (ii) subdivision of our sample into northern and southern populations; (iii) SNP imputation and (iv) read mapping quality (Supplementary Fig. 25).
Global and local selective sweeps
Standard methods were used to describe the pattern of polymorphism within and between populations (Supplementary Note) and to confirm previously published results concerning population structure and the distinctiveness of the northern Swedish population (Supplementary Figs. 7–9, 15 and Supplementary Table 5). Two lines were identified as likely contaminants (Supplementary Note). We scanned the genome for signs of selective sweeps using five different statistics: (i) CLR, the composite likelihood ratio calculated by SweepFinder34, which is sensitive to perturbations of the allele frequency distribution, was run separately for lines from northern and southern Sweden; (ii) FST, which simply measures divergence between populations (for example, due to fixation of locally adaptive alleles), was calculated in non-overlapping 100-kb windows; (iii) nucleotide diversity, which is expected to be reduced following a selective sweep, was similarly estimated in windows (but separately for lines from northern and southern Sweden); (iv) XP-CLR35, which uses one population as a reference and searches for selective sweeps in the other, was used to look for sweeps in both lines from northern and southern Sweden; and (v) XP-EHH45, which looks for extended haplotype sharing, was used search for evidence of selective sweeps in either population. Detailed results can be found on the project download site.
The chromosome 1 transposition was discovered via manual inspection of split reads in unswept lines. Distantly mapping read pairs were consistent with the transposition arrangement depicted in Supplementary Figure 16. PCR and Sanger sequencing of 5 unswept and 15 swept lines (including Col-0) confirmed the expected breakpoint arrangements. De novo assembly also correctly identified transposed breakpoint 4 (chromosome 1: 20,270,307 to 20,548,624) in five of six unswept accessions, breakpoint 5 (chromosome 1: 20,270,429 to 21,034,717) in five of six unswept accessions and breakpoint 6 (chromosome 1: 20,548,624 to 21,034,773) in two of six unswept accessions. The selective sweep was dated on the basis of divergence between swept haplotypes (Supplementary Note).
Supplementary Material
ACKNOWLEDGMENTS
We thank O. Mittelsten Scheid for comments on the manuscript, J. Dolezel for providing a size standard for flow cytometry, G. Schmauss for technical assistance with flow cytometry, N. Lettner for help with sample preparation, A. Sommer for help with sequencing and the Gregor Mendel Institute IT team (in particular, P. Forai) for excellent cluster support. This work was supported by European Research Council grant 268962 MAXMAP and European Community Framework Programme 7 grant 283496 transPLANT to M.N., by the Austrian Science Fund (Vienna Graduate School of Population Genetics, FWF W1225) to I.H. and by Czech Science Foundation grants P501/12/G090 and P506/12/0668 to M.A.L.
Footnotes
URLs. 1001 Genomes Project, http://1001genomes.org/; interactive map of the lines used, http://goo.gl/2n6wp; download site for data from this paper, http://downloads.gmi.oeaw.ac.at; NCBI Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/Traces/sra/.
Accession codes. Flat files of all polymorphism data as well as various lists and tables can be downloaded from the project website. Raw data have been deposited in the NCBI SRA under accession SRP012869. Seeds of all 180 lines have been submitted to the Arabidopsis Biological Resources Center stock center and will be available under accession CS78885.
Note: Supplementary information is available in the online version of the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Fournier-Level A, et al. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- 2.Hancock AM, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- 3.Platt A, et al. The scale of population structure in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000843. doi: 10.1371/journal.pgen.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 5.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 2012;44:212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weigel D, Mott R. The 1001 Genomes Project for Arabidopsis thaliana. Genome Biol. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneeberger K, et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. USA. 2011;108:10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 11.Schmitz RJ, et al. Patterns of population epigenomic diversity. Nature. 2013;495:193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmuths H, Meister A, Horres R, Bachmann K. Genome size variation among accessions of Arabidopsis thaliana. Ann. Bot. 2004;93:317–321. doi: 10.1093/aob/mch037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison J, Tyagi A, Comai L. Large-scale polymorphism of heterochromatic repeats in the DNA of Arabidopsis thaliana. BMC Plant Biol. 2007;7:44. doi: 10.1186/1471-2229-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Copenhaver GP, Pikaard CS. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J. 1996;9:259–272. doi: 10.1046/j.1365-313x.1996.09020259.x. [DOI] [PubMed] [Google Scholar]
- 16.Brown DD, Dawid IB. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- 17.Tartof KD. Increasing the multiplicity of ribosomal RNA genes in Drosophila melanogaster. Science. 1971;171:294–297. doi: 10.1126/science.171.3968.294. [DOI] [PubMed] [Google Scholar]
- 18.Yao MC, Kimmel AR, Gorovsky MA. A small number of cistrons for ribosomal RNA in the germinal nucleus of a eukaryote, Tetrahymena pyriformis. Proc. Natl. Acad. Sci. USA. 1974;71:3082–3086. doi: 10.1073/pnas.71.8.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pontvianne F, et al. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev. 2012;26:945–957. doi: 10.1101/gad.182865.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo HR, Richards EJ. Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana. BMC Plant Biol. 2008;8:92. doi: 10.1186/1471-2229-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riddle NC, Richards EJ. The control of natural variation in cytosine methylation in Arabidopsis. Genetics. 2002;162:355–363. doi: 10.1093/genetics/162.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casper AM, Mieczkowski PA, Gawel M, Petes TD. Low levels of DNA polymerase α induce mitotic and meiotic instability in the ribosomal DNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000105. doi: 10.1371/journal.pgen.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto A, et al. Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and γ-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell. 2003;15:2042–2057. doi: 10.1105/tpc.012369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 2006;66:134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 25.Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platt A, Vilhjálmsson BJ, Nordborg M. Conditions under which genome-wide association studies will be positively misleading. Genetics. 2010;186:1045–1052. doi: 10.1534/genetics.110.121665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilhjálmsson BJ, Nordborg M. The nature of confounding in genome-wide association studies. Nat. Rev. Genet. 2013;14:1–2. doi: 10.1038/nrg3382. [DOI] [PubMed] [Google Scholar]
- 28.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meer MV, Kondrashov AS, Artzy-Randrup Y, Kondrashov FA. Compensatory evolution in mitochondrial tRNAs navigates valleys of low fitness. Nature. 2010;464:279–282. doi: 10.1038/nature08691. [DOI] [PubMed] [Google Scholar]
- 30.Nordborg M, et al. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 2002;30:190–193. doi: 10.1038/ng813. [DOI] [PubMed] [Google Scholar]
- 31.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 33.Platzer A. Visualization of SNPs with t-SNE. PLoS ONE. 2013;8:e56883. doi: 10.1371/journal.pone.0056883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen R, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 37.Fransz PF, et al. Integrated cytogenetic map of chromosome arm 4S of A. thaliana: structural organization of heterochromatic knob and centromere region. Cell. 2000;100:367–376. doi: 10.1016/s0092-8674(00)80672-8. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. The Sequence Alignment/Map format and SAM-tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platzer A, Nizhynska V, Long Q. TE-Locate: a tool to locate and group transposable element occurrences using paired-end next-generation sequencing data. Biology. 2012;1:395–410. doi: 10.3390/biology1020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohta T. Linkage disequilibrium due to random genetic drift in finite subdivided populations. Proc. Natl. Acad. Sci. USA. 1982;79:1940–1944. doi: 10.1073/pnas.79.6.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cockram J, et al. Genome-wide association mapping to candidate polymorphism resolution in the un-sequenced barley genome. Proc. Natl. Acad. Sci. USA. 2010;107:21611–21616. doi: 10.1073/pnas.1010179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mangin B, et al. Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity. 2012;108:285–291. doi: 10.1038/hdy.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.