Abstract
Multiple mutations are required for cancer development, and genome sequencing has revealed that several cancers, including breast, have somatic mutation spectra dominated by C-to-T transitions1–9. Most of these mutations occur at hydrolytically disfavored10 non-methylated cytosines throughout the genome, and are sometimes clustered8. Here, we show that the DNA cytosine deaminase APOBEC3B (A3B) is a likely source of these mutations. A3B mRNA is up-regulated in the majority of primary breast tumors and breast cancer cell lines. Tumors that express high levels of A3B have twice as many mutations as those that express low levels and are more likely to have mutations in TP53. Endogenous A3B protein is predominantly nuclear and the only detectable source of DNA C-to-U editing activity in breast cancer cell line extracts. Knockdown experiments show that endogenous A3B correlates with elevated levels of genomic uracil, increased mutation frequencies, and C-to-T transitions. Furthermore, induced A3B over-expression causes cell cycle deviations, cell death, DNA fragmentation, γ-H2AX accumulation, and C-to-T mutations. Our data suggest a model in which A3B-catalyzed deamination provides a chronic source of DNA damage in breast cancers that could select TP53 inactivation and explain how some tumors evolve rapidly and manifest heterogeneity.
Most humans encode a total of eleven polynucleotide cytosine deaminase family members that could contribute to mutation in cancer – APOBEC1, activation-induced deaminase (AID), APOBEC2, APOBEC3s (A3A, A3B, A3C, A3D, A3F, A3G, and A3H), and APOBEC4. APOBEC2 and APOBEC4 have not shown activity. APOBEC1 and AID are expressed tissue specifically and implicated in cancers of those tissues, hepatocytes and B lymphocytes, respectively11,12. We therefore hypothesized that one or more of the seven APOBEC3s may be responsible for C-to-T mutation in other human cancers. This possibility is consistent with hybridization13 and expression studies14 (Fig. S1).
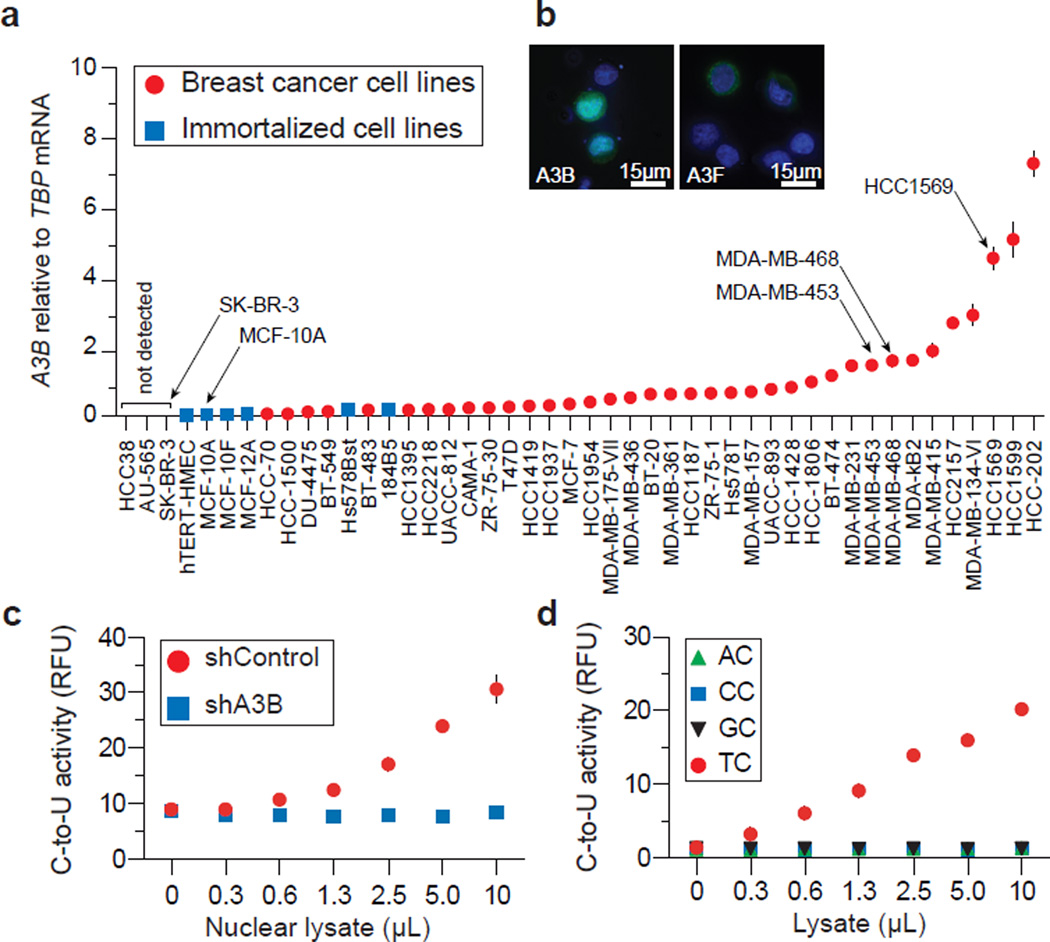
To identify the contributing APOBEC3, we quantified mRNA levels for each of the 11 family members in breast cancer cell lines (Fig. S2). Surprisingly, only A3B mRNA trended toward up-regulation. This analysis was expanded to include a total of 38 independent breast cancer cell lines. A3B was up-regulated by ≥3 s.d. relative to controls in 28/38 lines, with levels exceeding 10-fold in 12/38 lines (Fig. 1a & Table S1). MDA-MB-453, MDA-MB-468, and HCC1569, representative lines used below, showed 20-, 21-, and 61-fold up-regulation, respectively. These results correlate with cell line microarray data (Fig. S3, Tables S2-S9 & Supplementary Discussion). A3B up-regulation is most likely due to an upstream signal transduction event because it is not a frequent site of rearrangement or copy number variation (http://dbCRID.biolead.org) and sequencing failed to reveal promoter activating mutations or CpG islands indicative of epigenetic regulation (Fig. S4).
Figure 1. A3B up-regulation and activity in breast cancer cell lines.
a, A3B levels in indicated cell lines. Each point represents the mean of 3 reactions presented relative to TBP (s.d. shown unless smaller than symbol).
b, A3B-eGFP or A3F-eGFP localization in MDA-MB-453 (nuclei are blue).
c, Nuclear DNA C-to-U activity in extracts from MDA-MB-453 transduced with shControl or shA3B lentiviruses (n=3; s.d. shown unless smaller than symbol).
d, Intrinsic dinucleotide DNA deamination preference of endogenous A3B in extracts from MDA-MB-453 (n=3; s.d. smaller than symbols).
Epitope-tagged A3B localizes to the nucleus of several transfected cell types15. To ask whether this is also a property of breast cancer lines, an A3B-eGFP construct was transfected into MDA-MB-453, MDA-MB-468, and HCC1569. Live cell images of A3B-eGFP showed nuclear localization, in contrast to the cytoplasmic A3F-eGFP (Fig. 1b & S5). Corroborating data were obtained for HA-tagged proteins (Fig. S5). To study endogenous A3B subcellular compartmentalization and activity, we used a fluorescence-based DNA C-to-U assay. We first found that nuclear fractions of several breast cancer cell lines contain a robust DNA editing activity, which could be ablated by A3B knockdown (Fig. 1c, S6 & S7). Similar results were obtained with an independent knockdown construct (not shown). Protein extracts were then used to assess endogenous A3B’s local dinucleotide deamination preference. Similar to retroviral hypermutation signatures caused by A3B over-expression16, endogenous A3B showed a strong preference for editing cytosines in the TC dinucleotide context (Fig. 1d & S6). No deaminase activity was observed for extracts from MCF10A (A3Blow) or SK-BR-3 (A3Bnull), although it could be conferred by transient A3B transfection (Fig. S8). Both A3B-HA and A3A-HA could elicit measurable TC-to-TU activity in lysates from transfected HEK293T cells (Fig. S9). However, because A3A is myeloid lineage-specific17 and non-detectable in breast cancer cell lines (Fig. S1 & S2), our expression and activity studies indicated that A3B may be the only enzyme poised to deaminate breast cancer genomic DNA.
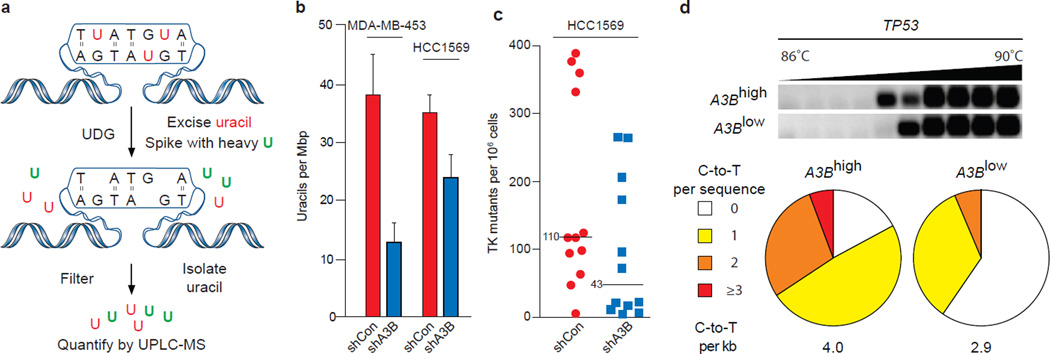
To address whether endogenous A3B damages genomic DNA, we employed a combination of biophysical and genetic assays. We first used a mass spectrometry-based approach to quantify levels of genomic uracil in MDA-MB-453 and HCC1569 with high levels of endogenous A3B vs. knockdown levels of A3B (shControl vs. shA3B) (Fig. 2a & S10). Genomic uracil loads decreased by 30% in HCC1569 expressing shA3B and by 70% in MDA-MB-453, where knockdown was stronger (Fig. 2b & S10). Although these relative differences may seem modest, 10 and 20 uracils per Mbp, respectively, this equates to 30,000 and 60,000 A3B-dependent uracils per haploid genome. The actual number of pro-mutagenic uracils may be even higher because several repair pathways may concurrently function to limit this damage.
Figure 2. A3B-dependent uracil lesions and mutations in breast cancer genomic DNA.
a, Workflow for genomic uracil quantification by UPLC-MS.
b, Average uracil loads in the indicated cell lines (n=3; errors, s.d.).
c, Dot plots representing TK mutant frequencies of HCC1569 subclones expressing shControl or shA3B. Each dot corresponds to one subclone. Medians are labeled.
d, Agarose gel and mutation analysis of TP53 3D-PCR amplicons from HCC1569 cells expressing shControl or shA3B (n≥35 sequences per condition). See Fig. S10 for additional data.
Second, we used a thymidine kinase-positive (TKplus) to TKminus fluctuation analysis17 to determine whether up-regulated A3B and elevated uracil loads lead to higher levels of mutation. MDA-MB-453 and HCC1569 cells were engineered to express TK, which confers sensitivity to the drug ganciclovir. TKplus lines were transduced with shA3B or shControl constructs and limiting dilution was used to generate single cell sub-clones. Expanded sub-clones were subjected to ganciclovir selection and resistant cells were grown to visible colonies, which revealed that cells with up-regulated A3B accumulate 3-to-5-fold more mutations (Fig. 2c & S10).
Third, 3D-PCR17,18 was used to ask whether C-to-T transition mutations accumulate differentially at three genomic loci in A3Blow and A3Bhigh pools of HCC1569 cells. This technique enables qualitative estimates of genomic mutation within a population of cells because DNA sequences with higher A/T content amplify at lower denaturation temperatures than parental sequences. Lower temperature amplicons were observed for TP53 and c-MYC, but not CDKN2B (Fig. 2d & S10). These amplicons were cloned and sequenced and more C-to-T transition mutations were observed in A3Bhigh versus A3Blow samples (Fig. 2d & S10). TP53 and c-MYC appeared more mutable than CDKN2B suggesting that all genomic regions are not equally susceptible to enzymatic deamination. Other base substitution mutations were rare, and some C-to-T transitions were still evident in the A3Blow samples possibly due to residual deaminase activity and/or amplification of spontaneous events.
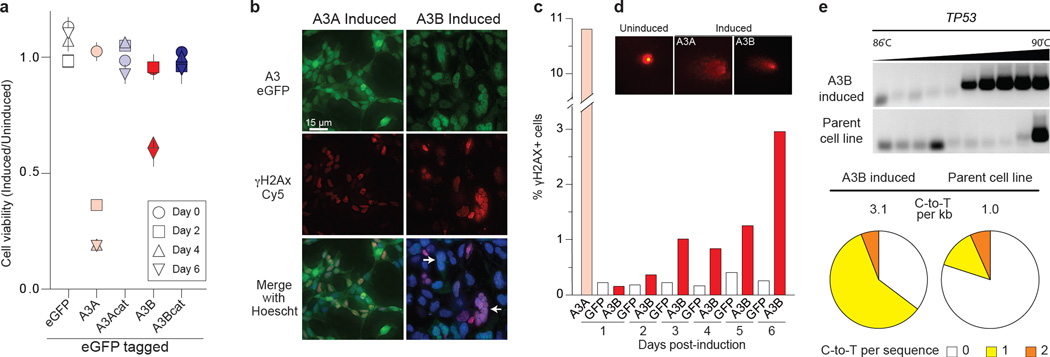
To ask whether A3B triggers additional cancer hallmarks19, we tried and failed to stably express A3B in several epithelial cell lines. We therefore constructed a panel of HEK293 clones with doxycycline (Dox)-inducible A3B, A3B-E68A-E255Q, A3A, or A3A-E72A eGFP fusions. As measured by flow cytometry, A3-eGFP levels were barely detectable without Dox and induced in nearly 100% of cells with Dox (Fig. S11). A3A over-expression caused rapid S-phase arrest, cytotoxicity, and γ-H2AX focus formation, as reported20 (Fig. 3a-c & S11). In comparison, A3B induction caused a delayed cell cycle arrest, a more pronounced formation of abnormal anucleate and multinucleate cells, and eventual cell death (Fig. 3a-b & S11). A3B induction also caused γ-H2AX focus formation, DNA fragmentation, as evidenced by visible comets, and C-to-T mutations (Fig. 3c-e). A3B catalytic activity, as evidenced by the glutamate mutants, was required for the induction of these cancer phenotypes.
Figure 3. Cancer phenotypes triggered by inducing A3B over-expression.
a, Cell viability at indicated times post-induction (mean and s.d. for n=3 per condition).
b & c, Representative fields of cells imaged for γ-H2AX and A3A-eGFP (1 day) or A3B-eGFP (3 days) post-induction, and γ-H2AX quantification. Abnormal, multinuclear clusters are typical of induced A3B-eGFP (white arrows).
d, Representative images of A3-induced DNA comets.
e, C-to-T mutations in TP53 detected by sequencing 3D-PCR products 4 days post-induction (n>12 sequences per condition).
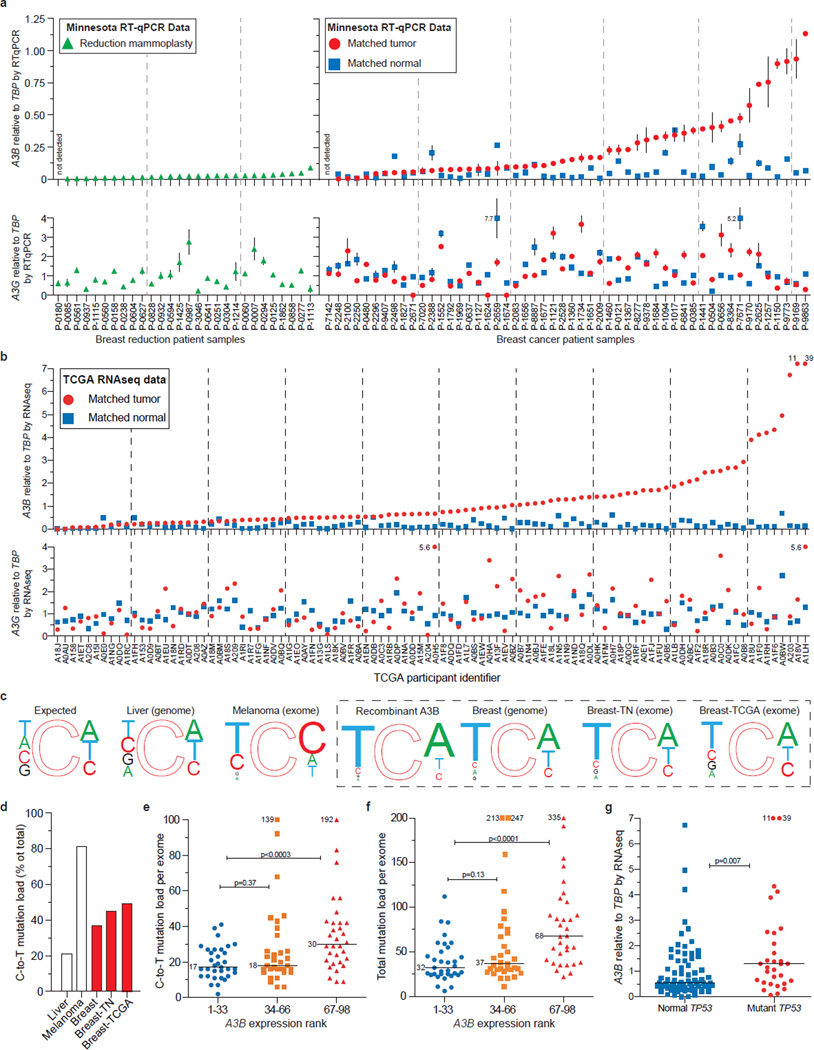
We next asked whether our cell-based results could be extended to primary tumors. First, we quantified mRNA levels for each of the 11 family members in 21 randomly chosen breast tumor specimens, in parallel with matched normal tissue procured simultaneously from an adjacent area or the contralateral breast. Only A3B was expressed preferentially in tumors (p=0.0003) (Fig. S12). We confirmed this analysis by measuring A3B levels in 31 additional tumor/normal matched tissue sets. In total, A3B was up-regulated by ≥3 s.d. in 20/52 tumors in comparison to the patient-matched normal tissue mean, and in 44/52 tumors in comparison to the reduction mammoplasty tissue mean (Fig. 4a, p=7.1×10−7 and p=2×10−5; patient information in Table S10). These are underestimates because tumor specimens have varying fractions of non-A3B expressing normal cells. Some of the matched ‘normal’ samples may also be contaminated by tumor cells, as judged by mean levels in mammoplasty samples (Fig. 4a; p=0.002). The related deaminase, A3G, was not expressed differentially in these samples, indicating that these observations are not due to immune cells known to express multiple APOBEC3s14 (p=0.591). Similar results were obtained by quantifying RNA sequencing data for independent matched tumor and normal pairs21, with ~50% showing up-regulated A3B (defined as tumors with A3B levels >3 s.d. above the mean of the normal matched samples; p<0.0001) (Fig. 4b).
Figure 4. A3B up-regulation and mutation in breast tumors.
a, A3B and A3G mRNA levels in the indicated tissues. Each symbol represents the mean mRNA level of three RTqPCR reactions presented relative to TBP (s.d. shown unless smaller than symbol; off-scale values are indicated numerically).
b, RNAseq data for A3B and A3G in the indicated samples (off-scale values are indicated numerically).
c, Local sequence contexts for all genomic cytosines (expected), cytosines deaminated by recombinant A3B (Fig. S13), and observed C-to-T transitions in the indicated cancers. Font size is proportional to each nucleotide frequency.
d, Percent C-to-T mutations in the indicated tumors.
e & f, C-to-T and total mutation counts for tumors in (b) grouped into lower, middle, and upper thirds based on A3B levels (medians are labeled; p values from Mann Whitney U test; off-scale values are indicated numerically).
g, Relationship between A3B level and TP53 status for tumors in (b) (p values from Mann Whitney U test; off-scale values are indicated numerically).
Finally, we assessed the impact of A3B on the breast tumor genome by correlating A3B’s deamination signature in vitro and the somatic mutation spectra accumulated during tumor development in vivo. Using a series of single-stranded DNA substrates varying only at the immediate 5’ or 3’ position relative to the _target cytosine, we found that recombinant A3B prefers TC>CC>GC=AC and CA>CT=CC (Fig. S13). These local sequence preferences were then compared to the expected distribution of cytosine in the human genome and the reported C-to-T mutation profiles for melanoma22, liver23, and breast8,9,21 tumors. Consistent with a spontaneous origin, the C-to-T frequency is low in liver tumors (~20%) and mutational events appear random (Fig. 4c-d). As expected, C-to-T frequencies are high in melanomas (~80%) and focused at di-pyrimidines consistent with UV-induced lesions and subsequent error-prone lesion bypass synthesis (Fig. 4c-d). Interestingly, the C-to-T frequency was intermediate in 3 independent breast tumor data sets (~40%) and largely focused at trinucleotides that mimic the preferred sites for A3B-dependent DNA deamination in vitro (Fig. 4c-d & S13). The availability of both RNAseq and somatic mutation data21 also enabled the establishment of strong positive correlations between A3B expression levels and C-to-T mutation load, overall base substitution mutation load, and TP53 inactivation (Fig. 4e-g). Importantly, tumors expressing high A3B have twice as many mutations (Fig. 4e-f & S14). This equates to 10 C-to-T and 30 total mutations per exome, or approximately 1000 and 3000 mutations per genome, attributable to A3B.
Taken together, we conclude that A3B is a significant mutational source in breast cancer accounting for C-to-T mutation biases and increased mutational loads. Moreover, the disproportional increase in overall base substitutions indicates that some of these other patterns may be due to further processing of U/G mispairs by ‘repair’ enzymes into transitions, transversions, and DNA breaks that could precipitate chromosomal rearrangements (model in Fig. S15 with similarities to AID-dependent antibody diversification mechanisms24). Future work is needed to understand A3B regulation and the potential interplay with other oncogenes and tumor suppressors. For example, although several common breast cancer markers do not correlate with A3B up-regulation, a mechanistic linkage between elevated A3B and inactivated TP53 is evident in primary tumor data and cell lines (Fig. 4g & S16). TP53 inactivation may be required to allow cells to bypass DNA damage checkpoints triggered by A3B.
This is the first study to demonstrate up-regulation of the DNA deaminase A3B in breast cancer and reveal it as a significant source of enzymatic mutation. Conceptually supportive of the original mutator hypothesis25, A3B-catalyzed genomic DNA deamination could provide genetic fuel for cancer development, metastasis, and even therapy resistance. We propose that A3B is a dominant underlying factor that contributes to tumor heterogeneity by broadly affecting multiple pathways and phenotypes. A3B may represent a new marker for breast cancer and a strong candidate for _targeted intervention, especially given its non-essential nature26. A3B inhibition may decrease the rate of tumor evolution and stabilize the _targets of existing therapeutics.
METHODS
RNA isolation, cDNA synthesis, and RTqPCR
Matched tumor/normal breast tumors and mammary reduction samples from the University of Minnesota Tissue Procurement Facility and breast cancer cell lines 30-4500K from the ATCC were used for RNA isolation, cDNA synthesis and qPCR as described14. Tissue RNA was from 100 mg flash-frozen tissue disrupted by a 2 h water bath sonication in 1 mL of Qiazol Lysis Reagent (RNeasy, Qiagen). Cell RNA was made using Qiashredder (RNeasy, Qiagen). qPCR was performed on a Roche Lightcycler 480 instrument. The housekeeping gene TBP was used for normalization. Statistical analyses for matched tissues were done using the Wilcoxson signed-rank test, and unmatched sets with the Mann-Whitney U-test (Graphpad Prism). Primer and probe sequences are listed in Table S11.
Knockdown constructs
A3B shRNA and shControl lentiviral constructs were from Open Biosystems (TRCN0000157469, TRCN0000140546, and scramble). Knockdown levels ranged from 80-95% by RTqPCR. Helper plasmids pdelta-NRF, containing HIV-1 gag, pol, rev, and tat genes, and pMDG, containing the VSV-G env gene, were co-transfected in HEK293T cells. Cell-free supernatants were harvested and concentrated by centrifugation (14,000 g×2 h). Stable transductants were selected with puromycin (1 µg/ml).
Cell fractionation and DNA deaminase activity assays
Cellular fractionation was performed as described by syringe treatment of 107 cells in 0.5 mL of hypotonic buffer28. Nuclei were lysed by sonication in lysis buffer (25 mM Hepes, pH7.4, 250 mM NaCl, 10% glycerol, 0.5% Triton X-100, 1 mM EDTA, 1 mM MgCl2, 1 mM ZnCl2). Anti-histone H3 (1:2000; Abcam) and anti-tubulin (1:10,000; Covance) followed by anti-mouse 800 or anti-rabbit 680 (1:5000; Licor) immunoblots were used to assess fractionation. Lysates were tested in a fluorescence-based deaminase activity assay17. Dilutions were incubated 2 h at 37°C with a DNA oligonucleotide 5’-(6-FAM)-AAA-TTC-TAA-TAG-ATA-ATG-TGA-(TAMRA). Fluorescence was measured on SynergyMx plate reader (BioTek). Local dinucleotide preferences in extracts were analyzed similarly using 5’-AC, CC, GC, or TC at the NN position of 5’-(6-FAM)-ATA-ANN-AAA-TAG-ATA-AT-(TAMRA).
Genomic uracil quantifications
Genomic DNA was prepared from shA3B of shControl cells transduced and cultured for 21 days. Samples were spiked with heavy (+6)-labeled uracil (C13 and N15; Cambridge Isotopes) and treated with UDG (NEB). Uracil was purified using 3,000 MWCO columns (Pall Scientific) and SPE (Carbograph, Grace). Samples were resuspended in water containing 0.1% formic acid. Analyses were performed on a capillary HPLC-ESI+-MS/MS (Thermo-Finnigan Ultra TSQ mass spectrometer, Waters nanoACQUITY HPLC). The MS was operated in positive ion mode, with 3.0 kV typical spray voltage, 250°C capillary temperature, 67 V tube lens offset, and nitrogen sheath gas (25 counts). Argon collision gas was used at 1.1 mTorr. MS/MS analyses were performed with a scan width of 0.4 m/z and scan time of 0.1 s. The Hypercarb HPLC column (0.5 mm x100 mm, 5 μm, Thermo Scientific) was maintained at 40°C and a flowrate of 15 μL/min. Solvents were 0.1% formic acid and acetonitrile. A linear gradient of 0% to 8% acetonitrile in 8 min was used, followed by an increase to 80% acetonitrile over 7 min. Uracils eluted at 11.5 min. Selected reaction monitoring was conducted with collision energy of 20V using the transitions: m/z 113.08 [M+H+]→70.08 [M-CONH]+ and m/z 96.08 [M-NH2]+ for uracil, while the internal standard ([15N-2, 13C-4]-uracil) was monitored by the transitions m/z 119.08 [M+H+]→m/z 74.08 [M-CONH]+ and m/z 101.08 [M-NH2]+ respectively. Internal standards were used for quantification.
TK fluctuations
TK-neo was introduced into MDA-MB-453 and HCC1569 cells as described17. TKplus cells were transduced with shA3B or shControl lentiviruses and subcloned by limiting dilution. 106 cells from each expanded subclone population were subjected to ganciclovir and incubated until colonies outgrew. Frequencies were determined by applying the method of the median29.
3D-PCR and sequencing
DNA was harvested from Ugi-expressing30 T-REx-293 clones or HCC1569 cells transduced with shA3B or shControl lentiviruses. 3D-PCR was done using Taq (Denville Scientific) as described17. Primers sequences available upon request. PCR products were analyzed by gel electrophoresis with ethidium bromide, PCR purified (Epoch), blunt-end cloned into pJET (Fermentas), sequenced with T7 primer (BMGC), and aligned and analyzed with Sequencher software (Gene Codes Corporation).
Cell cycle experiments
T-REx-293 cells (Invitrogen) were transfected with pcDNA5/TO A3-GFP using TransIT-LT1 (Mirus) followed by clone selection using hygromycin. Cells were induced with 1 µg/mL Dox (MP Biomedicals 198955) for the indicated times then trypsinized and fixed with 4% paraformaldehyde in PBS. Cell pellets were resuspended in 0.1% Triton X 100, 20 µg/mL propidium iodide and 40 µg/mL RNase A (Qiagen) in PBS for 30 min and the DNA content and GFP induction measured by flow cytometry (BD Biosciences FACS Canto II) and analyzed with FlowJo and GraphPad Prism.
Cell viability assays
Cells were plated into multiple 96 well plates (2500 cells/well) and measured at the days indicated. The MTS reagent and PMS reagents were used as directed (Promega, Celltiter Aq 96). Absorbance was measured at 490 nm (PerkinElmer 1420 Victor 3V). The results were normalized to untreated cells. For crystal violet staining wells of a six well plate were plated with 2×105 cells. Half of the wells were induced with 1 ug/mL Dox. A crystal violet (0.5%), methanol (49.5%), water (50%) solution was used to stain cells after seven days.
DNA damage experiments
Flow cytometric analysis of γ-H2AX foci was adapted31. Fixed cells were incubated overnight in 0.2% Triton-X 100, 1 % BSA in PBS (blocking buffer) with 1:100 rabbit anti-γ-H2AX (Bethyl A300-081A). Secondary incubation was with goat anti-rabbit TRITC (Jackson 111025144) for 3 hrs before flow cytometry (BD Biosciences FACS Canto II) and analysis (FloJo and GraphPad). For microscopy, HEK293 cells were induced with 1 ug/mL of Dox before fixation with 4% paraformaldehyde and incubation with 1:50 anti-γ-H2AX conjugated to Alexa 647 (Cell Signaling 20E3) in blocking buffer for 3 hours. The cells were stained with 0.1% Hoechst dye and imaged at 20× or 60× (Deltavision) and deconvolved (SoftWoRx, Applied Precision).
Comet Assays
As described32, microscope slides were coated with 1.5% agarose and dried. Low melting agarose (0.5% in PBS) was combined 1:1 with HEK293T cells transfected with A3A-eGFP (1 d) or A3B-eGFP (6 d). 10,000 cells were added to coated slides and the cells were lysed overnight in 10 mM Tris, 100 mM EDTA, 2.5 M NaCl, 1% Triton X-100. Slides were incubated for 10 min in running buffer (300 mM NaOH, 1 mM EDTA pH 13.1) then run at 0.75 V/cm 30 min. Gels were neutralized with 0.4 M Tris-Cl pH 7.5 and treated with RNase A (Qiagen). The microgels were allowed to dry and comets were visualized using propidium iodide.
Bioinformatic Analyses
Primary tumor genomic, exomic, or RNA sequencing data were obtained from public sources8,9,21–23. Liver tumor genomes had 654879, melanoma exomes had 2798, breast tumor genomes had 183916, breast tumor triple negative exomes had 6964, and TCGA breast tumor exomes had 5559 total single base substitution mutations. Local contexts were tabulated and presented as weblogo schematics. Complex mutational events and CpG motifs were excluded.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Hultquist and R. Vogel for statistics, T. Hwang for bioinformatic assistance, V. Polunovsky for hTERT-HMEC, V. Simon for a shRNA, S. Kaufmann, C. Lange, and D. Largaespada for consultation, and the Masonic Cancer Center Breast Cancer Research Fund for purchasing the ATCC breast cancer panel. Tissues were obtained from the Masonic Cancer Center Tissue Procurement Facility, which is part of BioNet, supported by the Academic Health Center and NIH grants P30 CA77598 (D. Yee), P50 CA101955 (D. Buchsbaum), and KL2 RR033182 (B. Blazar). M.B.B. was supported in part by a Cancer Biology Training Grant (NIH NCI T32 CA009138) and a Department of Defense Breast Cancer Research Program Predoctoral Fellowship (BC101124). L. Lackey was supported in part by an NSF Predoctoral Fellowship and by a position on the Institute for Molecular Virology Training Grant NIH T32 AI083196. A.M. Land was supported by a CIHR Postdoctoral Fellowship. Computational analyses (N.A.T, D.E.D) were supported by federal funds from the National Cancer Institute, National Institutes of Health, CBIIT/caBIG ISRCE yellow task #09-260. Harris laboratory operational support was provided in part by the Children’s Cancer Research Fund (Minneapolis, MN) and a seed grant from the University of Minnesota Clinical and Translational Science Institute (supported by NIH 1UL1RR033183).
Footnotes
AUTHOR CONTRIBUTIONS
R.S.H. conceived and managed the overall project. M.B.B. assisted R.S.H. with experimental design, project management, and manuscript preparation. M.B.B., E.W.R., and B.L. generated mRNA expression profiles, L.L. and E.L. performed microscopy, L.L. and A.R. performed biochemical fractionations and DNA deaminase assays, M.B.B. performed uracil quantifications, A.M.L. performed TK fluctuations, A.R. generated 3D-PCR sequences, and L.L., A.L., A.R., and M.A.C. determined the impact of induced A3B over-expression. M.A.C. performed deaminase assays with recombinant protein, and M.A.C., D.K., and N.T. assisted with UPLC-MS set-up. J.B.N. conducted the search and performed the bioinformatic analysis of the microarray data and developed the normalization algorithm for this analysis. N.A.T., D.E.D., and M.B.B. contributed bioinformatic analyses. All authors contributed to manuscript revisions.
REFERENCES
- 1.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 4.Kumar A, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087–17092. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrlich M, Norris KF, Wang RY, Kuo KC, Gehrke CW. DNA cytosine methylation and heat-induced deamination. Biosci Rep. 1986;6:387–393. doi: 10.1007/BF01116426. [DOI] [PubMed] [Google Scholar]
- 11.Pavri R, Nussenzweig MC. AID _targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka S, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 14.Refsland EW, et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lackey L, et al. APOBEC3B and AID have similar nuclear import mechanisms. J Mol Biol. 2012;419:301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suspène R, et al. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A. 2011;108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. International Cancer Genome Consortium Data Portal—a one-stop shop for cancer genomics data. Database. 2011;2011 doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 25.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 26.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter MA, et al. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shlyakhtenko LS, et al. Atomic force microscopy studies provide direct evidence for dimerization of the HIV restriction factor APOBEC3G. J Biol Chem. 2011;286:3387–3395. doi: 10.1074/jbc.M110.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. Journal of Genetics. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 30.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, Darzynkiewicz Z. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol Biol. 2006;314:73–80. doi: 10.1385/1-59259-973-7:073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fairbairn DW, Olive PL, O'Neill KL. The comet assay: a comprehensive review. Mutat Res. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.