Figure 5.
Global DNA Methylation Profiles of TSCs Grown in TX Compared to Standard TS Medium
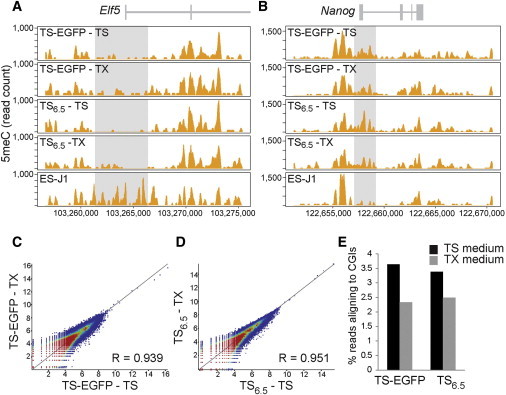
(A and B) DNA methylation patterns determined by meDIP-Seq at the Elf5 locus, which is hypomethylated in TSCs but methylated in ESCs, and Nanog, which exhibits the opposite methylation profile. The distribution and enrichment of methylation at these key transcription factors remains unchanged in TX medium.
(C and D) Scatter plots displaying Pearson’s correlation of global methylation between cells grown in TS versus TX medium. Each point corresponds to a 5 kb in silico probe spaced 20 kb apart. Values were normalized for total read count and converted to a Log2 scale. R values very close to one indicate that the methylation profiles of TSCs grown in TX and TS media are extremely similar to each other on a global scale.
(E) CpG islands (CGIs) displayed the most striking methylation differences between the two culture conditions with less reads mapping to CGIs in TX medium.
See also Figure S5.
