Abstract
One of the most fundamental questions in the control of gene expression in mammals is how epigenetic methylation patterns of DNA and histones are established, erased, and recognized. This central process in controlling gene expression includes coordinated covalent modifications of DNA and its associated histones. This article focuses on structural aspects of enzymatic activities of histone (arginine and lysine) methylation and demethylation and functional links between the methylation status of the DNA and histones. An interconnected network of methyltransferases, demethylases, and accessory proteins is responsible for changing or maintaining the modification status of specific regions of chromatin.
DNA and histone methylation are mechanistically linked. For example, there is an inverse correlation between DNA methylation and histone H3K4 methylation, and Dnmt3L induces DNA methylation if H3K4 is unmethylated.
Overview
All cells face the problem of controlling the amounts and timing of expression of their various genes. In some cases, this control involves relatively long-term and heritable modifications to the chromatin, albeit nonpermanent. Such modifications that do not change the DNA sequence are referred to as “epigenetic.” The resulting epigenetic effects maintain the various patterns of gene expression in different cell types. Epigenetic modifications include DNA methylation and histone posttranslational modifications (PTMs).
Nucleosomes consist of ∼146 bp of DNA wrapped approximately 1.8 times around a histone octamer and are evolutionarily conserved across all eukaryotes. The combinatorial pattern of DNA and histone modifications constitutes an epigenetic code that shapes transcriptional patterns and genomic stability. The coding modification is “written” by sequence- and site-specific enzymes, and “interpreted” or “read” by effector (or reader) molecules that mediate the assembly of higher-order chromatin structures, involving remodeling complexes, histone variants, and noncoding RNAs (see articles by Becker and Workman 2013; Allis et al. 2014; Henikoff and Smith 2014). Representative enzymes responsible for histone methylation (SET domain proteins, Dot1, and protein arginine methyltransferases), histone lysine demethylation (LSD1 and Jumonji proteins), and DNA methylation (Dnmt1 and Dnmt3) are discussed in this article. DNA methylation is an epigenetic modification that has long been known to repress transcription. Histone methylation, depending on the histone and residue of modification, contributes to either active or repressive chromatin configurations. The functional implications of the structural determination of enzymes in linking histone modifications to that of DNA in mammalian cells are also discussed. This article is complemented by subsequent articles discussing the structure and function of proteins that read DNA and histone methylation (Patel 2014), and previous articles discussing histone acetylation writers (HATs), histone deacetylation (HDAC) “eraser” enzymes, and readers of these epigenetic marks (Marmorstein and Zhou 2014; Seto and Yoshida 2014).
1. Histone methylation and demethylation
1.1. Histone Lysine (K) Methyltransferases (HKMTs)
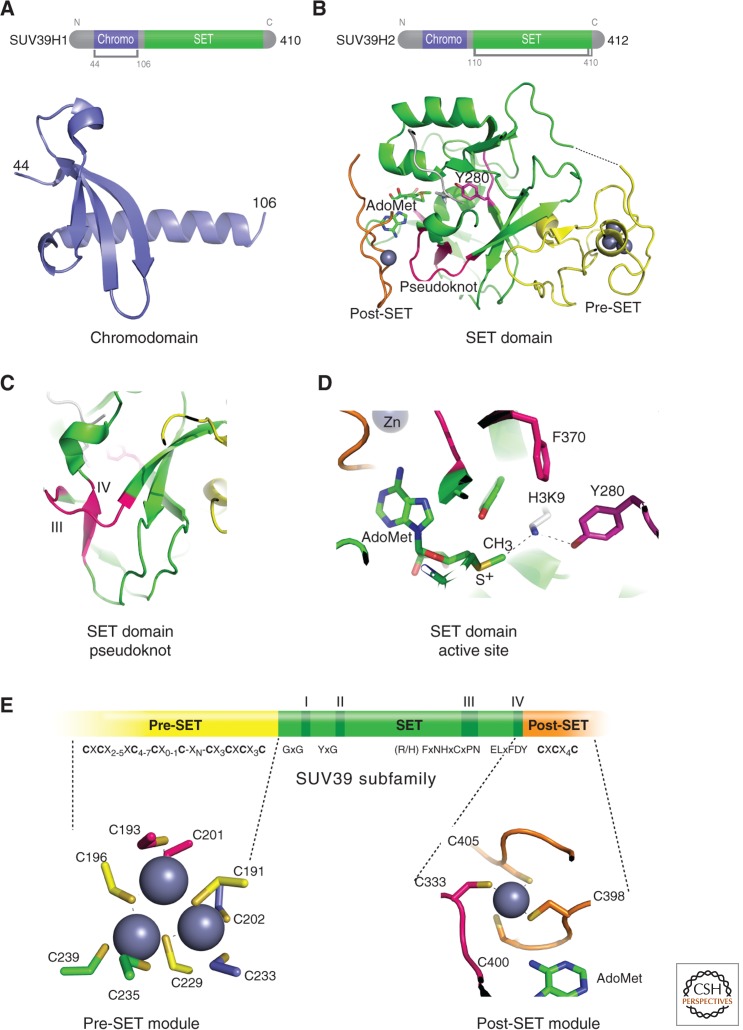
All known HKMTs (referred to in some of the literature as PKMTs or KMTs, as many of the substrates are nonhistone proteins) contain an evolutionarily conserved SET domain comprised of 130 amino acids (reviewed in Cheng et al. 2005), except Dot1 (see Sec. 1.2). The SET domain was first identified as a shared sequence motif in three Drosophila proteins: suppressor of variegation (Su(var)3-9), enhancer of zeste (E(z)), and homeobox gene regulator trithorax (Trx; Jenuwein et al. 1998). Mammalian homologs of the Drosophila Su(var)3-9 protein, SUV39H1 in humans, and Suv39h1 in mouse were the first characterized HKMTs involved in H3K9 methylation (Fig. 1A) (Rea et al. 2000). Since then, more than 50 SET domain-containing proteins have been identified in humans, with a proven or predicted enzymatic role in performing lysine methylation on histone tails (reviewed in Volkel and Angrand 2007).
Figure 1.
Structural features of SUV39H1 and SUV39H2. (A) Ribbon diagram of amino-terminal chromodomain of SUV39H1 (PDB 3MTS). (B) Carboxyl-terminal SET domain structure of SUV39H2 (PDB 2R3A). (C) Formation of the pseudoknot by motifs III and IV. (D) Formation of the active site showing the methyl donor (S-adenosyl-l-methionine [AdoMet]), _target H3K9 lysine, catalytic Y280 residue, and F370 Phe/Tyr Switch (Collins et al. 2005). AdoMet-dependent methyltransferases (including HKMTs) share a reaction mechanism in which the nucleophile acceptor (NH2) attacks the electrophilic carbon of AdoMet in an SN2 displacement reaction. (E) SUV39H1 and H2 have a pre-SET segment containing nine invariant cysteines, the SET region containing four signature motifs, and the post-SET region containing three invariant cysteines. An enlargement of the pre-SET Zn3Cys9 triangular zinc cluster structure is illustrated on the bottom left and the post-SET zinc center on the bottom right panel.
The majority of the SET-containing HKMTs contain at least one additional protein module in their protein sequence (e.g., a chromodomain in SUV39H1; Fig. 1A). SET-containing HKMTs are grouped into six different subfamilies based on sequence homology within and around the catalytic SET domain, homology with other protein modules, and their structures. The six subfamilies include SET1, SET2, SUV39, EZH, SMYD, and PRDM (Volkel and Angrand 2007). A number of SET-containing HKMTs, however, do not fall into the above six subfamilies because of lacking sequences or conservation flanking their SET domains. Examples of such proteins include Set8 (also known as PR-Set7; Couture et al. 2005; Xiao et al. 2005), which monomethylates H4K20 (H4K20me1), and SUV4-20H1 and SUV4-20H2, which di- and trimethylate H4K20 (H4K20me2 and me3). Set7/9 can monomethylate H3K4 (H3K4me1; Xiao et al. 2003) and many other nonhistone substrates, whereas SetD6 monomethylates only the nonhistone substrate, RelA, a subunit of NF-κB (Levy et al. 2011).
Structures of many SET domains from different subfamilies have been solved in various combinations, including bound to peptide substrates and methyl donor (S-adenosyl-l-methionine, also known as AdoMet or SAM) or reaction product (S-adenosyl-l-homocysteine, also known as AdoHcy or SAH). The SET domain adopts a unique structure formed by a series of β-strands folded into three sheets surrounding a knot-like structure (Fig. 1B). The knot-like structure (or pseudoknot) is formed by the carboxyl-terminal segment of the SET domain, which passes through a loop formed by the preceding stretch of sequences. Formation of this pseudoknot structure brings the two conserved SET domain sequence motifs, III and IV (Fig. 1C), in close proximity to the AdoMet-binding region and peptide-binding channel (Fig. 1D).
Available crystal structures of the SUV39 HKMT subfamily, which methylates H3K9 (DIM-5, Clr4, GLP/EHMT1, G9a/EHMT2, and SUV39H2), show the presence of two closely packed cysteine-rich modules in both the pre-SET and post-SET (before and after the SET) domains (Fig. 1E). These two modules are important for maintaining structural stability (pre-SET) and forming part of the active site lysine channel (post-SET) (Zhang et al. 2002; Zhang et al. 2003). The pre-SET module contains nine conserved cysteines, which coordinate three Zn2+ atoms in a triangular geometry. The post-SET module contains three conserved cysteines, which along with a cysteine from the conserved motif III sequence, (R/H)F(I/V)NHxCxPN, tetrahedrally coordinate the fourth Zn2+ atom near the active site. Binding of the fourth Zn2+ at the active site is essential for the activity of the SUV39 subfamily (Zhang et al. 2003).
1.2. Dot1p: Non-SET Domain HKMT
Histone H3 Lys-79 (H3K79) is methylated by Dot1p (reviewed in Frederiks et al. 2011), a protein originally identified as a disruptor of telomeric silencing in Saccharomyces cerevisiae (Singer et al. 1998). Methylation of H3K79 in S. cerevisiae is important for the proper localization of the silent information regulator complex and DNA damage signaling (see Grunstein and Gasser 2013).
Dot1p is a Class-I methyltransferase, as suggested by the presence of the AdoMet-binding sequence motifs (Dlakic 2001; Schubert et al. 2003), similar to those found in protein arginine methyltransferases and DNA methyltransferases (see Secs. 1.3 and 2). Class-I methyltransferases such as Dot1p are distinct from most other HKMTs because they do not contain the SET domain. Thus, they have an entirely different structural scaffolding and unrelated local active-site spatial arrangement that catalyzes AdoMet-dependent methyl transfer to a protein lysine side chain.
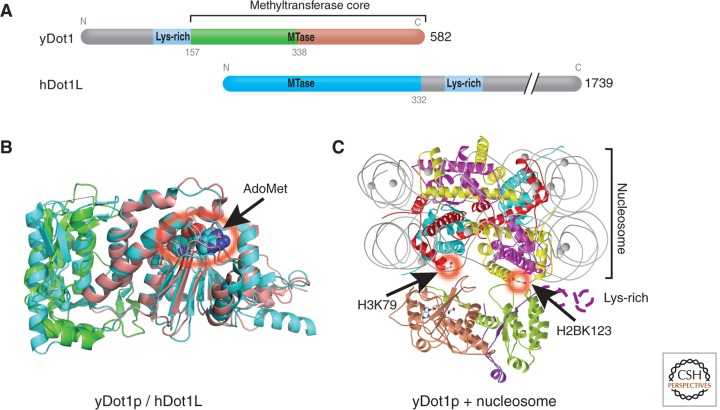
Yeast Dot1p contains a core region (indicated in Fig. 2A) conserved between Dot1p homologs in human, Caenorhabditis elegans, Drosophila, and the mosquito, Anopheles gambiae. The length of these Dot1 proteins varies from 582 amino acids in yeast to 2237 amino acids in Drosophila. The conserved Dot1p core is located at the carboxyl terminus in yeast, but is at the amino terminus in human, C. elegans, Drosophila, and Anopheles gambiae Dot1p homologs. The Dot1p conserved core contains an amino-terminal helical domain and a seven-stranded catalytic domain that harbors the binding site for the methyl donor and an active-site pocket sided with conserved hydrophobic residues (Fig. 2B).
Figure 2.
Dot1p family (non-SET HKMTs). (A) Schematic representation of Dot1 homologs from yeast and human, indicating the conserved methyltransferase core regions. (B) Superimposition of the conserved core regions of yeast Dot1p (residues 176-567; PDB 1U2Z), colored in green and brown, and human Dot1L (residues 5-332; PDB 1NW3), colored in cyan. The amino-terminal helical domains are shown on the left side of the panel in green for yDot1p or cyan for hDot1L. The carboxyl-terminal catalytic domain is shown on the right side of the panel with the bound methyl donor, AdoMet, as spheres (circled in red with an arrow). (C) A model of yDot1p docked with a nucleosome, adapted from Sawada et al. (2004). The structure of the nucleosome core particle is shown as ribbons (red, H3; green, H4; magenta, H2A; yellow, H2B; gray lines, DNA). The model was put together by aligning the _target H3K79, located on the nucleosome disk surface, with the active site pocket of Dot1p.
Dot1p has several unique biochemical properties. Yeast Dot1p and its human homolog Dot1L methylate only nucleosomal substrates, but not free histone H3 protein (reviewed in Frederiks et al. 2011). A stretch of positively charged residues (i.e., Lys-rich) at the carboxyl-terminal end of the human Dot1L core or the amino-terminal end of the yeast Dot1p core were critical for nucleosome binding (Fig. 2C) and therefore for enzymatic activity (Min et al. 2003; Sawada et al. 2004; Oh et al. 2010). Given in S. cerevisiae, H3K79 methylation requires ubiquitination of H2B K123 in vivo (Briggs et al. 2002), and both histone residues are located on the same nucleosome disk surface ∼30 Å apart, Dot1p may interact specifically with nucleosomes containing ubiquitinated H2B (Fig. 2C) (Oh et al. 2010). Such an interaction could be significant in vivo because Dot1p could be recruited to specific high-order chromatin in which ubiquitinated histone H2B might serve as a spacer between adjacent nucleosome disk surfaces (Sun and Allis 2002), allowing Dot1p access to its _target H3K79 lysine (see also Fig. 10 in Allis et al. 2014).
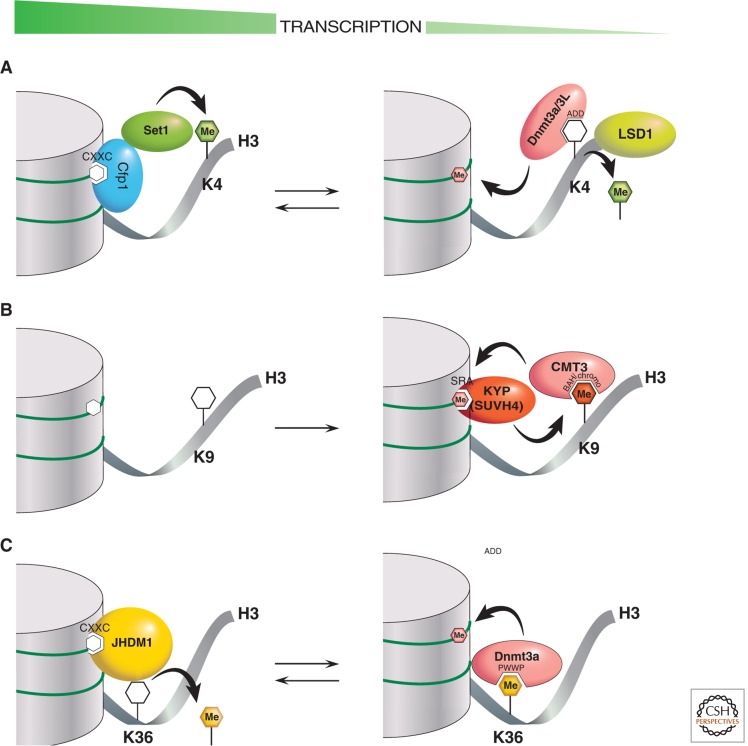
Figure 10.
Cartoons of interactions that regulate DNA methylation and associated histone H3 modifications. There are three examples of PTM cross talk between histone and DNA methylation. Chromatin on the left represents transcriptionally active states, whereas chromatin on the right represents transcriptionally repressive states. The “Me”-labeled filled hexagons indicate one or more methyl groups in DNA (pink) or protein lysine residues (K). The catalytic action of methylation writers and erasers are indicated by curved black arrows. Methylation readers interact via specific labeled domains that fit in a lock-and-key fashion to methylated (filled hexagons) or unmethylated CpGs or lysines (unfilled hexagons). (A) Enzymatic reactions by the Dnmt3a-Dnmt3L complex and the SET1/CFP1 complex regulate the inverse correlation of DNA methylation and H3K4 methylation. (B) In plants, enzymatic reactions by CMT3 and KYP reinforce the correlation between DNA CHG methylation and H3K9 methylation. (C) Enzymatic reactions by Dnmt3a and JHDM1 positively regulate the association of DNA methylation with H3K36 methylation.
Mis_targeted hDOT1L function has been implicated in the leukemogenesis of mixed lineage leukemia with the MLL-AF10 fusion (Okada et al. 2005). It does this by interacting with the AF10 protein and up-regulating genes such as Hox9a. Recently, the in vivo inhibition of hDot1L led to the increased survival of mice that had a xenograft model of mixed lineage leukemia (Daigle et al. 2011). This is, notably, the first example of selective inhibition of an HKMT that has efficacy in a cancer model.
1.3. Protein Arginine Methylation
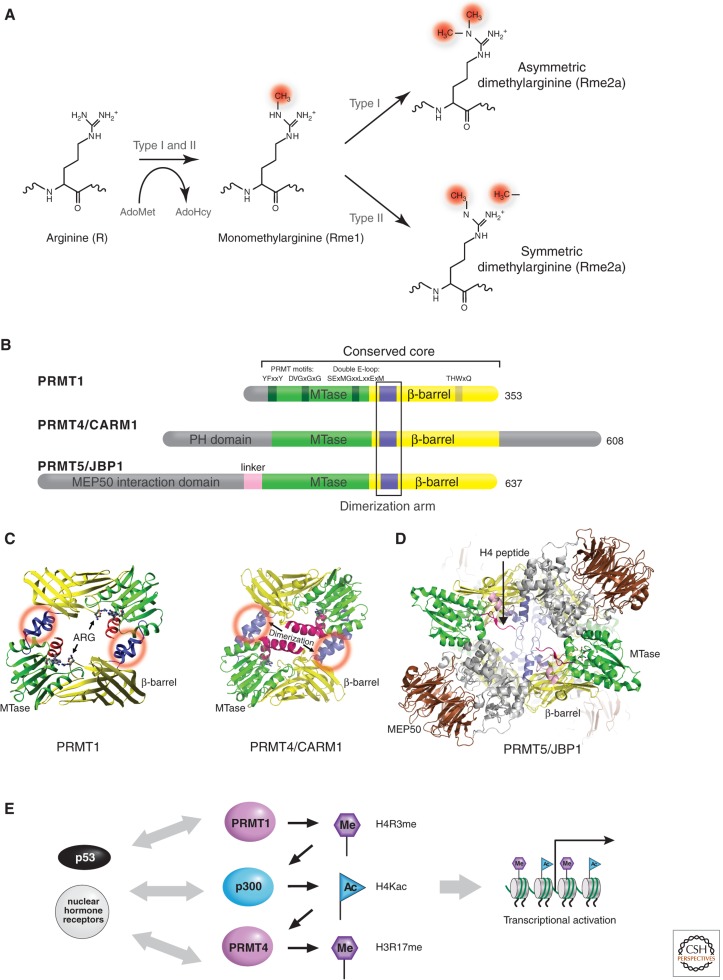
Protein arginine methylation is a common posttranslational modification in eukaryotes. There are two major types of protein arginine (R) methyltransferases (PRMTs) that transfer the methyl group from AdoMet to the guanidino group of arginines in protein substrates (Lee et al. 1977), called type I and type II PRMTs (Fig. 3A). Both catalyze the formation of monomethylarginine (Rme1) as an intermediate, but type I PRMTs also form asymmetric dimethylarginine (Rme2a), whereas type II PRMTs form symmetric dimethylarginine (Rme2s). Among the nine canonical members of PRMT family (Herrmann et al. 2009; Table 1), only PRMT5 (also known as JBP1 for Jak-binding protein 1; Branscombe et al. 2001), and possibly PRMT7 and PRMT9 are type II PRMTs (Lee et al. 2005; Cook et al. 2006); they symmetrically dimethylate-specific arginines not only on histones, but also other proteins such as myelin basic protein (Kim et al. 1997), spliceosomal Sm proteins (Friesen et al. 2001), and Piwi proteins (Vagin et al. 2009). Highly relevant to the focus of this article, PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, the de novo DNA methyltransferase, coupling histone arginine, and DNA methylation in gene silencing (Zhao et al. 2009).
Figure 3.
PRMT family. (A) Reactions catalyzed by the two major types of protein arginine methylation. (B) Representatives of PRMT members (type I: PRMT1 and PRMT4; type II: PRMT5). The conserved methyltransferase (MTase) domain is in green and the unique β-barrel domain in yellow. (C) Dimeric structures of PRMT1 (PDB 1OR8) and PRMT4 (PDB 3B3F). Dimerization arms are indicated by red circles. (D) Type II PRMT5-MEP50 tetramer complex (DPB 4GQB), formed by the stacking of two dimers. The second dimer is faded in the background. MEP50 (colored in brown) interacts with the amino-terminal domain (gray) of PRMT5. Bound H4 peptide is colored in red. (E) An example of three coactivators acting synergistically for p53-mediated transcription.
Table 1.
Members of human PRMT family
| Enzyme | Type | Activity | Chromosome | EST | Coding exon | Genomic size (kb) | Protein accession number | Protein size (residues) |
|---|---|---|---|---|---|---|---|---|
| PRMT1 | I | +++ | 19q13 | +++ | 9–10 | 10 | CAA71764 | 361 |
| PRMT2 | I | - | 21q22 | ++ | 10 | 30 | P55345 | 433 |
| PRMT3 | I | + | 11p15 | + | 13 | 50 | AAH64831 | 531 |
| PRMT4(CARM1) | I | + | 19p13 | ++ | 16 | 50 | NP_954592 | 608 |
| PRMT5(JBP1) | II | + | 14q11 | ++ | 17 | 8.5 | AAF04502 | 637 |
| PRMT6 | I | + | 1p13 | +/– | 1 | 2.5 | AAK85733 | 375 |
| PRMT7 | II? | + | 16q22 | ++ | 17 | 41 | Q9NVM4 | 692 |
| PRMT8 | I | ? | 12p13 | + | 9 | 52 | AAF91390 | 334 |
| PRMT9 | II? | ? | 4q31 | + | 10 | 40 | AAH64403 | 845 |
The PRMT proteins vary in length from 353 amino acids for PRMT1 to 637 amino acids for PRMT5/JBP1, but they all contain a conserved core region of approximately 310 amino acids (Fig. 3B). The sequences beyond the conserved PRMT core region are all amino-terminal additions; however, PRMT4 also has a carboxyl-terminal addition. The size of the amino-terminal additions varies between ∼30 amino acids in PRMT1 to ∼300 amino acids in PRMT5. The variation in amino termini allows each PRMT to be subject to a different mode of regulation. An interesting feature of PRMT7 and PRMT9 is that they seem to have arisen from a gene duplication event and contain two conserved core regions, each with a putative AdoMet-binding motif (Miranda et al. 2004; Cook et al. 2006).
Several crystal structures of type I PRMTs are currently available (Cheng et al. 2005; Troffer-Charlier et al. 2007; Yue et al. 2007). These structures reflect a striking structural conservation of the PRMT catalytic core (Fig. 3B). The overall monomeric structure of the PRMT core can be divided into three parts: a methyltransferase domain, a β-barrel, and a dimerization arm. The methyltransferase domain has a consensus fold conserved in Class-I AdoMet-dependent methyltransferases (like that of Dot1p) that harbor an AdoMet-binding site (Schubert et al. 2003). The β-barrel domain is unique to the PRMT family (Zhang et al. 2000). Dimer formation is a conserved feature in the type I PRMT family, validated by crystal structure studies, an example of which is illustrated in Figure 3C (Cheng et al. 2005). Dimerization may be required to correctly engage the residues in the AdoMet-binding site, so that they bind AdoMet and/or generate dimethylated (Rme2) products processively, akin to the spread of H3K9 trimethylation by DIM-5 (Zhang et al. 2003). Indeed, phosphorylation of PRMT4 at a conserved serine residue in the dimer interface results in inefficient AdoMet binding, and reduced histone methylation activity (Higashimoto et al. 2007). Similarly, an allosteric inhibitor that binds in the dimer interface of PRMT3 results in reduced binding of AdoMet and methylation activity (Siarheyeva et al. 2012).
The type II enzyme, PRMT5, functions as part of various high molecular weight protein complexes that also contain the WD repeat protein MEP50 (methylosome protein 50) (Friesen et al. 2002). The structure of the human PRMT5-MEP50 complex revealed that PRMT5 was organized into a tetramer through the stacking of two primary dimers and the amino-terminal domain of PRMT5 interacting with the seven-bladed β-propeller MEP50 (Fig. 3D) (Antonysamy et al. 2012).
Two well-studied enzymes, PRMT1 and PRMT4, methylate histones H2B, H3, and H4 (reviewed in Bedford and Clarke 2009), in addition to many nonhistone substrates including the carboxyl-terminal domain of RNA polymerase II (Sims et al. 2011). Histone arginine methylation is a component of the “histone code” that directs a variety of processes involving chromatin. For example, methylation of H4R3 by PRMT1 facilitates H4 acetylation and enhances transcriptional activation by nuclear hormone receptors. It acts synergistically with PRMT4 because PRMT4 prefers acetylated histone tails to generate methylated H3R17 (Wang et al. 2001; Daujat et al. 2002). Similarly, the synergistic action in vitro of PRMT1, PRMT4, and p300 for p53-mediated transcription is the greatest when all three coactivators are present, whether added sequentially or at the same time (Fig. 3E) (An et al. 2004). Even preincubating a chromatin template with p53 and PRMT1 significantly stimulated the histone acetyltransferase activity of p300, as did preincubation of the template with p53 and p300 to stimulate H3 arginine methylation by PRMT4. Indeed, PRMT4 was initially discovered as a transcriptional coactivator-associated arginine (R) methyltransferase 1 (CARM1; Chen et al. 1999). PRMT4/CARM1 also acts synergistically with the p160 coactivator to stimulate gene activation by nuclear receptors (Chen et al. 1999; Lee et al. 2002). These results provide compelling evidence that histones are relevant _targets for PRMT4/CARM1, PRMT1, and p300, and that the resulting histone modifications are directly important for transcription.
1.4. Lysine Demethylation by Oxidation: LSD1
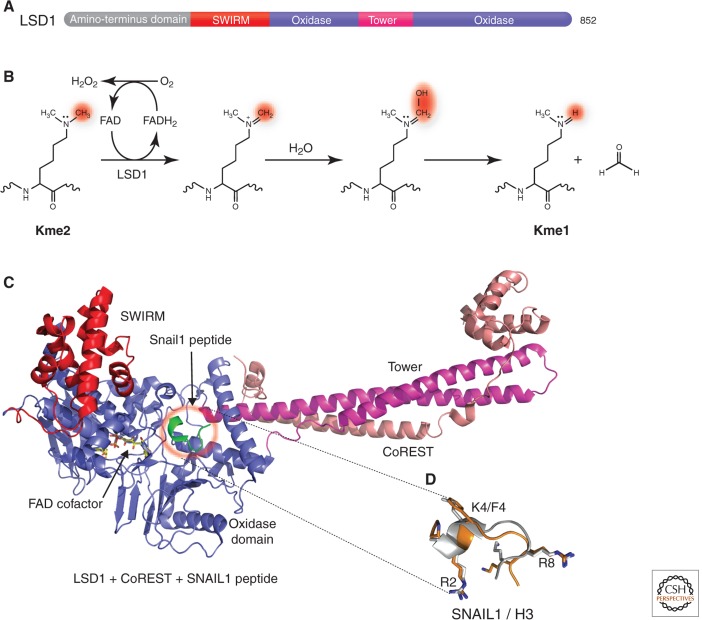
To the surprise of the research community, protein lysine methylation was finally shown to be a reversible posttranslational modification in 2004 (described in Shi and Tsukada 2013). Before this, Bannister et al. had hypothesized that methyl groups from both lysine and arginine side chains could be oxidatively removed using a FAD (flavin adenine dinucleotide) cofactor as the electron acceptor (Bannister et al. 2002). Then came the discovery of the lysine-specific demethylase 1 (LSD1) protein (Fig. 4A) (Shi et al. 2004). LSD1 is a flavin-dependent amine oxidase that demethylates H3K4me2/me1 (Shi et al. 2004), H3K9me2/me1 in an androgen receptor-mediated pathway (Metzger et al. 2005), and even p53, a nonhistone protein (Huang et al. 2007). The closely related LSD2 demethylates H3K4me2/me1 (Karytinos et al. 2009). Both LSD1 and LSD2 demethylate methyl lysines by forming an imine intermediate that undergoes hydrolysis in aqueous buffer to complete the demethylation process (Fig. 4B). LSD1 and LSD2, however, cannot demethylate trimethylated lysines because of the mechanistic requirement for a protonated amine in this demethylation pathway.
Figure 4.
Histone demethylation by oxidation. (A) Schematic representation of human LSD1 domain organization: the amino-terminal putative nuclear localization signal, followed by a SWIRM (Swi3p, Rsc8p, and Moira) domain, and the catalytic oxidase domain. The oxidase domain contains an atypical insertion of the Tower domain not found in other oxidases. (B) Scheme of the demethylation reaction catalyzed by LSD1. (C) Crystal structure of LSD1-CoREST in complex with the SNAIL1 peptide (PDB 2Y48). LSD1 includes residues 171–836 in red, blue, and magenta. CoREST shows residues 308–440 in orange. The SNAIL1 peptide is in green, and the FAD cofactor is shown as a yellow ball-and-stick. (D) Superposition between SNAIL1 (orange) and histone H3 (gray) peptides. (Adapted from Baron et al. 2011).
Thus far, crystal structures of LSD1 in various configurations have been determined (Fig. 4C) (reviewed in Hou and Yu 2010). In one study, the first 16 residues of histone H3 was observed in a complex structure with LSD1-CoREST (Forneris et al. 2007), in perfect agreement with biochemical data that LSD1 is active on peptide substrates longer than 16 amino acids (Forneris et al. 2005). Interestingly, the amino-terminal extremity of the transcription factor SNAIL1 has sequence similarity with the amino-terminal tail of histone H3 (Lin et al. 2010), and binds to the LSD1 catalytic site in the same way as a histone H3 peptide substrate (Fig. 4D) (Baron et al. 2011). Specifically, the binding positions of the amino-terminal Arg2, Phe4, and Arg7 residues of the SNAIL1 peptide correspond to the amino-terminal Arg2, Lys4, and Arg8 residues of histone H3. Thus, the Snail1-LSD1-CoREST complex effectively inhibits LSD1 enzymatic activity (Baron et al. 2011), and is found in certain cancer cells. The fact that LSD1 recognizes the first amino-terminal amino group (a conserved positive charge), customizing the orientation of the fourth amino acid side chain (i.e., H3K4me2/me1) to point toward and be in direct contact with the flavin ring of the cofactor, raises the question of how LSD1 demethylates methyllysines further away from the amino terminus (e.g., H3K9me2/me1), in an androgen receptor-dependent manner (Metzger et al. 2005).
1.5. Lysine Demethylation by Hydroxylation: Jumonji-Containing Demethylases
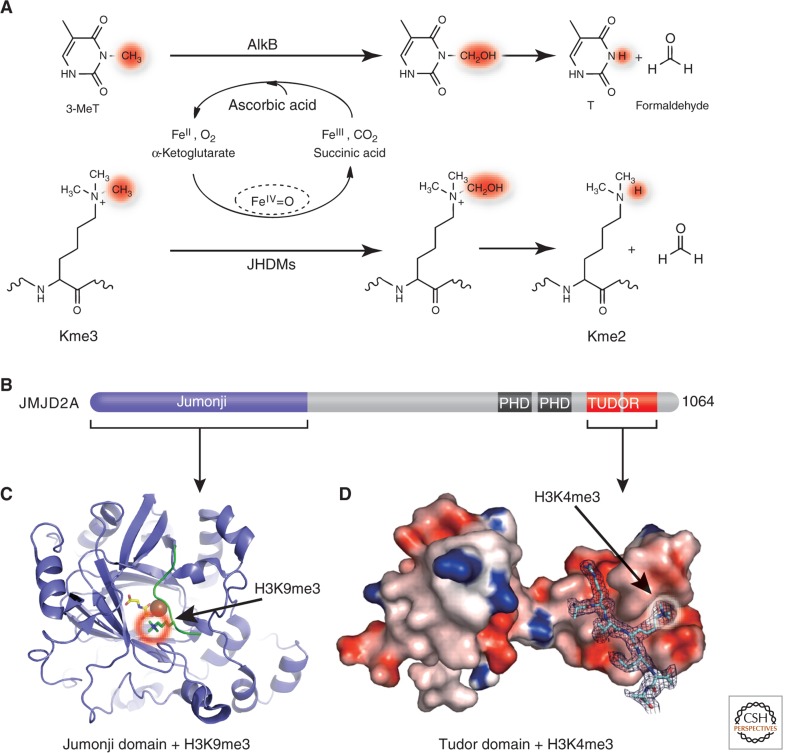
In search of enzymes capable of reversing methylated lysines, Trewick et al. hypothesized that Jumonji domain-containing Fe(II)- and α-ketoglutarate-dependent dioxygenases can reverse lysine methylation via a similar mechanism as the bacterial AlkB family of DNA repair enzymes (Fig. 5A) (Trewick et al. 2005). This hypothesis was quickly verified by the discovery of JHDM1 as the Jumonji domain-containing histone demethylase 1, using a biochemical assay based on the detection of formaldehyde, one of the predicted reaction products (Tsukada et al. 2006; described in Shi and Tsukada 2013). Jumonji-containing proteins are members of the cupin super family (Clissold and Ponting 2001), including the Tet proteins involved in conversion of 5-methylcytosine to 5-hydroxymethylcytosine (Kriaucionis and Tahiliani 2014; Li and Zhang 2014). Demethylation reactions catalyzed by Jumonji enzymes follow a hydroxylation pathway involving a reactive Fe(IV) intermediate. As they do not require a lone pair of electrons on the _target nitrogen atom, thereby they can demethylate mono-, di-, and trimethylated lysines (Fig. 5A) (Hoffart et al. 2006; Ozer and Bruick 2007).
Figure 5.
Demethylation by hydroxylation. (A) Mechanisms of demethylation of 3-methylthymine by AlkB (top) and of methyllysine by Jumonji-domain proteins (bottom). (B) Schematic representation of JMJD2A domain organization. (C) Structure of the amino-terminal Jumonji (ribbons) in complex with H3K9me3 (PDB 2OX0). (D) Structure of the carboxyl-terminal double Tudor domain (surface representation) in complex with H3K4me3 (PDB 2GFA). (Adapted, with permission, from Huang et al. 2006.)
JMJD2A contains carboxyl-terminal PHD and Tudor domains, which typically act as methyl-binding proteins, called readers, in addition to the amino-terminal Jumonji domain (Fig. 5B). The JMJD2A Jumonji domain alone is capable of demethylating H3K9me3/me2 and H3K36me3/me2. Structural studies revealed that the JMJD2A Jumonji domain predominantly recognizes the backbone of the histone peptides (unusual for a sequence-specific enzyme), which allows the enzyme to demethylate both H3K9 (Fig. 5C) and H3K36 (reviewed in Hou and Yu 2010; McDonough et al. 2010). The Tudor domain binds both H3K4me3 (Fig. 5D) and H4K20me3 (Huang et al. 2006; Lee et al. 2008). The functional connection between the methyl mark reader and eraser in JMJD2A, however, is currently not clear.
2. DNA methylation
In mammals and other vertebrates, DNA methylation occurs at the C5 position of cytosine, generating 5-methylcytosine (5mC), mostly within CpG dinucleotides. This methylation, together with histone modifications, plays an important role in modulating chromatin structure, thus controlling gene expression and many other chromatin-dependent processes (Cheng and Blumenthal 2010; reviewed in Li and Zhang 2014). The resulting epigenetic effects maintain the various patterns of gene expression in different cell types (reviewed in De Carvalho et al. 2010). In mammals, DNA methyltransferases (Dnmts) include three proteins belonging to two families that are structurally and functionally distinct. Dnmt3a and Dnmt3b establish the initial CpG methylation pattern de novo, whereas Dnmt1 maintains this pattern during chromosome replication and repair (see Fig. 2 of Li and Zhang 2014).
2.1. Maintenance Methyltransferase Dnmt1
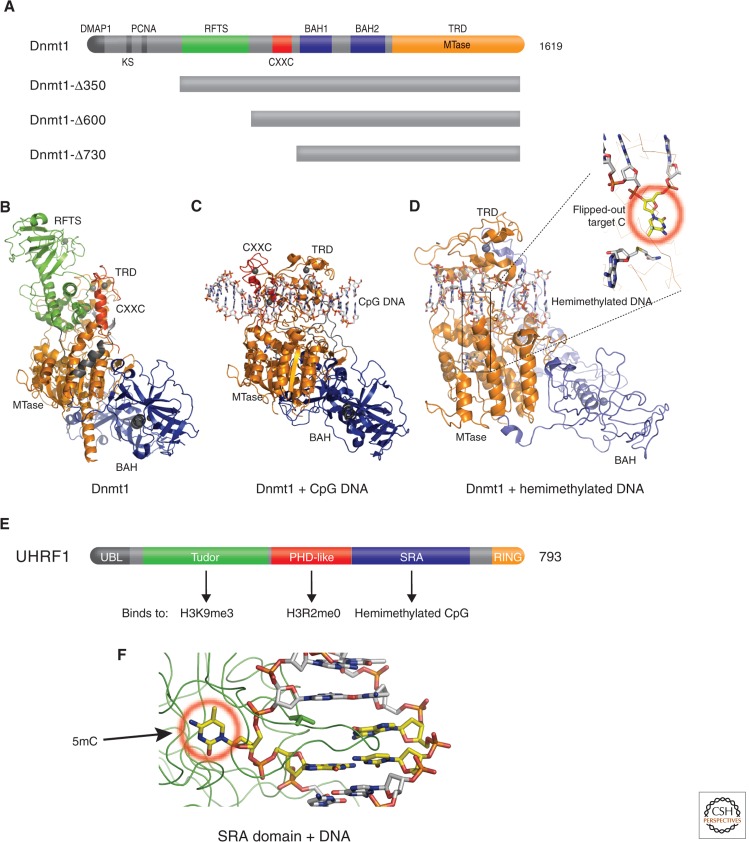
The so-called “maintenance” methyltransferase, Dnmt1, contains multiple functional domains (Fig. 6A) (Yoder et al. 1996). Structures are currently available for three amino-terminal deletions of mouse Dnmt1: an amino-terminal deletion of 350 residues (Δ350) in complex with AdoMet or its product AdoHcy (Takeshita et al. 2011), a larger deletion (Δ600) bound to DNA-containing an unmethylated CpG site (Song et al. 2011), and an even larger deletion (Δ730) bound to a hemimethylated CpG site (Song et al. 2012). When Dnmt1 is not bound to DNA, the amino-terminal replication focus _targeting sequence (RFTS) domain of Δ350 is inserted into the DNA-binding surface cleft of the carboxyl-terminal MTase domain (Fig. 6B), indicating that this domain must be removed for methylation to occur. The RFTS domain is required for _targeting Dnmt1 to replication foci, in which hemimethylated DNA is transiently generated. When the isolated RFTS domain is added in trans to an RFTS-lacking Dnmt1 protein, the RFTS domain acts as a DNA-competitive inhibitor of Dnmt1 (Syeda et al. 2011). In the structure of the Δ600 fragment, lacking the RFTS domain, the DNA bound CXXC domain (which specifically binds nonmethylated DNA) positions itself in the catalytic domain and prevents aberrant de novo methylation of CpG sequences (Fig. 6C). Only after physical removal of both the RFTS and CXXC domains can the carboxyl-terminal half of Dnmt1 (Δ730) bind to hemimethylated CpG DNA by flipping the _target cytosine out of the double-stranded DNA helix into the active site (Fig. 6D). Thus, a multistep process, accompanied by structural changes, must occur during the _targeting of full-length Dnmt1 to replication foci, which undergoes maintenance methylation of hemimethylated CpG DNA. This involves the removal of both RFTS and CXXC domains away from the catalytic center.
Figure 6.
Structures of maintenance Dnmt1 and UHRF1. (A) Schematic representation of mouse Dnmt1 domain organization and available Dnmt1 amino-terminal deletion mutants. An amino-terminal region interacts with Dnmt1-associated protein(s) (DMAP1; Rountree et al. 2000). Then an adjacent lysine and serine are subject to a methylation and phosphorylation switch that determines Dnmt1 stability (Esteve et al. 2011), a PCNA (proliferating cell nuclear antigen) interacting sequence (Chuang et al. 1997), and an RFTS (Leonhardt et al. 1992) that interacts with the SET- and RING-associated (SRA) domain of UHRF1 (Achour et al. 2008). This is followed by a CpG-interacting CXXC domain (Song et al. 2011), a tandem BAH (bromo-adjacent homology) domain (Callebaut et al. 1999), and the catalytic DNA methyltransferase domain that includes the _target-recognizing domain (Lauster et al. 1989) at the carboxyl terminus. (B) Structure of Dnmt1 in the absence of DNA (PDB 3AV4). (C) Structure of Dnmt1 in the presence of unmethylated CpG (PDB 3PT6). (D) Structure of Dnmt1 with hemimethylated CpG DNA oligonucleotides (PDB 4DA4). (E) UHRF1 harbors at least five recognizable functional domains: an ubiquitin-like domain at the amino terminus, followed by a tandem tudor domain recognizing H3K9me3 (Rothbart et al. 2012), a plant homeodomain (PHD) recognizing H3R2me0 (Rajakumara et al. 2011), an SRA domain recognizing hemimethylated CpG, and really interesting new gene (RING) domain at the carboxyl terminus that may endow UHRF1 with E3 ubiquitin ligase activity to histones (Citterio et al. 2004). (F) Structure of SRA-DNA complex illustrates 5mC flipped out from the DNA helix and bound in a cage-like pocket (circled in red; PDB 2ZO1). (Adapted from Hashimoto et al. 2009).
Dnmt1 alone, however, is insufficient for proper maintenance methylation. In vivo, maintenance methylation of hemimethylated CpG dinucleotides by Dnmt1 at DNA replication forks requires an accessory protein called UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1; Bostick et al. 2007; Sharif et al. 2007). UHRF1, a multidomain protein (Fig. 6E), binds both hemimethylated CpG site (Fig. 6F), the substrate of Dnmt1, and histone H3 (reviewed in Hashimoto et al. 2009). Somehow Dnmt1 must displace UHRF1 from the site to allow methylation. In the coming years, it will be important to understand how these multiple binding events are coordinated and whether they are cooperative for faithful mitotic inheritance of genomic methylation patterns.
2.2. De Novo Methyltransferase Dnmt3 Family
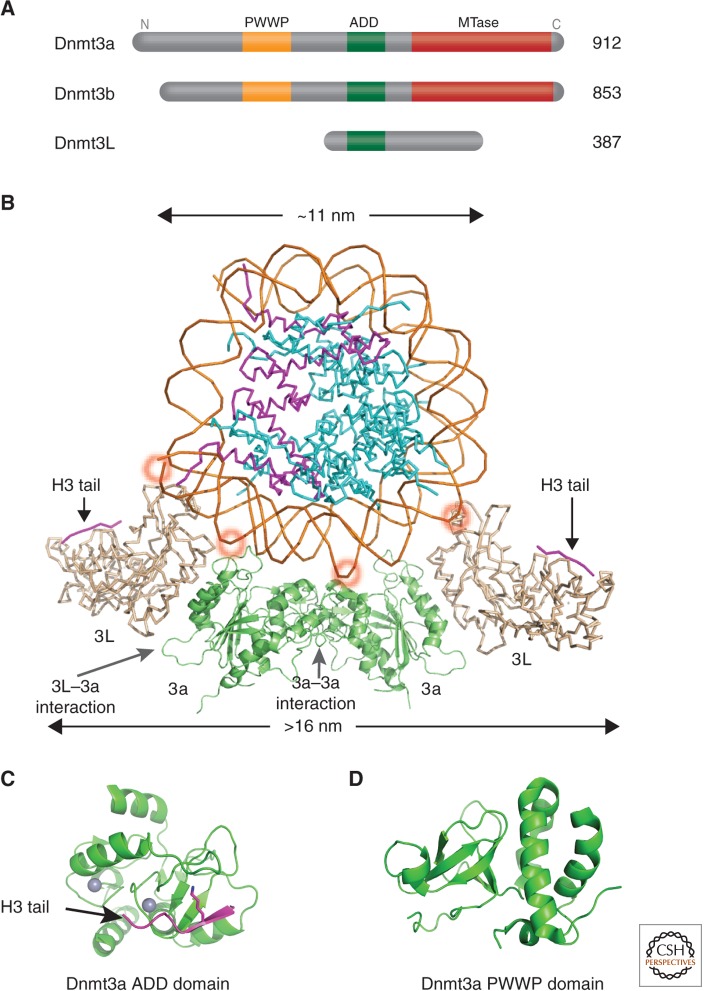
The Dnmt3 family includes two active de novo Dnmts, Dnmt3a and Dnmt3b, and one regulatory factor, Dnmt3-Like protein (Dnmt3L; Fig. 7A) (Goll and Bestor 2005). Dnmt3a and Dnmt3b have similar domain arrangements: a variable region at the amino terminus, followed by a PWWP (Pro-Trp-Trp-Pro) domain that may be involved in the nonspecific DNA binding for Dnmt3b (Qiu et al. 2002) or binding to histone H3K36me3 by Dnmt3a (Dhayalan et al. 2010), the Cys-rich 3-Zn-binding ADD (ATRX-DNMT3-DNMT3L) domain, and a carboxyl-terminal catalytic domain. The amino acid sequence of Dnmt3L is very similar to that of Dnmt3a and Dnmt3b in the ADD domain, but it lacks the conserved residues required for DNA methyltransferase activity in the carboxy-terminal domain. Structures are available for a complex between the carboxy-terminal domains of Dnmt3a and Dnmt3L (Jia et al. 2007) and the intact Dnmt3L in complex with a histone H3 amino-tail peptide (Fig. 7B) (Ooi et al. 2007). There is also structural data for an isolated ADD domain of Dnmt3a (Fig. 7C) (Otani et al. 2009), and isolated PWWP domains of Dnmt3a (Fig. 7D) and Dnmt3b (Qiu et al. 2002).
Figure 7.
Structures of the de novo Dnmt3a-Dnmt3L complex. (A) Domain architecture of Dnmt3a, 3b, and 3L. (B) A nucleosome is shown docked to a Dnmt3L-3a-3a-3L tetramer (Dnmt3a is colored in green and Dnmt3L in gray; PDB 2QRV). (Adapted from Cheng and Blumenthal 2008). The position of the histone H3 amino-terminal tail (purple) bound to Dnmt3L is shown, taken from a cocrystal structure (PDB 2PVC). By wrapping the Dnmt3a/3L tetramer around the nucleosome, the two Dnmt3L molecules are able to bind both histone tails emanating from one nucleosome. The ∼10 bp periodicity of binding to the major DNA groove is indicated by red circles. (C) Structure of the Dnmt3a ADD domain (PDB 3A1B) possibly interacting with histone tails from neighboring nucleosomes. (D) Structure of the Dnmt3a PWWP domain (PDB 3L1R).
The phenotype of the Dnmt3L knockout mouse is indistinguishable from that of a Dnmt3a germ cell-specific conditional knockout—both have dispersed retrotransposons and aberrant germ cell patterns of de novo DNA methylation at loci that usually set parent-specific imprints (Bourc’his et al. 2001; Bourc’his and Bestor 2004; Kaneda et al. 2004; Webster et al. 2005). The interaction between Dnmt3L and Dnmt3a occurs in a minimal region located at the carboxy-terminal domain of both proteins, and is necessary for catalytic activity (Chedin et al. 2002; Suetake et al. 2004). The overall size of the Dnmt3a/Dnmt3L carboxy-terminal complex is ∼16 nm long, which is greater than the diameter of an 11-nm core nucleosome (Fig. 7B). This complex contains two monomers of Dnmt3a and two of Dnmt3L, forming a tetramer with two 3L–3a interfaces and one 3a–3a interface (3L–3a–3a–3L). Substituting key, although noncatalytic residues at the Dnmt3a–3L or Dnmt3a–3a interfaces eliminates enzymatic activity, indicating that both interfaces are essential for catalysis (Jia et al. 2007).
The structure of the 3a–3a dimer interface suggests that the two active sites are located in adjacent DNA major grooves, facilitating the methylation by Dnmt3a of two CpGs separated by one helical turn, in one binding event (Fig. 7B). Methylation of CpG sites on long DNA substrates, by Dnmt3a, occurs with a periodicity of ∼10 bp, suggesting a structural model in which Dnmt3a forms an oligomer docked to the DNA (Jia et al. 2007). This periodicity is also observed on maternally imprinted mouse genes (Jia et al. 2007). CpG methylation patterns on human chromosome 21 also correlate with a ∼10-bp methylated CpG periodicity (Zhang et al. 2009). Interestingly, the ∼10-bp methylation periodicity was evident in embryonic stem cells, however, often at non-CpG sites (which are substrates of Dnmt3a as well) that occur mostly in gene bodies as opposed to regulatory regions (Lister et al. 2009). Non-CG methylation is specific to the embryonic stem cell stage, as it disappears on induced differentiation and is restored in induced pluripotent stem cells. In the plant, Arabidopsis thaliana 10-bp periodic non-CpG DNA methylation by DRM2 (which is related to mammalian Dnmt3a) has similarly been observed (Cokus et al. 2008).
3. Interplay between DNA methylation and histone modification
3.1. Dnmt3L Connects Unmethylated H3K4 to De Novo DNA Methylation
Genome-scale DNA methylation profiles suggest that DNA methylation is correlated with histone methylation patterns (Meissner et al. 2008). Specifically, DNA methylation is correlated with the absence of H3K4 methylation and the presence of H3K9 methylation. Considering the inverse relationship between H3K4 methylation and DNA methylation, it is important to note that for the mammalian LSD histone demethylases, whose substrates include H3K4me2/me1, LSD1 is absolutely essential in maintaining global DNA methylation (Wang et al. 2009a) and LSD2 in establishing maternal DNA genomic imprints (Ciccone et al. 2009). Indeed, disruption of LSD1 results in earlier embryonic lethality and a more severe hypomethylation defect than disruption of the Dnmts themselves (Wang et al. 2009a).
The mammalian Dnmt3L-Dnmt3a de novo DNA methylation machinery can translate patterns of H3K4 methylation into heritable patterns of DNA methylation that mediate transcriptional silencing of the affected sequences (Ooi et al. 2007). Peptide interaction assays showed that Dnmt3L specifically interacts with the extreme amino terminus of histone H3; this interaction was strongly inhibited by H3K4 methylation, but was insensitive to modifications at other positions (Ooi et al. 2007). Cocrystallization of Dnmt3L with the amino tail of H3 showed this tail bound to the ADD domain of Dnmt3L (Fig. 7B), and substitution of key residues in the binding site eliminated the H3–Dnmt3L interaction. The main in vivo interaction partners of epitope-tagged Dnmt3L are Dnmt3a2, a shorter isoform of Dnmt3a (Chen et al. 2002), Dnmt3b, and the four core histones (Ooi et al. 2007). Given Dnmt3a and Dnmt3b bind nucleosomal DNA (Sharma et al. 2011), the data suggest that Dnmt3L is a probe of H3K4 methylation, and if the methylation is absent, then Dnmt3L induces de novo DNA methylation by docking activated Dnmt3a to the nucleosome.
Histone-Dnmt3L-Dnmt3a–DNA interactions have been studied in the budding yeast S. cerevisiae (Hu et al. 2009), which has no detectable DNA methylation (Proffitt et al. 1984) and lacks Dnmt orthologs. Introduction of the murine methyltransferases Dnmt1 or Dnmt3a leads to detectable, but extremely low levels of DNA methylation (Bulkowska et al. 2007). In contrast, a substantially higher level of de novo methylation can be achieved in yeast by coexpressing murine Dnmt3a and Dnmt3L (Hu et al. 2009). This induced DNA methylation was found preferentially in heterochromatic regions where H3K4 methylation is rare. When genes for components of the H3K4-methylating complex were disrupted in the context of Dnmt3a/3L overexpression, a greater level of genomic DNA methylation was observed. Deletions or _targeted mutations in the ADD domain of Dnmt3L inhibited both global DNA methylation and the ability of Dnmt3L to associate with an H3K4me0 peptide. These same Dnmt3L mutants failed to restore normal DNA methylation to a specific promoter when introduced into embryonic stem cells from Dnmt3L–/– mice (Hu et al. 2009).
The above data has led to a model in which Dnmt3L binds to H3K4me0 (via its ADD domain) and recruits Dnmt3a to regions of chromatin where H3K4 is unmethylated. Such a model could explain part of the puzzle of how DNA methylation patterns are established de novo during embryonic and germ cell development, windows of time in which both proteins are expressed (Kato et al. 2007). However, whereas Dnmt3a and 3b expression is retained in somatic cells, Dnmt3L is expressed poorly, if at all, in differentiated cell types. This raises the question of how de novo DNA methylation is restricted in somatic cells, whether Dnmt3a and 3b alone are capable of discriminating H3K4 methylation status, and (if so) the structural basis for that discrimination. The key probably lies in the fact that, in vitro, the ADD domains of Dnmt3a or Dnmt3b possess the same H3 tail-binding specificity as that of Dnmt3L (Zhang et al. 2010), and a structure of the Dnmt3a ADD domain in complex with an amino-terminal-tail peptide from histone H3 indicates that the ADD domain is sufficient to recognize H3K4me0 (Fig. 7C) (Otani et al. 2009). Furthermore, Jeong et al. showed that in nuclei from HCT116 human colon cancer cells (which do not express DNMT3L) almost all of the cellular DNMT3a and 3b (but not DNMT1) was associated with nucleosomes (Jeong et al. 2009). Chromatin binding of DNMT3a and 3b required an intact nucleosomal structure, although no other chromatin factors, suggesting that DNMT3a and 3b alone are capable of direct interaction (via the ADD domain) with chromatin components (H3K4me0) in addition to DNA.
3.2. MLL1 Links H3K4 Methylation to Unmethylated CpGs
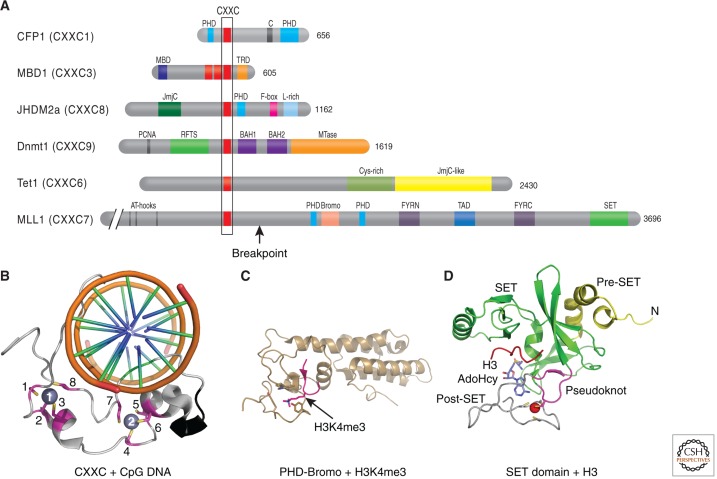
In humans there are at least eight HKMTs with specificity for H3K4. These include the mixed lineage leukemia (MLL) genes, MLL1–MLL5, hSET1a, hSET1b, and ASH1. MLL1/SET1-associated methyltransferase activity appears to be functional only in the context of multiprotein complexes; characterization of these reveals distinct multiprotein complexes for each with several shared components (reviewed in Cosgrove and Patel 2010). The MLL family plays an important role in embryonic development and is necessary for methylation of H3K4 at a subset of genes in the human and mouse genomes, particularly the HOX gene clusters (Ansari and Mandal 2010). Translocations involving MLL genes are involved in the etiology of myeloid and lymphoid leukemias. Considering the inverse relationship between H3K4 methylation and DNA methylation, it is interesting to note that disruption of the MLL1 gene in mice results in loss of H3K4 methylation and de novo DNA methylation at some Hox gene promoters (Milne et al. 2002; Terranova et al. 2006), suggesting that MLLs, directly or indirectly (through H3K4 methylation), prevent DNA methylation or perhaps stabilize unmethylated DNA. In fact, MLL proteins contain a CXXC domain, an evolutionarily conserved domain that mediates selective binding to unmethylated CpGs (Fig. 8A). The CXXC interaction with unmethylated CpGs was confirmed by a solution structure of an MLL1-CXXC domain complexed with unmethylated DNA (Fig. 8B) (Cierpicki et al. 2010) and an X-ray structure of DNMT1 in complex with unmethylated DNA (Fig. 6C) (Song et al. 2011). Structurally, the CXXC domain has a novel fold arranged in an elongated shape (Fig. 8B). The CXXC domain binds DNA in a clamp-like manner with the long axis of the structure linking the two Zn ions nearly perpendicular to the DNA axis (Fig. 8B) (Cierpicki et al. 2010).
Figure 8.
CpG-interacting proteins including MLL1. (A) Domain architecture of CXXC domain-containing proteins. (B–D) The structures of three isolated MLL1 domains in complex with interacting histones or DNA: (B) the amino-terminal CXXC domain in complex with CpG DNA (PDB 2KKF; Cierpicki et al. 2010), (C) the central PHD-bromodomain in complex with H3K4me3 peptide (PDB 3LQJ; Wang et al. 2010), and (D) the carboxyl-terminal SET domain (PDB 2W5Z; Southall et al. 2009).
The Set1 H3K4 methyltransferase also appears to interact with unmethylated DNA, although in this case it is via the Cfp1 accessory protein, which contains a CXXC domain (Fig. 8A) (Lee and Skalnik 2005; Lee et al. 2007). High throughput sequencing of Cfp1-bound chromatin identified a notable concordance between H3K4me3 and Cfp1 at unmethylated CpG islands in the mouse brain (Thomson et al. 2010). Also, Cfp1 binds specifically to the unmethylated allele of loci that are allele-specifically DNA methylated (e.g., imprinted loci, Xist gene). Depletion of Cfp1 results in a marked reduction in H3K4me3 genome-wide. The _targeting of Cfp1 to CpG islands is independent of promoter activity as the insertion of an untranscribed, unmethylated CpG-dense construct into the genome of embryonic stem cells is sufficient to nucleate Cfp1 binding and H3K4me3. This suggests that unmethylated CpGs recruit Cfp1 and the associated methyltransferase Set1 creates new marks of H3K4me3 on the local chromatin (illustrated in Fig. 1 of Blackledge et al. 2013).
CXXC domains are also found in Dnmt1 (Fig. 6), the methyl-CpG-binding protein MBD1 (Jorgensen et al. 2004), and Tet1, a Jumonji-like enzyme that catalyzes the conversion of 5mC to 5-hydroxymethylcytosine (Fig. 8A) (Tahiliani et al. 2009). Interestingly, the recurrent translocation, t(10;11)(q22;q23), has been described in acute myelogenous leukemias, and results in a fusion transcript that juxtaposes the first six exons of MLL1 (containing the AT hook and CXXC) to the carboxyl-terminal one third of TET1, thus “replacing” the TET1 CXXC with the MLL1 CXXC (labeled as the breakpoint in MLL1; Fig. 8A) (Ono et al. 2002; Lorsbach et al. 2003). Whether this leads to altered _targeting of methyl hydroxylation remains to be determined.
3.3. JHDM1 Binds CpG DNA and Demethylates H3K36me2
Like the histone H3K4 methyltransferases of the MLL/SET1 family, the Jumonji domain-containing histone demethylases, JHDM1A (also known as CXXC8 or KDM2A; Fig. 8A) and JHDM1B (CXXC2 or KDM2B) have CXXC DNA-binding domains (Tsukada et al. 2006). Like the Set1-Cfp1 complex, JHDM1A is recruited to unmethylated CpG islands on a genome-wide scale via its CXXC domain (Blackledge et al. 2010). Like Cfp1, its localization to CpG islands is independent of promoter activity and gene-expression levels, and correlated with the selective depletion of H3K36me2/me1 within the CpG island, but not surrounding regions or the bodies of genes (see Fig. 1 of Blackledge et al. 2013); knockdown of JHDM1A/KDM2A results in the selective accumulation of H3K36me2 in these regions. Consistent with the idea that DNA methylation restricts the localization of CXXC proteins, JHDM1A/KDM2A becomes mislocalized to DNA hypomethylated pericentric heterochromatin in DNA methyltransferase Dnmt1–/– mice. Although in vitro studies suggest that the CXXC domains can bind a single CpG site with micromolar affinity, both the Set1-Cfp1 and JHDM1A/KDM2A studies suggest that the _targeting of CXXC proteins in vivo is dependent on CpG density as well as its methylation status. It could be possible that these proteins oligomerize and form nucleoprotein filaments on CpG-dense DNA, in a manner similar to that described for the DNA methyltransferase Dnmt3a-3L complex (Jurkowska et al. 2008). To reinforce the correlation between DNA methylation and H3K36 methylation, the PWWP domain of Dnmt3a is capable of binding H3K36me3 (Dhayalan et al. 2010) and directs DNA methylation (Chen et al. 2004).
3.4. Linkage between H3K9 Methylation and DNA Methylation
Methylation at H3K9 is positively correlated with DNA methylation, in contrast to its negative correlation with H3K4 methylation. There is in vivo evidence that the H3K9-linked DNA methylations represent an evolutionarily conserved silencing pathway. Studies in Neurospora and Arabidopsis have shown a strict dependence of DNA methylation on the H3K9 methyltransferases Dim-5 and KRYPTONITE (KYP; Tamaru and Selker 2001; Jackson et al. 2002; Tamaru et al. 2003). The SRA domain of KYP (also known as SUVH4) binds directly to methylated CHG-containing oligonucleotides (Johnson et al. 2007), whereas a plant-specific DNA CHROMOMETHYLASE3 (CMT3), responsible for CHG methylation, binds H3K9me2-containing nucleosomes via its associated BAH and chromodomains within the same polypeptide (Du et al. 2012), resulting in a self-reinforcing loop between H3K9me2 and CHG methylation in plants.
With regard to mammals, G9a, and GLP, two related euchromatin-associated H3K9 methyltransferases form heterodimers and have been implicated in DNA methylation at various loci, including imprinting centers, retrotransposons and satellite repeats, a G9a/GLP _target promoter, an Oct4 promoter, and a set of embryonic genes (reviewed in Collins and Cheng 2010; Shinkai and Tachibana 2011). Furthermore, G9a is required for de novo DNA methylation and the establishment of silencing of newly integrated proviruses in murine embryonic stem cells (Leung et al. 2011). The G9a/GLP heterodimer interacts with a chromodomain protein MPP8, which in turn interacts with DNA methyltransferase Dnmt3a and/or methylated H3K9 (Kokura et al. 2010; Chang et al. 2011). Together, these findings provide a molecular explanation, at least in part, for the co-occurrence of DNA methylation and H3K9 methylation in chromatin.
The functional relationships between DNA methylation and two other H3K9 methyltransferases in mammals are complex and context dependent, as deletion of Suv39h1/h2 (Lehnertz et al. 2003) or SETDB1 (Matsui et al. 2010) has only a minor impact on DNA methylation at constitutive heterochromatin or endogenous retroelements, respectively. A particularly interesting observation is that the methyl-CpG-binding domain protein MBD1 (Fig. 8A) forms a stable complex with SETDB1 (Sarraf and Stancheva 2004; Lyst et al. 2006) as well as the Suv39h1/HP1 complex (Fujita et al. 2003), constituting the heterochromatin-specific H3K9me3 writer and reader. The methyl-CpG-binding domain (MBD) is present in a family of proteins conserved throughout the eukaryotic lineage. This domain, in some but not all cases, confers the ability to bind fully methylated CpGs. Mammals have five well-characterized members of this family, each with unique biological characteristics (reviewed in Dhasarathy and Wade 2008; Li and Zhang 2014). SETDB1 also contains an intrinsic putative MBD domain with two conserved DNA-interacting arginine residues known to make direct contact with DNA in the structures of the MBD domain (reviewed in Hashimoto et al. 2010). It remains to be seen whether the putative MBD domain of SETDB1 is similarly able to selectively bind methylated DNA. The intrinsic or associated coupling of a DNA methylation “reader” (MBD) with an H3K9me3 “writer” (SETDB1) implies an interdependent mechanism for the propagation or maintenance of these marks.
Finally, the identification of UHRF1 and its potential role in modulating the specificity of Dnmt1 for hemimethylated CpG sites and binding of histones provides another layer to the mechanism that ensures the faithful transmission of epigenetic information during DNA replication. Given that UHRF1 has the potential to interact with both hemimethylated CpGs (via the SRA domain) and H3K9me3 (via the Tudor domain), and is known to interact with a wide variety of epigenetic regulators, including Dnmt1, the H3K9 methyltransferase G9a and a histone acetyltransferase Tip60, it is possible that UHRF1 and the proteins in this larger complex play a more central role in coupling the transmission of DNA and histone methylation (H3K9, in particular) during mitotic cell division. It is interesting to know that, like plant CMT3, the mammalian Dnmt1 (and its homologs in Neurospora DIM2 and plant MET1) contains a BAH domain(s) within the same polypeptide (Fig. 6A). It remains to be seen whether the BAH domains of Dnmt1 are similarly able to bind methylated histones, either methylated H3K9, as recognized by the BAH domain of CMT3, or methylated H4K20, as recognized by the BAH domain of human ORC1 (origin of replication complex; Kuo et al. 2012).
3.5. PHF8 Binds H3K4me3 and Demethylates H3K9me2
The examples discussed so far provide a molecular explanation, at least in part, for the inverse correlation of DNA methylation and H3K4 methylation, as well as the co-occurrence of DNA methylation and H3K9 methylation in chromatin. The functional linkage of DNA methylation, H3K4 methylation, and H3K9 methylation is further illustrated by the finding that treatment with 5-aza-2′-deoxycytidine (5-aza), a DNA-demethylating drug (Yoo et al. 2007), leads to depletion of DNA methylation and H3K9 methylation, and a corresponding increase in H3K4 methylation (Nguyen et al. 2002). How is the inverse correlation of H3K4 and H3K9 methylation maintained?
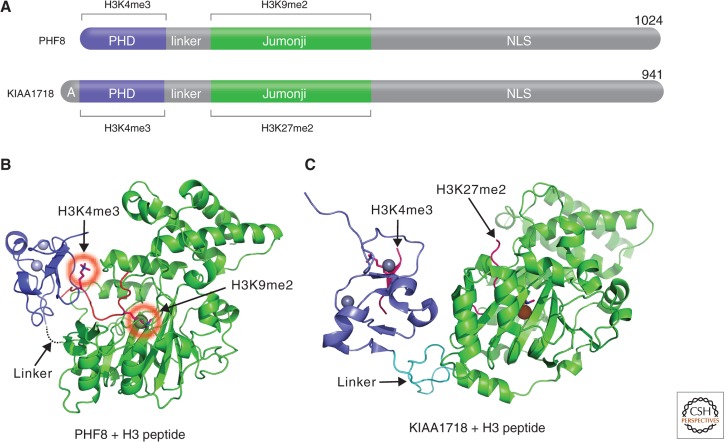
PHF8 belongs to a small family of Jumonji proteins with three members in mice and human (PHF2, PHF8, and KIAA1718; Klose et al. 2006). Mutations in the PHF8 gene lead to X-linked mental retardation (Loenarz et al. 2010), and knockdown of KIAA1718 (also known as JHDM1D) and PHF8 homologs in zebrafish cause brain defects (Qi et al. 2010; Tsukada et al. 2010). These proteins harbor two domains in the amino-terminal half (Fig. 9A): a PHD domain that binds H3K4me3 and a Jumonji domain that demethylates H3K9me2, H3K27me2, or H4K20me1. However, the presence of H3K4me3 on the same peptide as H3K9me2 makes the doubly methylated peptide a significantly better substrate of PHF8 (Feng et al. 2010; Fortschegger et al. 2010; Horton et al. 2010; Kleine-Kohlbrecher et al. 2010). In contrast, the presence of H3K4me3 has the opposite effect in that it diminishes the H3K9me2 demethylase activity of KIAA1718 with no adverse effect on its H3K27me2 activity (Horton et al. 2010). Differences in substrate specificity between the two enzymes are explained by a bent conformation of PHF8 (Fig. 9B), allowing each of its domains to engage their respective _targets, and an extended conformation of KIAA1718, which prevents the access to H3K9me2 by its Jumonji domain when its PHD domain engages H3K4me3 (Fig. 9C). Thus, the structural linkage between the PHD domain binding to H3K4me3 and the placement of the catalytic Jumonji domains relative to this active epigenetic mark determines which repressive marks (H3K9me2 or H3K27me2) are removed by these demethylases. Thus, the data indicate that the PHF8 and KIAA1718 Jumonji domains on their own are promiscuous enzymes; the PHD domains and linker—a determinant for the relative positioning of the two domains—are mainly responsible for substrate specificity.
Figure 9.
Coordinated methyllysine erasure between a Jumonji and a PHD. (A) Schematic representations of PHF8 and KIAA1718 domain structure. (B) A bent conformation of PHF8 bound to a histone H3 peptide (in red) containing K4me3 and K9me2 (circled in red; PDB 3KV4). (C) An extended conformation of KIAA1718 (PDB 3KV6). The position of the histone H3 peptide is taken from a cocrystal structure of Caenorhabditis elegans KIAA1718 (PDB 3N9P).
Another structural study on C. elegans KIAA1718 suggested that the PHD and Jumonji domains might enable a trans-histone peptide-binding mechanism, in which the substrate peptide associated with the PHD domain and the peptide bound to the Jumonji domain could be coming from two separate histone molecules of the same nucleosome or two neighboring nucleosomes (Yang et al. 2010). The trans-binding mechanism is an attractive model for PHF8 and could explain the finding that PHF8 also functions in vivo as an H4K20me1 (histone H4 monomethylated at lysine 20) demethylase, whereas its PHD domain interacts with H3K4me3 in the context of nucleosome (Liu et al. 2010; Qi et al. 2010). But one has to explain why PHF8 is only active on monomethylated H4K20 (H4K20me1), whereas it is active on dimethylated H3K9 and H3K27. One possibility is that only H4K20me1 coexists with H3K4me3 in vivo.
4. Summary
Combinatorial readout of multiple covalent chromatin modifications (including DNA methylation) is an explicit prediction of the “histone code” hypothesis (Strahl and Allis 2000; Jenuwein and Allis 2001; Turner 2007). Although it is well-accepted that DNA methylation patterns are replicated in a semiconservative fashion during cell division via a Dnmt1-dependent mechanism discussed (see also Li and Zhang 2014), one of the fundamental unresolved questions is how, and indeed whether, histone modifications are similarly “inherited.” Considering that the well-studied lysine methylation events reside on histone H3 (K4, K9, K27, K36, and K79) or H4 (K20), this evokes a model in which “old” histone methylation patterns may be retained (possibly by UHRF1) and copied onto newly deposited tetramers from neighboring parental nucleosomes. Indeed, many of the SET domain histone methyltransferases contain intrinsic or associated reader domains that recognize the same mark that they generate, allowing for the copying of these marks from old to new nucleosomes. For example, G9a/GLP catalyzes H3K9me1/2 and contains an ankyrin-repeat domain that binds H3K9me1/2 (Collins et al. 2008). Likewise, SUV39H1/2, the H3K9me3 writer, interacts with HP1, the H3K9me3 reader (reviewed in Grewal and Jia 2007). Similarly, yeast Clr4 methylates H3K9 and contains a chromodomain that binds H3K9me3 (Zhang et al. 2008). This interdomain cross talk provides a possible mechanism for propagating a methyl mark. Thus, higher organisms have evolved coordinated mechanisms of deposition and transmission of repressive chromatin marks to both DNA and histones. Enzymes that affect more complex cross talk include PHF8, which contains modules within the same polypeptide for both recognizing (PHD) and removing (Jumonji domain) two opposing methyl marks. This cross talk provides a possible mechanism for removing a repressive methyl mark (H3K9me2 or H4K20me1) based on an existing active methyl mark (H3K4me3). An even more complex situation is JARID1A, which contains multiple PHD domains for recognizing the substrate (H3K4me3) and product (H3K4me0) of its catalytic Jumonji domain, respectively (Wang et al. 2009b).
We have also discussed enzyme complexes that cross talk between DNA methylation (or lack thereof) and histone marks that are probably on the same nucleosome. These include the Dnmt3a-Dnmt3L complex, containing a reader domain for H3K4me0 coupled to DNA methyltransferase activity, whereas MLL1 (or Set1-Cfp1 complex) contains reader domains for DNA CpG and a SET domain for making methylated H3K4 (Fig. 10A). The mammalian Dnmt1-UHRF1 complex (Rothbart et al. 2012) and plant-specific CMT3 (Du et al. 2012) contain reader domains for H3K9me3/2 and DNA methyltransferase activity (Fig. 10B). The function of the Jumonji H3K36me3-specific demethylase JHDM1 is linked to the CXXC domain, which associates with unmethylated CpG DNA (Fig. 10C) (Blackledge et al. 2010).
Another intriguing observation involving DNA methylation is its mutually antagonistic relationship with histone variant H2A.Z (Zilberman et al. 2008; Conerly et al. 2010). How the exclusion is specifically established remains largely unknown. One possibility is that the histone variant H2A.Z is preferentially deposited by the remodeling ATPase complexes to regions lacking DNA methylation. Another scenario is that nucleosomes that contain the histone variant H2A.Z are no longer the substrates of DNA methyltransferases. Future experiments are needed to uncover the mechanisms of correct assembly of machinery required to accurately modify chromatin. Although the field still faces a number of critical questions, it is clear that structural analyses will continue to play a central and synergistic role, together with biochemical and genetic studies to address them.
ACKNOWLEDGMENTS
The author wishes to thank the former and current members of his laboratory, Xing Zhang for preparing Table 1, and support from the U.S. National Institutes of Health (GM049245).
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Achour M, Jacq X, Ronde P, Alhosin M, Charlot C, Chataigneau T, Jeanblanc M, Macaluso M, Giordano A, Hughes AD, et al. 2008. The interaction of the SRA domain of ICBP90 with a novel domain of DNMT1 is involved in the regulation of VEGF gene expression. Oncogene 27: 2187–2197 [DOI] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739 [DOI] [Google Scholar]
- An W, Kim J, Roeder RG 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117: 735–748 [DOI] [PubMed] [Google Scholar]
- Ansari KI, Mandal SS 2010. Mixed lineage leukemia: Roles in gene expression, hormone signaling and mRNA processing. FEBS J 277: 1790–1804 [DOI] [PubMed] [Google Scholar]
- Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, Han B, Jungheim LN, Qian Y, Rauch C, et al. 2012. Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci 109: 17960–17965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T 2002. Histone methylation: Dynamic or static? Cell 109: 801–806 [DOI] [PubMed] [Google Scholar]
- Baron R, Binda C, Tortorici M, McCammon JA, Mattevi A 2011. Molecular mimicry and ligand recognition in binding and catalysis by the histone demethylase LSD1-CoREST complex. Structure 19: 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Becker PB, Workman JL 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG 2009. Protein arginine methylation in mammals: Who, what, and why. Mol Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Blackledge N, Thomson JP, Skene PJ 2013. CpG island chromatin is shaped by recruitment of ZF-CxxC proteins. Cold Spring Harb Perspect Biol 5: a018648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764 [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539 [DOI] [PubMed] [Google Scholar]
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S 2001. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem 276: 32971–32976 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD 2002. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Bulkowska U, Ishikawa T, Kurlandzka A, Trzcinska-Danielewicz J, Derlacz R, Fronk J 2007. Expression of murine DNA methyltransferases Dnmt1 and Dnmt3a in the yeast Saccharomyces cerevisiae. Yeast 24: 871–882 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Courvalin JC, Mornon JP 1999. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett 446: 189–193 [DOI] [PubMed] [Google Scholar]
- Chang Y, Sun L, Kokura K, Horton JR, Fukuda M, Espejo A, Izumi V, Koomen JM, Bedford MT, Zhang X, et al. 2011. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun 2: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F, Lieber MR, Hsieh CL 2002. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci 99: 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR 1999. Regulation of transcription by a protein methyltransferase. Science 284: 2174–2177 [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277: 38746–38754 [DOI] [PubMed] [Google Scholar]
- Chen T, Tsujimoto N, Li E 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol 24: 9048–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM 2008. Mammalian DNA methyltransferases: A structural perspective. Structure 16: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM 2010. Coordinated chromatin control: Structural and functional linkage of DNA and histone methylation. Biochemistry 49: 2999–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Collins RE, Zhang X 2005. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct 34: 267–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a _target for p21WAF1. Science 277: 1996–2000 [DOI] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T 2009. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461: 415–418 [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH 2010. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol 17: 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM 2004. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol 24: 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clissold PM, Ponting CP 2001. JmjC: Cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem Sci 26: 7–9 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R, Cheng X 2010. A case study in cross-talk: The histone lysine methyltransferases G9a and GLP. Nucleic Acids Res 38: 3503–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X 2005. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem 280: 5563–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X 2008. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol 15: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S 2010. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res 20: 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S 2006. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun 342: 472–481 [DOI] [PubMed] [Google Scholar]
- Cosgrove MS, Patel A 2010. Mixed lineage leukemia: A structure-function perspective of the MLL1 protein. FEBS J 277: 1832–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Brunzelle JS, Trievel RC 2005. Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase. Genes Dev 19: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. 2011. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20: 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol 12: 2090–2097 [DOI] [PubMed] [Google Scholar]
- De Carvalho DD, You JS, Jones PA 2010. DNA methylation and cellular reprogramming. Trends Cell Biol 20: 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhasarathy A, Wade PA 2008. The MBD protein family-reading an epigenetic mark? Mutat Res 647: 39–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A 2010. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285: 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M 2001. Chromatin silencing protein and pachytene checkpoint regulator Dot1p has a methyltransferase fold. Trends Biochem Sci 26: 405–407 [DOI] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. 2012. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, Pradhan S 2011. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol 18: 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Yonezawa M, Ye J, Jenuwein T, Grummt I 2010. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol 17: 445–450 [DOI] [PubMed] [Google Scholar]
- Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A 2005. Human histone demethylase LSD1 reads the histone code. J Biol Chem 280: 41360–41365 [DOI] [PubMed] [Google Scholar]
- Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A 2007. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J Biol Chem 282: 20070–20074 [DOI] [PubMed] [Google Scholar]
- Fortschegger K, de Graaf P, Outchkourov NS, van Schaik FM, Timmers HT, Shiekhattar R 2010. PHF8 _targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol 30: 3286–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks F, Stulemeijer IJ, Ovaa H, van Leeuwen F 2011. A modified epigenetics toolbox to study histone modifications on the nucleosome core. Chembiochem 12: 308–313 [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 21: 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem 277: 8243–8247 [DOI] [PubMed] [Google Scholar]
- Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M 2003. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem 278: 24132–24138 [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- *.Grunstein M, Gasser SM 2013. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol 5: a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Cheng X 2009. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics 4: 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Vertino PM, Cheng X 2010. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2: 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Henikoff S, Smith MM 2014. Histone variants and epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann F, Pably P, Eckerich C, Bedford MT, Fackelmayer FO 2009. Human protein arginine methyltransferases in vivo—Distinct properties of eight canonical members of the PRMT family. J Cell Sci 122: 667–677 [DOI] [PubMed] [Google Scholar]
- Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W 2007. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci 104: 12318–12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffart LM, Barr EW, Guyer RB, Bollinger JM Jr, Krebs C 2006. Direct spectroscopic detection of a C-H-cleaving high-spin FeIV complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci 103: 14738–14743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X 2010. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17: 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Yu H 2010. Structural insights into histone lysine demethylation. Curr Opin Struct Biol 20: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JL, Zhou BO, Zhang RR, Zhang KL, Zhou JQ, Xu GL 2009. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci 106: 22187–22192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM 2006. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312: 748–751 [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. 2007. p53 is regulated by the lysine demethylase LSD1. Nature 449: 105–108 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD 2001. Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G 1998. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci 54: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA 2009. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol 29: 5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X 2007. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449: 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE 2007. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen HF, Ben-Porath I, Bird AP 2004. Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains. Mol Cell Biol 24: 3387–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowska RZ, Anspach N, Urbanke C, Jia D, Reinhardt R, Nellen W, Cheng X, Jeltsch A 2008. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res 36: 6656–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903 [DOI] [PubMed] [Google Scholar]
- Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A 2009. A novel mammalian flavin-dependent histone demethylase. J Biol Chem 284: 17775–17782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280 [DOI] [PubMed] [Google Scholar]
- Kim S, Lim IK, Park GH, Paik WK 1997. Biological methylation of myelin basic protein: Enzymology and biological significance. Int J Biochem Cell Biol 29: 743–751 [DOI] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Christensen J, Vandamme J, Abarrategui I, Bak M, Tommerup N, Shi X, Gozani O, Rappsilber J, Salcini AE, et al. 2010. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell 38: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y 2006. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Kokura K, Sun L, Bedford MT, Fang J 2010. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J 29: 3673–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kriaucionis S, Tahiliani M 2014. Expanding the epigenetic landscape: Novel modifications of cytosine in genomic DNA. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauster R, Trautner TA, Noyer-Weidner M 1989. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable _target-recognizing domains. J Mol Biol 206: 305–312 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 280: 41725–41731 [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim S, Paik WK 1977. S-adenosylmethionine: Protein-arginine methyltransferase. Purification and mechanism of the enzyme. Biochemistry 16: 78–85 [DOI] [PubMed] [Google Scholar]
- Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR 2002. Synergy among nuclear receptor coactivators: Selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol 22: 3621–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cook JR, Yang ZH, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S 2005. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem 280: 3656–3664 [DOI] [PubMed] [Google Scholar]
- Lee JH, Tate CM, You JS, Skalnik DG 2007. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem 282: 13419–13428 [DOI] [PubMed] [Google Scholar]
- Lee J, Thompson JR, Botuyan MV, Mer G 2008. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol 15: 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH 1992. A _targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71: 865–873 [DOI] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. 2011. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci 108: 5718–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, et al. 2011. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat Immunol 12: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Li E, Zhang Y 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP 2010. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 29: 1803–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, Ratcliffe PJ, Schofield CJ 2010. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nε-dimethyl lysine demethylase. Hum Mol Genet 19: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR 2003. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 17: 637–641 [DOI] [PubMed] [Google Scholar]
- Lyst MJ, Nan X, Stancheva I 2006. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J 25: 5317–5328 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- *.Marmorstein R, Zhou M-M 2014. Writers and readers of histone acetylation: Structure, mechanism and inhibition. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ 2010. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr Opin Struct Biol 20: 659–672 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437: 436–439 [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL 2002. MLL _targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Z, Zhang Y, Xu RM 2003. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112: 711–723 [DOI] [PubMed] [Google Scholar]
- Miranda TB, Miranda M, Frankel A, Clarke S 2004. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem 279: 22902–22907 [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res 62: 6456–6461 [PubMed] [Google Scholar]
- Oh S, Jeong K, Kim H, Kwon CS, Lee D 2010. A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem Biophys Res Commun 399: 512–517 [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y 2005. hDOT1L links histone methylation to leukemogenesis. Cell 121: 167–178 [DOI] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y 2002. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res 62: 4075–4080 [PubMed] [Google Scholar]
- Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin S-P, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M 2009. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep 10: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer A, Bruick RK 2007. Non-heme dioxygenases: Cellular sensors and regulators jelly rolled into one? Nat Chem Biol 3: 144–153 [DOI] [PubMed] [Google Scholar]
- *.Patel D 2014. A structural perspective on readout of epigenetic H & DM marks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt JH, Davie JR, Swinton D, Hattman S 1984. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol Cell Biol 4: 985–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. 2010. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol 9: 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al. 2011. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell 43: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. 2012. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 25: 269–277 [DOI] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I 2004. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 15: 595–605 [DOI] [PubMed] [Google Scholar]
- Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X 2004. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem 279: 43296–43306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X 2003. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem Sci 28: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912 [DOI] [PubMed] [Google Scholar]
- Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G 2011. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet 7: e1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Shi YG, Tsukada Y 2013. The discovery of histone demethylases. Cold Spring Harb Perspect Biol 5: a017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M 2011. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 25: 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siarheyeva A, Senisterra G, Allali-Hassani A, Dong A, Dobrovetsky E, Wasney GA, Chau I, Marcellus R, Hajian T, Liu F, et al. 2012. An allosteric inhibitor of protein arginine methyltransferase 3. Structure 20: 1425–1435 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ III, Eick D, Reinberg D 2011. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH, Patel DJ 2011. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Teplova M, Ishibe-Murakami S, Patel DJ 2012. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335: 709–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR 2009. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell 33: 181–191 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S 2004. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem 279: 27816–27823 [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Syeda F, Fagan RL, Wean M, Avvakumov GV, Walker JR, Xue S, Dhe-Paganon S, Brenner C 2011. The replication focus _targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1. J Biol Chem 286: 15344–15351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita K, Suetake I, Yamashita E, Suga M, Narita H, Nakagawa A, Tajima S 2011. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1). Proc Natl Acad Sci 108: 9055–9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283 [DOI] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU 2003. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet 34: 75–79 [DOI] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M 2006. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci 103: 6629–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, McLaughlin PJ, Allshire RC 2005. Methylation: Lost in hydroxylation? EMBO Rep 6: 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J 2007. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J 26: 4391–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Ishitani T, Nakayama KI 2010. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev 24: 432–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM 2007. Defining an epigenetic code. Nat Cell Biol 9: 2–6 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA 2009. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkel P, Angrand PO 2007. The control of histone lysine methylation in epigenetic regulation. Biochimie 89: 1–20 [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293: 853–857 [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. 2009a. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41: 125–129 [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD 2009b. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ 2010. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell 141: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster KE, O’Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle R, et al. 2005. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci 102: 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ 2003. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421: 652–656 [DOI] [PubMed] [Google Scholar]
- Xiao B, Jing C, Kelly G, Walker PA, Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ, et al. 2005. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev 19: 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hu L, Wang P, Hou H, Lin Y, Liu Y, Li Z, Gong R, Feng X, Zhou L, et al. 2010. Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans. Cell Res 20: 886–898 [DOI] [PubMed] [Google Scholar]
- Yoder JA, Yen RW, Vertino PM, Bestor TH, Baylin SB 1996. New 5′ regions of the murine and human genes for DNA (cytosine-5)-methyltransferase. J Biol Chem 271: 31092–31097 [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, Jones PA 2007. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res 67: 6400–6408 [DOI] [PubMed] [Google Scholar]
- Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH 2007. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J 26: 4402–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou L, Cheng X 2000. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J 19: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, Selker EU, Cheng X 2002. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell 111: 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X 2003. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell 12: 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, et al. 2009. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet 5: e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, et al. 2010. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 38: 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al. 2009. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol 16: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S 2008. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]