Embryonic stem cells are maintained in a self-renewing and pluripotent state by multiple regulatory pathways. Hirsch et al. performed a functional RNAi screen and identified components of the SAGA histone acetyltransferase complex, in particular Gcn5, as critical regulators of reprogramming initiation. In mouse pluripotent stem cells, Gcn5 strongly associates with Myc, and, upon initiation of somatic reprogramming, Gcn5 and Myc form a positive feed-forward loop that activates a distinct alternative splicing network and the early acquisition of pluripotency-associated splicing events.
Keywords: Gcn5, SAGA, Myc, iPSCs, reprogramming, alternative splicing
Abstract
Embryonic stem cells are maintained in a self-renewing and pluripotent state by multiple regulatory pathways. Pluripotent-specific transcriptional networks are sequentially reactivated as somatic cells reprogram to achieve pluripotency. How epigenetic regulators modulate this process and contribute to somatic cell reprogramming is not clear. Here we performed a functional RNAi screen to identify the earliest epigenetic regulators required for reprogramming. We identified components of the SAGA histone acetyltransferase complex, in particular Gcn5, as critical regulators of reprogramming initiation. Furthermore, we showed in mouse pluripotent stem cells that Gcn5 strongly associates with Myc and that, upon initiation of somatic reprogramming, Gcn5 and Myc form a positive feed-forward loop that activates a distinct alternative splicing network and the early acquisition of pluripotency-associated splicing events. These studies expose a Myc–SAGA pathway that drives expression of an essential alternative splicing regulatory network during somatic cell reprogramming.
Somatic cell reprogramming by ectopic expression of the transcription factors Oct4, Sox2, Klf4, and Myc (OSKM) offers limitless potential to capture patient-specific induced pluripotent stem cells (iPSCs) for therapeutic purposes as well as to facilitate drug screening and disease modeling (Takahashi and Yamanaka 2006; Inoue et al. 2014). However, the ability to harness this potential requires a greater understanding of the molecular mechanisms associated with reprogramming.
Primary reprogramming systems in which transgenes are introduced de novo into somatic cells are relatively inefficient, thus challenging the molecular dissection of reprogramming mechanisms, particularly in the early stages. However, secondary reprogramming systems that employ somatic cells derived from a primary iPSC generated using stably integrated, inducible OSKM transgenes yield higher-efficiency reprogramming. Induction of OSKM in secondary systems facilitated the identification of three tiered transcriptional phases during reprogramming, termed initiation, maturation, and stabilization (Woltjen et al. 2009; Samavarchi-Tehrani et al. 2010; Golipour et al. 2012; David and Polo 2014). Among these phases, initiation is largely characterized by a BMP-driven mesenchymal-to-epithelial transition (MET), increased cell growth, up-regulation of RNA processing factors, and the onset of a metabolic change toward a glycolytic state (Li et al. 2010; Samavarchi-Tehrani et al. 2010; Hansson et al. 2012; Polo et al. 2012). Conversely, the maturation phase is associated with the first wave of pluripotency gene expression and transient up-regulation of differentiation-associated genes. Complete activation of the endogenous self-sustaining pluripotency transcriptional network is ultimately achieved in the final stabilization phase that occurs upon suppression of transgene expression (Buganim et al. 2012; Golipour et al. 2012; David and Polo 2014).
The major alterations in gene expression profiles that occur in reprogramming cells are accompanied by reorganization of chromatin architecture as well as the patterns of DNA methylation (Polo et al. 2012; Apostolou and Hochedlinger 2013). Furthermore, the relative abundance of various post-translational histone modifications changes vastly from mouse embryonic fibroblasts (MEFs) to iPSCs, ultimately leading to a more euchromatic environment in pluripotent cells (Mattout et al. 2011; Sridharan et al. 2013). Decreased levels of heterochromatic histone modifications H3K9me2 and H3K9me3 are found in iPSCs, for example, relative to levels observed in MEFs (Mattout et al. 2011; Onder et al. 2012; Soufi et al. 2012; Sridharan et al. 2013). Histone-modifying enzymes that mediate these histone modification states often function in the context of large multimeric complexes that collectively modulate recruitment, substrate specificity, and enzymatic activity. The histone modifications in turn serve as platforms to recruit chromatin readers that harbor specific post-translational modification (PTM) recognition domains. Accordingly, multiple components of chromatin-modifying complexes (such as EZH2, SUZ12, and EED from the PRC2 polycomb-repressive complex) as well as readers (such as the heterochromatin protein 1 family member Cbx3) are all implicated in reprogramming (Onder et al. 2012; Sridharan et al. 2013). Moreover, knockdown of the H3K9 methyltransferases EHMT2, SUV39H1, SUV39H2, and SETDB1 (human) as well as Ehmt1, Ehmt2, and Setdb1 (mouse) improves reprogramming efficiency and facilitates activation of pluripotency genes such as Nanog (Onder et al. 2012; Soufi et al. 2012; Sridharan et al. 2013; Qin et al. 2014).
In addition to removal of heterochromatin marks, histone modifications such as histone acetylation that are associated with more open chromatin structures are gained at nearly all H3 and H4 lysines in iPSCs compared with MEFs (Sridharan et al. 2013). The histone acetyltransferase (HAT) enzymes responsible for these alterations have yet to be defined. Nonetheless, histone acetylation plays an important role in the transition of MEFs to iPSCs, as histone deacetylase inhibitors boost reprogramming efficiency in a Myc-dependent manner (Liang et al. 2010). In addition, Myc is thought to establish its transcriptional network much earlier in the reprogramming process than OSK by recruiting coactivators to enhance DNA accessibility (Sridharan et al. 2009; Polo et al. 2012). Loss of Myc in neural progenitor cells leads to histone hypoacetylation and nuclear condensation (Knoepfler et al. 2006), further suggesting that Myc is important for recruitment of HATs to induce or maintain stemness. Overall, while it is known that chromatin-modifying complexes as well as chromatin readers negotiate rearrangement of the epigenetic landscape, it is unclear how these regulatory components intersect with reprogramming factors to regulate transcriptional programs that dampen or fuel reprogramming.
In addition to changes in gene expression and histone modification patterns, cellular reprogramming is also accompanied by regulated changes in RNA splicing. Alternative splicing (AS) is associated with controlling lineage commitment, where pre-mRNA splice sites are selectively used to generate functionally disparate mature mRNA transcripts from the same gene (Irimia and Blencowe 2012). Furthermore, embryonic stem cells (ESCs) display splicing patterns that are distinct from differentiated cells and critical for maintenance of pluripotency. (Atlasi et al. 2008; Rao et al. 2010; Salomonis et al. 2010; Wu et al. 2010; Das et al. 2011; Gabut et al. 2011; Han et al. 2013; Ohta et al. 2013; Lu et al. 2014). Moreover, step-wise acquisition of ESC AS patterns is critical for successful somatic cell reprogramming (Gabut et al. 2011; Han et al. 2013; Ohta et al. 2013). Although some of the splicing factors that regulate these events—including MBNL, SFRS2, U2af1, and Srsf3—have been uncovered (Han et al. 2013; Ohta et al. 2013; Lu et al. 2014), how these AS regulatory networks are modulated during reprogramming remains to be elucidated.
In the present study, we used a doxycycline (Dox)-inducible mouse secondary reprogramming system to perform a focused RNAi screen directed toward uncovering the earliest epigenetic participants in somatic cell reprogramming. We identified Gcn5 and multiple components of SAGA as the primary HAT complex required for early reprogramming. Furthermore, our data reveal that Myc initiates a positive feed-forward loop by directly driving expression of Gcn5 as well as the SAGA component Ccdc101 within the first days of reprogramming. Myc and Gcn5 (SAGA) in turn stimulate a novel transcriptional network encoding factors associated with AS, which is distinct from the cell cycle-related genes that we show are controlled by Myc and Gcn5 in mouse ESCs (mESCs). This study thus highlights a novel interplay between epigenetic factors and transcriptional networks in early reprogramming that triggers Myc–SAGA-mediated rewiring of an AS network.
Results
A functional RNAi screen for epigenetic regulators of reprogramming initiation
We previously reported that cellular reprogramming is accompanied by a phased series of gene expression changes (Samavarchi-Tehrani et al. 2010). However, little is known about how epigenetic regulatory pathways initiate massive reorganization of the chromatin landscape that is required for the broad transcriptional alterations that underlie changes in cellular plasticity associated with reprogramming. To identify epigenetic regulators that function in the earliest stages of reprogramming, we performed a systematic RNAi screen during the initiation phase of reprogramming using a secondary MEF model. Our RNAi library contained all known histone-modifying enzymes, chromatin remodelers, histone chaperones, enzymes associated with DNA methylation, epigenetic readers, and additional components of epigenetic modifying complexes as well as family members closely related to the above (652 siRNAs) (Supplemental Fig. S1A). Control siRNAs _targeting Oct4, Sox2, Klf4, Myc, Nanog, and Smad1 were also utilized. For screening, secondary MEFs were transfected with siRNA 1 d prior to OSKM transgene induction with Dox. After 5 d, the cells were then fixed; stained for alkaline phosphatase (AP) activity, which is an early marker of pluripotency; counterstained with DAPI; and imaged by automated image analysis that quantified reprogramming based on AP and DAPI colony costaining (Fig. 1A). As expected, individual knockdown of each of the OSKM transgenes greatly impaired reprogramming, with the most significant reductions observed upon loss of Oct4, Sox2, or Myc (Fig. 1B). We performed two biological replicate screens of our epigenetic factor library and rank-ordered the average of the median center AP and DAPI colony areas (median rank variability ±5.1%) (Fig. 1C; Supplemental Fig. S1B). Given that the screen was tailored specifically to epigenetic regulators, we found that knockdown of many factors impacted reprogramming to some degree. Therefore, we set a stringent cutoff that classified only the lowest 15% from AP and DAPI colony staining (76 _targets) as key factors required for reprogramming, which is similar to the effect of Oct4 knockdown (Supplemental Fig. S1C; Supplemental Table S1). This stringently defined list concentrates factors that were previously identified as facilitators of reprogramming, such as Ezh2, Suz12, Wdr5, Sirt6, and Prmt5 (Ang et al. 2011; Nagamatsu et al. 2011; Onder et al. 2012; Sharma et al. 2013; Ding et al. 2014), thus validating our screen. However, it also included many factors that have not been previously implicated in reprogramming.
Figure 1.
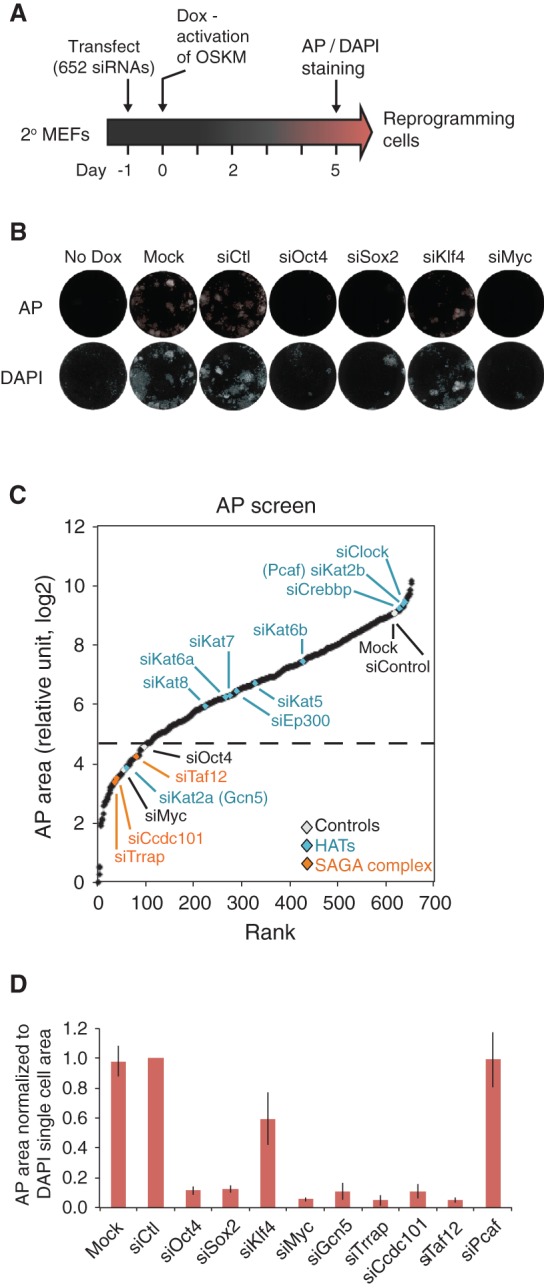
RNAi screen of epigenetic factors during the initiation phase of reprogramming. (A) Experimental representation of the functional RNAi screen. (B) Representative images from automated image analysis of AP-stained (top) and DAPI-stained (bottom) mock, siControl, siOct4, siSox2, siKlf4, and siMyc transfected cells. Stained cells are shown in white surrounded by red (AP; top) and blue (DAPI; bottom) colony masks used to quantify the stained area. (C) Result of the RNAi screen is displayed as a rank order plot of AP staining using log2 transformed values from the average area of two biological replicate experiments, each performed in duplicate. The control values are highlighted in white, while HATs are shown in blue, and hits from the SAGA complex are displayed in orange. The dotted gray line indicates the cutoff for _targets within the lowest 15%. (D) AP area normalized to DAPI single-cell area for various conditions in the RNAi screen. Normalized AP area is shown relative to siControl. Error bars indicate standard error from two biological replicate experiments performed in duplicate.
We focused our attention initially on histone-modifying enzymes with previously undefined roles in somatic cell reprogramming. Of particular note, loss of the HAT Gcn5 (encoded by Kat2a) greatly inhibited colony formation, while knockdown of the closely related enzyme Pcaf (encoded by Kat2b) had little effect (Fig. 1C; Supplemental Fig. S1B). Furthermore, loss of Tip60 (encoded by Kat5), which plays a role in ESC maintenance (Fazzio et al. 2008), or other HATs (Kat6a, Kat6b, Kat7, Kat8, or p300) had no appreciable effect on reprogramming (Fig. 1C; Supplemental Fig. S1B, blue), indicating a specific requirement for Gcn5 in this process. Gcn5 was the first transcription-related HAT enzyme identified (Brownell and Allis 1995; Brownell et al. 1996) and functions in the context of multisubunit complexes, including SAGA and ATAC (Grant et al. 1997; Martinez et al. 1998; Guelman et al. 2006, 2009; Koutelou et al. 2010). Importantly, consistent with a role for Gcn5 in reprogramming, our screen identified three other components of the SAGA complex—Trrap, Ccdc101, and Taf12—as regulators of reprogramming (Fig. 1C; Supplemental Fig. S1B, orange).
To assess whether reduced colony formation upon loss of the SAGA components was a consequence of decreased cell proliferation or survival during reprogramming, we measured the area of nonreprogramming DAPI single cells. Knockdown of Gcn5, Trrap, Ccdc101, and Taf12 as well as the reprogramming factors had variable effects on the DAPI single-cell area in reprogramming cells (Supplemental Fig. S1D) but minimal effects in nonreprogramming MEFs (Supplemental Fig. S1E), However, even after normalizing the AP area to the DAPI single-cell area, we observed a dramatic defect in reprogramming upon loss of Gcn5, Trrap, Cdcc101, and Taf12 (Fig. 1D). Furthermore, we manually validated these primary screen hits by knocking down Gcn5, Trrap, Ccdc101, and Taf12. We confirmed that at least two individual siRNAs from each pool (Supplemental Table S1) effectively suppressed expression of the _target gene that correlated with inhibition of reprogramming. We also confirmed that knockdown of Pcaf using either pools or individual siRNAs while efficiently decreasing levels of this factor did not affect reprogramming (Fig. 1D; Supplemental Table S1).
Importantly, knockdown of the SAGA components (Gcn5, Trrap, Ccdc101, and Taf12) 1 d after inducing reprogramming also compromised AP colony formation, while knockdown of these factors later in reprogramming (day 5 and day 9) and secondary iPSCs had a lesser effect (Supplemental Fig. S1F), indicating that these SAGA components have a selective function during early reprogramming. Supporting the generality and specificity of these effects, defective reprogramming caused by loss of Gcn5 was partially restored by transient expression of a siRNA-resistant Gcn5 cDNA construct (Supplemental Fig. S1G), and knockdown of Gcn5 in OSKML mRNA-mediated reprogramming of human BJ fibroblasts also suppressed AP colony formation (Supplemental Fig. S1H; Mandal and Rossi 2013). Together, these results indicate that successful transition through the initiation phase of reprogramming is dependent on Gcn5 and the SAGA complex and is conserved in mouse and human systems.
Gcn5 is a coactivator of the Myc transcriptional network in pluripotent cells
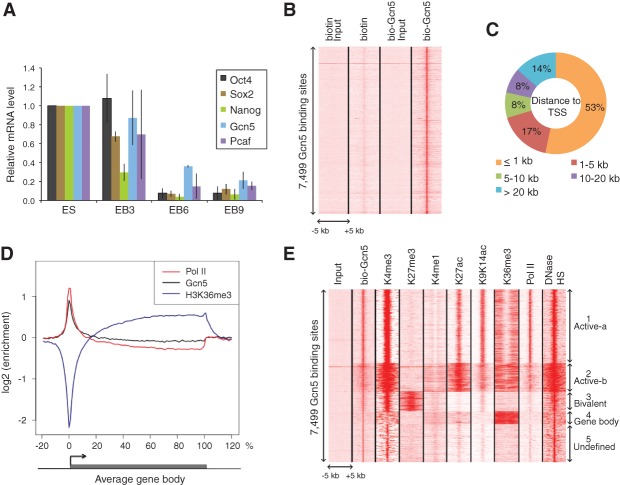
Gcn5 is essential for embryonic survival in mice (Xu et al. 2000; Yamauchi et al. 2000; Bu et al. 2007) and is highly expressed in mESCs compared with differentiating cells (Fig. 2A), but little is known of the gene expression programs regulated by Gcn5 in pluripotent cells and during development. To better address Gcn5 functions in mESCs, we preformed chromatin immunoprecipitations (ChIPs) to identify direct _target genes. Available Gcn5 antibodies proved unsuccessful in ChIP assays (data not shown). Therefore, we used an in vivo biotinylated and Flag-tagged form of Gcn5 expressed at levels similar to that of the endogenous protein (Supplemental Fig. S2A; Kim et al. 2008, 2009, 2010). Coupling biotin:streptavidin-mediated ChIPs with massive parallel sequencing (bioChIP-seq) identified 7499 common Gcn5-bound sites in duplicate experiments that were highly enriched relative to control cells (which expressed an unfused biotin construct) (Fig. 2B; Supplemental Table S2). The majority (53%) of Gcn5-bound sites were located within 1 kb of a transcription start site (TSS), and the average distribution of Gcn5 across the gene bodies correlated strongly with that of RNA polymerase II (Pol II) (Fig. 2C,D). Furthermore, when compared with publicly available mESC histone mark ChIP-seq data sets, unsupervised clustering revealed Gcn5 sites partitioned into five distinct clusters (Fig. 2E; Supplemental Table S2). The majority (59%) were enriched for modifications associated with active promoters, including H3K9/14ac, H3K27ac, H3K4me3, and Pol II, supporting a role for Gcn5 in their activation (Supplemental Fig. S2B,C). We termed these clusters Active-a and Active-b (Fig. 2E), with the latter displaying a broader distribution of active marks. Interestingly, cluster 3 was associated with bivalent marks (Supplemental Fig. S2D), while cluster 4 included transcript elongation, and cluster 5 comprised undefined intergenic sites. Altogether, these results are in accordance with a set of previous studies defining Gcn5 as a gene-specific coactivator (Lee et al. 2000; Krebs et al. 2011) rather than a global regulator of Pol II transcription (Bonnet et al. 2014).
Figure 2.
Gcn5 bioChIP-seq in pluripotent cells. (A) Gcn5 expression decreases upon mESC differentiation. Quantitative RT–PCR (qRT–PCR) was performed to measure the mRNA levels of Oct4, Sox2, Nanog, Gcn5, and Pcaf in mESCs and embryoid bodies (EBs) differentiated for 3, 6, or 9 d. Error bars indicate SD from the mean (n = 2). (B) Specific enrichment of bio-Gcn5. The heat map shows signal densities of biotin, bio-Gcn5, and their inputs at 100-bp resolution within ±5 kb of 7499 Gcn5-binding sites. (C) The majority of Gcn5 binds close to the TSS. The pie chart shows the fraction of Gcn5-binding sites within a defined distance to the TSS of the closest gene. (D) Average binding profile of Gcn5. ChIP-seq signal density enrichments over input were plotted across an average gene structure for Gcn5 (black), Pol II (red), and H3K36me3 (purple). (E) Gcn5-binding sites resolve into five distinct clusters. The heat map is based on ChIP-seq signal densities of selected histone marks (H3K4me3, H3K27me3, H3K4me1, H3K27ac, H3K9K14ac, and H3K36me3), Pol II, and DNase-hypersensitive sites (HS) at 100-bp resolution within ±5 kb of 7499 Gcn5 binding sites.
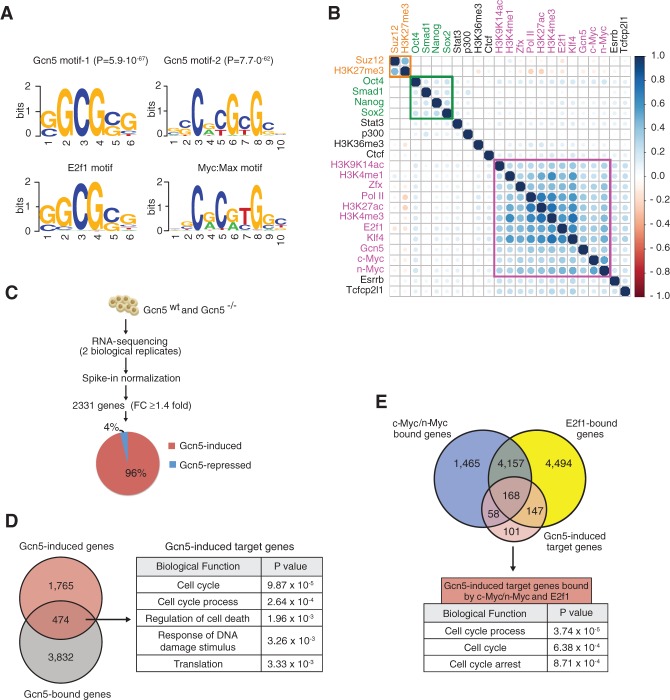
To investigate which transcription factors might recruit Gcn5 to promoters, we performed de novo motif enrichment analysis. This revealed remarkable concordance with E2f1-binding sites (P = 5.9 × 10−67) and Myc/Max-binding sites (P = 7.7 × 10−62) (Fig. 3A). No such enrichment was observed upon comparing the Gcn5-bound sites with random sequences of the same GC content and length (Supplemental Table S2). Furthermore, Gcn5-bound sites also revealed strong association with sites bound by n-Myc, c-Myc, E2f1, and H3K9/K14ac (Fig. 3B; Supplemental Table S2 for values) but not Oct4, Sox2, or Nanog (Chen et al. 2008; Rosenbloom et al. 2012). In addition, these sites primarily fell into the Active-a and Active-b clusters (Supplemental Fig. S3A,B), suggesting that Gcn5 coactivates Myc and E2f1 transcriptional networks in pluripotent cells. Indeed, when we analyzed altered gene expression by RNA sequencing (RNA-seq) in Gcn5flox/flox mESCs with and without Gcn5 deletion upon expression of Cre recombinase (Supplemental Fig. S3E,F), we identified 2239 genes that were down-regulated and only 92 genes that were up-regulated upon Gcn5 loss (Fig. 3C; Supplemental Table S3; Loven et al. 2012). Of these, 474 genes were direct _targets of Gcn5, as they were also identified as Gcn5-bound in our ChIP experiments. These genes are mainly involved in cell cycle regulation (Fig. 3D) and are also mostly Myc/E2f1 _targets (Fig. 3E; Supplemental Fig. S3G). Interestingly, Gcn5−/− mESCs display no obvious abnormalities and are capable of differentiating into three germ layers (Lin et al. 2007). Furthermore, they stably express pluripotency markers and show normal cellular morphology and growth kinetics (Supplemental Fig. S3H,I). Together with our functional screen (Fig. 1), these results indicate that Gcn5 is necessary to establish, but not maintain, pluripotency.
Figure 3.
Gcn5 is part of the Myc regulatory network in mESCs. (A) E2f1 and Myc:Max motifs are enriched at Gcn5-binding sites. (Top) The DNA-binding motifs identified within 300 base pairs (bp) of the top 1000 Gcn5 peak summits with associated P-values. (Bottom) Myc:Max and E2f1 consensus motif sequences obtained from the TRANSFAC database. (B) Gcn5 clusters with components of the Myc stem cell regulatory network (purple). Unsupervised hierarchical clustering of selected histone marks, transcription factors (TFs), and Gcn5. The Pearson correlation matrix is graphically displayed using the corrplot package in R and represents _target similarity. The circle area demonstrates the absolute value of the corresponding correlation coefficients. The color scale indicates whether the correlation is positive (blue) or negative (red). The green and orange boxes encompass components of the core pluripotency network and the polycomb network, respectively. (C) Schematic of RNA-seq analysis. Expression values in wild-type and Gcn5−/− mESCs were normalized to spike-in standards. Differentially expressed genes were identified based on fold change of normalized expression values in wild-type versus Gcn5−/− mESCs. The pie chart indicates the fraction of Gcn5-induced and Gcn5-repressed genes. (D) Genes directly regulated by Gcn5 in mESCs. (Left) The Venn diagram indicates the overlap between Gcn5-bound genes and Gcn5-induced genes. (Right) The top five gene ontology (GO) terms of Gcn5 direct _target genes. (E, top) The Venn diagram indicates that Gcn5-induced _target genes overlap strongly with c-Myc-bound or n-Myc-bound and E2f1-bound genes in mESCs. (Bottom) The top three GO terms of Gcn5-induced _target genes bound by c-Myc/n-Myc and E2f1.
Myc directly activates early expression of SAGA components during reprogramming
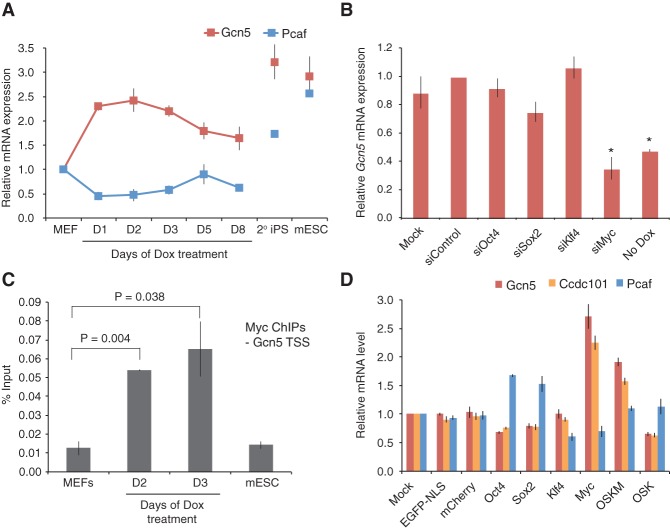
Our ChIP-seq analyses indicate that Gcn5 is recruited to Myc _target genes in mESCs, raising the possibility that Gcn5 might also engage with Myc during somatic cell reprogramming. To further address how Gcn5 contributes to reprogramming, we monitored Gcn5 expression during this process and found that Gcn5, but not Pcaf, mRNA levels were rapidly induced upon expression of OSKM (Fig. 4A). Furthermore, siRNA depletion of individual transgenes during the first 2 d of reprogramming revealed that Myc was primarily responsible for the early spike-in Gcn5 expression (Fig. 4B; Supplemental Fig. S4C), and ChIP analysis of Myc binding to the TSS of Gcn5 revealed a strong increase upon induction of reprogramming (Fig. 4C). In addition to Gcn5, Ccdc101 mRNA levels also increased abruptly after just 1 d of reprogramming (Supplemental Fig. S4A), and Myc binding to the TSS of this gene was increased similar to the increase in binding observed at Gcn5 (Supplemental Fig. S4B–D). In contrast, Trrap and Taf12 levels remained constant during reprogramming. To further determine whether OSK also had the potential to activate Gcn5 and Ccdc101 expression in the absence of Myc, we overexpressed the individual reprogramming factors as well as an OSK cocktail in wild-type MEFs by lentiviral infection (Supplemental Fig. S4E). In agreement with the above data, infection with Myc alone, but not OSK, was sufficient to up-regulate Gcn5 and Ccdc101 expression comparable with levels detected during early reprogramming (Fig. 4D; Supplemental Fig. S4F). Conversely, Myc inhibited Pcaf expression. Interestingly, our analysis in mESCs also revealed that Gcn5 as well as other components of the SAGA complex (Ccdc101, Taf12, and Atxn7l3) are direct _targets of Myc (see Supplemental Fig. S3C,D for examples of Gcn5 and Ccdc101). Collectively, these data suggest that Myc drives a Gcn5-SAGA feed-forward loop early in reprogramming by directly regulating expression of SAGA components and recruiting SAGA proteins for activation of Myc _target genes.
Figure 4.
Myc up-regulates Gcn5 expression levels during early somatic cell reprogramming. (A) Gcn5 mRNA expression increases during reprogramming. qRT–PCR quantification of Gcn5 mRNA levels across a time course of Dox-inducible reprogramming in secondary (2°) MEFs. (D) Days of Dox treatment. Error bars indicate SD from the average of four independent experiments. (B) Myc up-regulates Gcn5 mRNA expression during reprogramming. Secondary MEFs were transfected with siControl, siOct4, siSox2, siKlf4, and siMyc or under mock conditions 1 d prior to Dox exposure. Gcn5 mRNA levels were analyzed 2 d following Dox induction. Asterisks indicate t-test P value < 0.01 relative to siControl. Error bars indicate SD from the average of three independent experiments. (C) Myc binds the TSS of Gcn5. ChIP-qPCR was performed using c-Myc antibody and primers immediately upstream of the Kat2a (Gcn5) TSS in mESCs cells and secondary reprogramming MEFs cultured in the absence or presence of Dox for 2 or 3 d. (D) Days of Dox treatment. Error bars indicate SD from the average of two representative data sets. (D) Overexpression of Myc is sufficient to up-regulate Gcn5 and Ccdc101 mRNA levels. EGFP, mCherry, Oct4 (O), Sox2 (S), Klf4 (K), or c-Myc (M) was introduced into wild-type MEFs by lentiviral infection. Three days later, mRNA levels were analyzed. Error bars indicate SD from the average of three independent experiments.
Myc and Gcn5 cooperate to activate a network of RNA processing genes during initiation of reprogramming
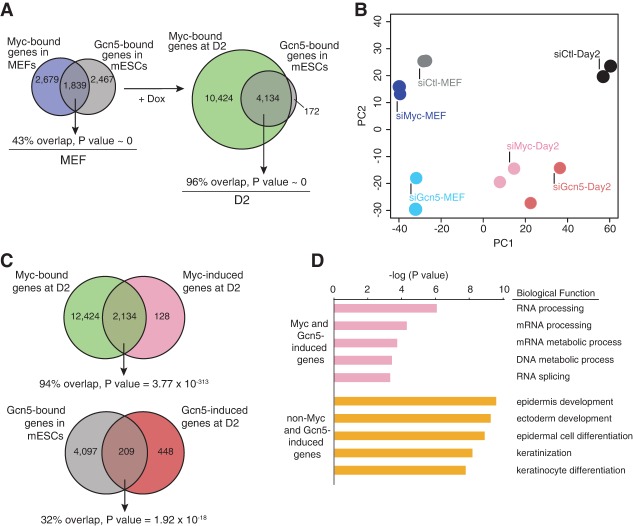
The requirement for Gcn5 and, more specifically, the SAGA complex during the initiation phase suggests that a Myc–SAGA-mediated regulatory module is one of the earliest epigenetic mechanisms activated during reprogramming. To directly evaluate the Myc–Gcn5 axis, we next performed Myc ChIP-seq on secondary MEFs grown for 2 d in the absence (MEFs) or presence (day 2 [D2]) of Dox. As expected, Myc bound to more genes in D2 reprogramming cells (∼3.2-fold more) than in MEFs and bound a number of genes comparable with that observed previously in early human reprogramming cells (Supplemental Fig. S5A; Soufi et al. 2012). Strikingly, the Myc-bound genes in D2 reprogramming cells (96%) incorporated virtually all genes bound by Gcn5 in mESCs (Fig. 5A). In stark contrast, the overlap was much less in nonreprogramming MEFs (43%). Furthermore, c-Myc coimmunoprecipitation experiments demonstrated a physical interaction between Gcn5 and c-Myc in both reprogramming D2 cells and mESCs (Supplemental Fig. S5B). These data suggest that Myc and Gcn5 cooperate to regulate gene expression profiles early in the reprogramming process through physical interaction. To determine how this interaction contributes to establishment of pluripotency, we depleted either Gcn5 or Myc in secondary MEFs and D2 reprogramming cells, performed duplicate RNA-seq analyses (Supplemental Fig. S5C,D; Supplemental Table S4), and globally compared gene expression profiles by principal component analysis (PCA). These comparisons revealed that the gene expression profiles in Myc versus Gcn5 knockdown MEFs were distinct (Fig. 5B, blue dots). In contrast, in D2 reprogramming MEFs, Myc or Gcn5 knockdown profiles were strikingly similar to each other but were distinct from D2 siControl cells (Fig. 5B, pink dots). Indeed, of 2246 genes that were dependent on Gcn5 during reprogramming, 78% (1760) were also Myc-responsive (Supplemental Fig. S5E). Collectively, these results indicate that in MEFs, Myc and Gcn5 have distinctive roles in maintaining the global gene expression program but that during reprogramming, they form a tightly coupled functional module that launches a transcriptional program required for acquisition of pluripotency.
Figure 5.
Gcn5 and Myc coregulate a group of RNA splicing and RNA processing genes during early reprogramming. (A) The overlap between Myc- and Gcn5-bound genes increases during reprogramming. (Left) The Venn diagram indicates the overlap between Myc-bound genes in MEFs and Gcn5-bound genes in mESCs. (Right) The Venn diagram depicts genes bound by Myc at D2 of reprogramming and Gcn5-bound genes in mESCs. (B) Loss of Gcn5 and Myc in reprogramming cells generates similar gene expression profiles. Principal component projections of MEFs and D2 reprogramming cells upon knockdown of Myc and Gcn5, colored by their specific conditions (n = 2). (C) Myc and Gcn5 directly regulate a significant portion of their _target genes during early reprogramming. (Top) The Venn diagram shows the overlap between Myc-induced genes and Myc-bound genes at D2 of reprogramming. (Bottom) The overlap between D2 Gcn5-induced genes and genes bound by Gcn5 in mESCs is displayed as a Venn diagram with the associated P-values. (D) Biological functional annotation of RNA-seq analysis for the top 200 Myc/Gcn5-induced and non-Myc/Gnc5-induced genes using the DAVID functional annotation tool.
We next identified reprogramming-specific transcriptional programs governed by the Myc–Gcn5 module. For this, we identified genes specifically ≥1.4-fold up-regulated by Myc and Gcn5 during reprogramming but not in MEFs. This revealed 2262 and 657 genes up-regulated by Myc and Gcn5, respectively. Of these Myc reprogramming-responsive genes, 94% were also directly bound by Myc, indicating that almost the entire program is a direct _target (Fig. 5C, top). Interestingly, although technical challenges of performing bioChIP in reprogramming cells prevented an analysis of Gcn5 genome occupancy, analysis of our mESC results revealed that 32% of Gcn5-dependent genes are occupied by Gcn5 in mESCs (Fig. 5C, bottom). Since almost all Gcn5-bound genes in mESCs are occupied by Myc during reprogramming (Fig. 5A), these data suggest that Myc and Gcn5 are tightly coupled early during reprogramming.
We next analyzed Myc–Gcn5 _targets in reprogramming cells by gene ontology (GO) analysis, which revealed a strong enrichment for RNA processing and RNA splicing among the top functional categories (Fig. 5D; Supplemental Table S4). Among these genes are several general splicing factors, including Snrpd1, U2af1, Isy1, Skiv2l2, Prpf4, Pnn, and Snrpg. In accordance, genes cobound by Myc and Gcn5 were also enriched for these categories (Supplemental Fig. S5F). Importantly, this group of genes was distinct from non-Myc/Gcn5-induced genes, which were enriched for MET genes (functional categories were epidermis development, ectoderm development, epidermal cell differentiation, and keratinization) that are regulated by Klf4 and BMP–Smad signaling (Fig. 5D; Supplemental Table S4; Mikkelsen et al. 2008; Li et al. 2010; Samavarchi-Tehrani et al. 2010; O'Malley et al. 2013). Furthermore, genes bound by Myc and up-regulated by Myc at D2 of reprogramming (Fig. 5C, top) were also incredibly enriched for RNA processing functions (P value = 1.62 × 10−89), while Myc-suppressed genes were enriched for skeletal system development and cell adhesion-related processes (Supplemental Table S4). These findings imply that Myc may play a more direct role in stimulating RNA processing events than cellular proliferation during early reprogramming. We also assessed whether disruption of the other SAGA components identified in our screen interfered with expression of a panel of splicing and/or RNA processing-associated genes affected by Myc/Gcn5 induction. Indeed, loss of either Trrap, Ccdc101, or Taf12 disrupted induction of the majority of genes tested (Supplemental Fig. S5G). These results demonstrate that Myc and Gcn5 cooperate to control the expression of key splicing and RNA processing genes during reprogramming.
Myc–Gcn5-regulated splicing factors are necessary for reprogramming
To determine whether any of the RNA splicing factors directly regulated by Myc and Gcn5 are functionally required for reprogramming, we performed a small-scale RNAi screen _targeting these factors in early reprogramming cells and performed a parallel screen in mESCs to distinguish factors required for establishment versus maintenance of pluripotency. Our library included siRNAs _targeting 20 RNA processing genes expressed in MEFs and mESCs and directly coregulated by Myc and Gcn5 as well as three additional RNA processing genes that were not identified as _targets of Myc and Gcn5 (Larp4, Celf1, and Hnrnpa1). Also, siRNAs _targeting Gcn5, Myc, and Oct4 were used as controls, along with a non_targeting siControl. The establishment screen was performed in reprogramming cells similar to the epigenetic regulator screen (Fig. 1A), while, in the maintenance screen, mESCs were transfected with siRNAs and fixed after 2 d in culture (Fig. 6A). We performed three biological replicates and analyzed the average relative AP and DAPI colony area as above. As expected, knockdown of Gcn5 or Myc suppressed colony formation selectively in reprogramming cells compared with mESCs, whereas Oct4 knockdown prevented colony formation in both reprogramming cells and mESCs (Fig. 6B; Supplemental Fig. S6A–C). To define RNA processing genes that were required for the initiation phase of reprogramming, similar to Gcn5, a cutoff for the lowest 15% based on AP area was set (Fig. 6B, bottom dotted black line). This analysis revealed a total of 13 splicing/RNA processing genes coregulated by Myc and Gcn5 as mediators of reprogramming and hence associated with the establishment of pluripotency. The requirement for each of these genes was further validated using the four individual siRNAs that made up the pools, and all except Naa38 were confirmed as hits based on at least two individual siRNAs inhibiting expression of the _target gene and abrogating reprogramming (Supplemental Table S5). Conversely, the cutoff for the mESC screen was set at 25% AP area reduction (similar to siOct4) compared with siControl transfected cells (Fig. 6B, top dotted black line). This revealed that six of the RNA processing factors implicated in early reprogramming were also necessary to maintain pluripotent cell growth (Utp6, U2af1, Isy1, Snrpd1, Phf5a, and Snrpg). These proteins thus play multifaceted roles in pluripotency, whereas the six remaining RNA processing factors identified as Myc–Gcn5 _targets are primarily associated with inducing pluripotency (Tra2b, Prpf4, Snrnp70, Hnrnpc, Skiv2l2, and Pnn). Together, these data reveal that the Myc–SAGA module regulates a network of RNA splicing and processing factors that is required for reprogramming during early initiation.
Figure 6.
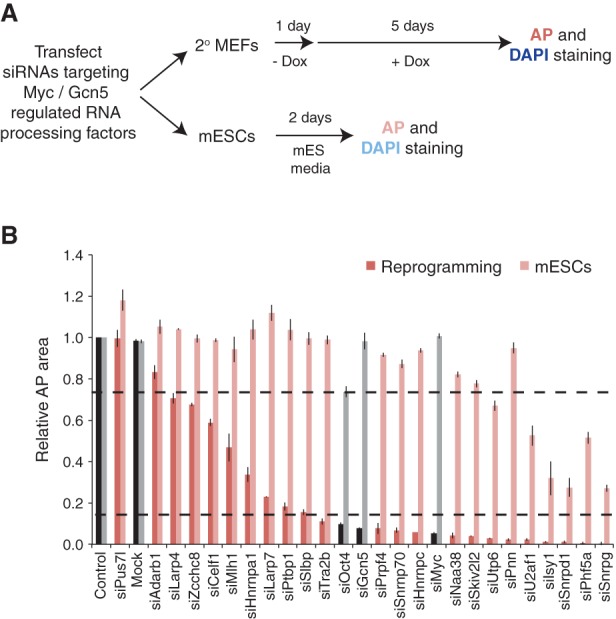
RNA processing factors regulated by Myc and Gcn5 are needed for reprogramming. (A) Schematic of RNAi screens performed in secondary (2°) reprogramming MEFs and mESCs. (B) Results of the RNAi screens performed in reprogramming cells (dark red) and mESCs (light red) are displayed and plotted by rank order of relative AP area compared with siControl cells. All controls samples are displayed as black bars for the reprogramming screen and gray for the mESC screen. The black dotted lines indicate cutoffs for the reprogramming (bottom) and mESC (top) screens. Error bars indicate the SD from three independent experiments.
The Myc-SAGA regulated splicing program _targets cell migration
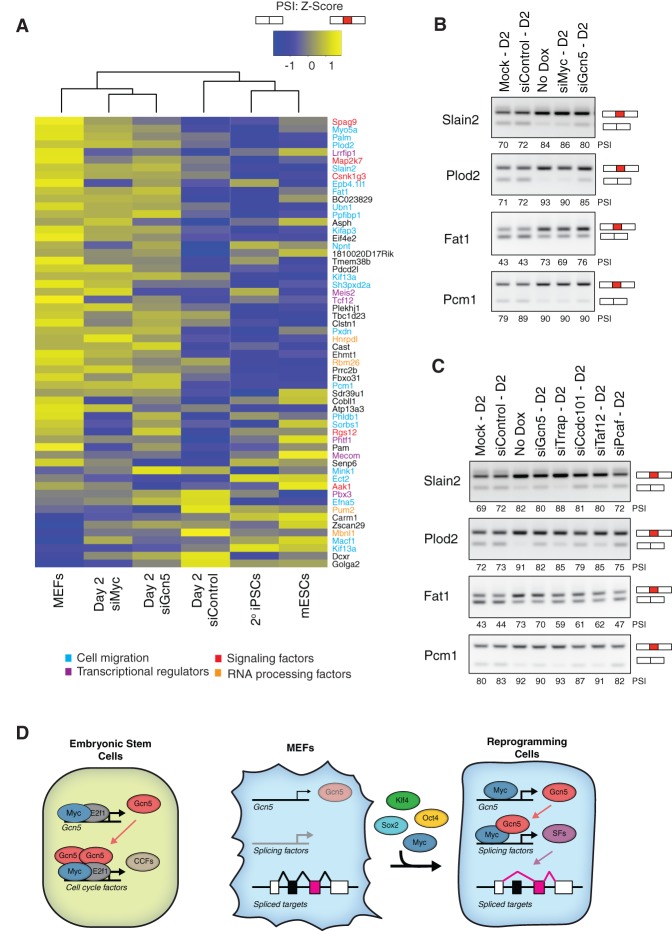
Regulated AS has recently emerged as an important event in the control of stem cell pluripotency and somatic cell reprogramming (Gabut et al. 2011; Han et al. 2013; Ohta et al. 2013; Venables et al. 2013; Lu et al. 2014). Our data suggest that Myc–SAGA-driven expression of a splicing-associated network is critical early in reprogramming to modulate AS events. To explore this possibility, we first assessed whether knockdown of Gcn5 or Myc impacted alternative exon splicing within the first 2 d of reprogramming. For this, we quantified AS events using our RNA-seq data from secondary MEFs and D2 reprogramming cells in which Myc or Gcn5 was knocked down by siRNAs (Supplemental Fig. S5C). Splicing levels were quantified by measuring the percentage of transcripts with the alternative exon spliced in (percent spliced in [PSI]). In addition, AS patterns were monitored in mESCs and matched secondary iPSCs. We identified 59 differential AS events that changed by a PSI value ≥15% by D2 of reprogramming when compared with MEFs, with the majority of events (48) associated with exon exclusion during reprogramming (Fig. 7A; Supplemental Fig. S7A; Supplemental Table S6). In particular, genes that mediate cell migration—including such processes as cytoskeletal organization, cellular polarization, and cell adhesion (Fig. 7A, blue)—were identified among these early reprogramming AS events, while signaling factors (Fig. 7A, red), transcriptional regulators (Fig. 7A, purple), and RNA processing factors (Fig. 7A, orange) were less represented. Myc and/or Gcn5 modulated 27 out of the 59 AS events within 2 d of reprogramming (Supplemental Fig. S7B), and, by unsupervised clustering, we noted that Myc and Gcn5 knockdown reprogramming cells corresponded better with nonreprogrammed secondary MEFs than D2 reprogramming cells (Fig. 7A). Interestingly, the latter clustered with mESCs and secondary iPSCs, indicating that Myc–Gcn5 induces a pluripotency-associated AS program at an early stage during reprogramming. Furthermore, we observed that Myc and Gcn5 coregulated a number of reprogramming-specific AS events associated with exon exclusion (Fig. 7A; Supplemental Fig. 7A; Supplemental Table S6), six of which are associated with genes involved in cell migration and are necessary for reprogramming (Slain2, Plod2, Fat1, Pxdn, Myo5a, and Pcm1) (Supplemental Fig. S7C).
Figure 7.
Gcn5 and Myc mediate an AS program during early reprogramming. (A) Heat map of Z-scores from PSI values in MEFs and D2 reprogramming cells transfected with siRNAs _targeting Myc and Gcn5 and a non_targeting control. The average of two samples sets is plotted for 59 splicing events. The scale indicates high (yellow) to low (blue) PSI values. The biological functions of select genes are highlighted by color. (B,C) RT–PCR assays monitoring mRNA splicing levels of Slain2, Plod2, Fat1, and Pcm1 in MEFs and D2 reprogramming cells after knockdown of Myc or Gcn5 (B) or knockdown of Gcn5, Trrap, Ccdc101, Taf12, or Pcaf (C). Semiquantitative PSI values are displayed. The presence of the red exon denotes exon inclusion. Representative images from three independent experiments are shown. (D) Gcn5 and Myc cooperate to initiate a sequence of events centered on RNA splicing in reprogramming cells. (Left) In mESCs, Myc and E2f1 stimulate a feed-forward circuit by enhancing Gcn5 levels so that Gcn5 may occupy cell cycle factor (CCF)-related genes with Myc and E2f1. (Middle and right) At the onset of reprogramming, Myc interacts with the TSS of Gcn5 to stimulate Gcn5 expression and facilitate a positive feed-forward loop. Myc associates with Gcn5 in reprogramming cells to up-regulate the expression levels of splicing factors (SFs). The splicing factors mediate AS, primarily exon exclusion, to advance somatic cell reprogramming.
AS of the Slain2 and Plod2 genes was recently described in somatic cell reprogramming and has been implicated in cytoplasmic microtubule elongation and collagen cross-link stabilization, respectively (van der Slot et al. 2003; van der Vaart et al. 2011; Han et al. 2013; Ohta et al. 2013; Venables et al. 2013). However, the mechanism that contributes to AS of these genes has not been described. Interestingly, we identified Slain2 and Plod2 as the two most differentially spliced genes coregulated by Myc and Gcn5 that functioned in reprogramming. To validate the involvement of Gcn5 and Myc in these AS events, RT–PCR was performed using primers bracketing the AS exons in Slain2 and Plod2 transcripts, and PSI values were quantified. Furthermore, we monitored splicing of two additional genes linked to cell migration with previously undescribed roles in reprogramming: Fat1 and Pcm1 (Moeller et al. 2004; Ge et al. 2010). Comparison of PSI values for these four genes confirmed that exon skipping was more prevalent in D2 reprogramming cells compared with MEFs and was altered to variable degrees upon loss of Myc or Gcn5 (Fig. 7B). To determine whether exon exclusion at these genes was also affected by loss of other components of the SAGA complex identified in our screen as regulators of reprogramming (Fig. 1C,D), we knocked down Trrap, Ccdc101, and Taf12. Similar to Gcn5, these SAGA components also influenced exon exclusion of the Slain2, Plod2, Fat1, and Pcm1 genes during early reprogramming, although their ability to modulate these splicing events was quite variable, with knockdown of Pcaf having little effect (Fig. 7C). Knockdown of Trrap generally yielded the most striking changes, while Ccdc101 and Taf12 knockdowns were less severe, implying that Trrap-mediated recruitment of the SAGA complex is vital for initiating the AS network, although SAGA may retain some residual activity in the absence of the Ccdc101 and Taf12 subunits. Additionally, we confirmed that different combinations of the RNA processing factors directly regulated by Myc and SAGA also had the capacity to control these AS events (Supplemental Fig. S7D). However, RNA processing factors that control AS in reprogramming cells generally did not regulate the same events in mESCs (Supplemental Fig. S7E), suggesting that the Myc/Gcn5 axis may not be maintained in pluripotent cells but rather may serve as an immediate means to activate RNA processing in early reprogramming. Together, these results thus demonstrate that Myc cooperates with Gcn5 in the context of the SAGA complex to control expression of key genes required for regulated splicing events that _target cell migration during the first days of reprogramming.
Discussion
Reprogramming occurs through temporally distinct gene expression phases, known as initiation, maturation, and stabilization (Samavarchi-Tehrani et al. 2010; Golipour et al. 2012), which involve large-scale changes in the chromatin environment and in patterns of AS (Gabut et al. 2011; Polo et al. 2012; Han et al. 2013; Ohta et al. 2013; Sridharan et al. 2013; Venables et al. 2013). This study uncovers Gcn5 as the primary HAT required during the initiation phase of reprogramming and further implicates three components of the Gcn5-containing SAGA complex (Trrap, Ccdc101, and Taf12) in this process. Moreover, our findings reveal a positive feed-forward loop where Gcn5 and Ccdc101 are direct _targets of Myc during early reprogramming and then form a Myc–Gcn5 functional module that controls expression of genes involved in RNA processing. Downstream AS events ultimately affect genes associated with cell migration, signaling, transcriptional regulatory networks, and RNA processing factors. Surprisingly, most of these AS events are linked to genes associated with pluripotency. Of note, previous studies revealed that up-regulation of pluripotency-associated genes occurs in the later phases of reprogramming (Samavarchi-Tehrani et al. 2010; Golipour et al. 2012). Our findings here demonstrate that regulation of splicing factors and rewiring of AS networks by the Myc–Gcn5 module are among the earliest pluripotency-specific events in reprogramming (Fig. 7D).
SAGA is a gene-specific coactivator complex for Myc
Our data further demonstrate that Gcn5, but not Pcaf, is critical for somatic cell reprograming. These two highly related HATs clearly have both shared (Jin et al. 2014a,b) and unique functions (Xu et al. 2000; Yamauchi et al. 2000). Why particular SAGA components, including Gcn5, Trrap, Ccdc101, and Taf12, are necessary for reprogramming but others are not is intriguing. The SAGA complex comprises distinct multisubunit structural modules that mediate histone acetylation (Gcn5, Ada2b, Ada3, and Ccdc101), transcription activation (Trrap, Spt3, Spt7l, and Spt20 as well as the general transcription factors Taf5l, Taf6l, Taf9, Taf10, and Taf12), and histone deubiquitination (Usp22, Atxn7, Atxn7l3, and Eny2) (Koutelou et al. 2010; Samara and Wolberger 2011; Spedale et al. 2012). Based on our observations, it seems unlikely that the SAGA deubiquitination module has an impact on reprogramming. However, recruitment of the SAGA complex with an intact HAT module appears to be key. Trrap and Gcn5 have both been reported to interact directly with Myc (McMahon et al. 1998, 2000; Zhang et al. 2014), consistent with our findings (Supplemental Fig. S5B), so these factors likely negotiate recruitment of the SAGA complex to Myc _target genes. In addition, the bromodomain of Gcn5 and/or the tudor domain of Ccdc101 may aid in docking the complex to acetylated lysine residues and H3K4me2/3 modifications, respectively (Li and Shogren-Knaak 2009; Bian et al. 2011). Previous studies in yeast and mammalian cells indicated that SAGA is recruited to promoter regions by sequence-specific binding proteins, where it increases acetylation of H3 and other factors to drive expression of select genes (Wang et al. 1997; Nagy et al. 2010). More recent work has suggested a general role for the SAGA complex in transcription, as loss of ADA3 or ATXN7L3 leads to alterations in H3K9ac or H2Bub, respectively, at all active Pol II-driven genes in HeLa cells (Bonnet et al. 2014). However, Bonnet et al. (2014) did not directly examine GCN5 or PCAF recruitment. Our ChIP-seq and RNA-seq data in mESCs clearly indicate that Gcn5 is recruited to and activates a subset of active genes. Rather than acting as a general transcription factor, most Gcn5-binding sites were located near the TSS of select genes and were colocalized with active histone modifications specifically at those sites.
The role of Myc in gene activation has also been controversial. Although Myc and its partners clearly bind to E-box sequences, global gene expression studies suggested that Myc serves as an amplifier of all active genes (Lin et al. 2012; Nie et al. 2012). A more recent study concluded that global amplification of gene expression might be secondary to the effects of Myc in regulating specific gene _targets, especially those related to cell growth and division (Sabo et al. 2014). Our data are consistent with previous studies that indicated a role for Myc and E2f1 in SAGA complex recruitment to specific _target genes in transformed cells (McMahon et al. 1998, 2000; Zhang et al. 2014). Furthermore, Myc facilitates global maintenance of active acetylated chromatin and recruits Gcn5 to cell cycle-related genes in neural stem cells (Knoepfler et al. 2006; Martinez-Cerdeno et al. 2012). Altogether, these studies demonstrate the close connections between Gcn5 and Myc in both normal and transformed cells.
Gcn5 is required for establishing pluripotency
Our studies also uncovered a specific requirement for Gcn5 in the early initiation of a pluripotency-associated AS program but little if any role in the maintenance of pluripotency, as Gcn5−/− mESCs remain pluripotent with no obvious morphological or growth defects (Lin et al. 2007). This apparent contradiction is quite interesting. There is precedence for such effects, as knockout of Utx or the Tet dioxygenases has no effect on pluripotency or self-renewal of mESCs, but depletion of these factors in MEFs compromises reprogramming (Mansour et al. 2012; Welstead et al. 2012; Hu et al. 2014). Pcaf might at least partially compensate for Gcn5 loss in mESCs, as deletion of Pcaf in Gcn5-null mice leads to more severe developmental defects than deletion of Gcn5 alone (Xu et al. 2000; Yamauchi et al. 2000). Alternatively, Gcn5 may simply play a more pivotal role in mediating cellular plasticity and hence pluripotent induction rather than the pluripotent state per se.
Myc–SAGA regulate a distinct RNA processing network during reprogramming
Temporal proteomic profiling of reprograming cells indicates that RNA processing factors are robustly up-regulated early in reprogramming, but the mechanism underlying this increase is not clear (Hansson et al. 2012). Subsequently, the RNA-binding proteins MBNL1, MBNL2, U2af1, and Srsf3 have emerged as important regulators of AS during reprogramming (Han et al. 2013; Ohta et al. 2013). Our studies now reveal that the Myc–SAGA module directly activates expression of U2af1. Moreover, we identified six additional RNA processing/splicing factor genes (Tra2b, Prpf4, Snrnp70, Hnrnpc, Skiv2l2, and Pnn) similarly coregulated by Myc and SAGA that are necessary for inducing pluripotency, further indicating that Myc and SAGA nucleate regulation of a RNA splicing pathway that is distinct from that found in MEFs. Moreover, we found that genes induced by Myc and Gcn5-SAGA regulate AS of a number of genes associated with cell migration, including Slain2, Plod2, Fat1, Pxdn, Myo5a, and Pcm1. These results are consistent with previous reports that Plod2 and Slain2 are alternatively spliced during reprogramming (Han et al. 2013; Ohta et al. 2013). Interestingly, the longer isoform of Plod2 decreased during reprogramming increases collagen cross-links (Mercer et al. 2003; Walker et al. 2005). Therefore, it is tempting to speculate that Myc and SAGA regulate an AS gene network that ultimately mediates reorganization of cellular structures and migration to facilitate pluripotency. Future studies will be aimed at dissecting the functionality of these AS events and their mechanistic roles in establishing pluripotency.
The ability of Myc to trigger a positive feed-forward loop and couple with SAGA to modulate a novel set of AS events defines a second key pathway within the initiation phase that is entirely distinct from the BMP-driven MET mediated by Klf4 and Smad1 (Li et al. 2010; Samavarchi-Tehrani et al. 2010). These studies not only reveal a new role for Myc in boosting reprogramming potential but also convey that reprogramming factors carry out distinct functions that are collectively required for successful reprogramming to a pluripotent state.
In summary, our studies highlight a previously undefined role for a Myc–Gcn5 feed-forward loop in driving Myc _target genes important for self-renewal in stem cells and for RNA processing and splicing during the earliest stage of somatic cell reprogramming. This pathway is distinct from but parallel to the induction of MET genes during the initiation phase of reprogramming. Moreover, our findings imply that the Myc–Gcn5 RNA processing module identified here may also play a fundamental role in Myc-driven oncogenesis. Forthcoming studies will address this important question as well as the role of other epigenetic regulators at different stages of reprogramming.
Materials and methods
Generation of stable mESCs and bioChIP assays
AB1 ESCs stably expressing in vivo biotinylated Gcn5 (BirAV5-FLBioGcn5) were generated, and bioChIPs were performed as previously described with minor modifications (Kim et al. 2009).
siRNA screening
RNAi-mediated knockdown was performed with 40 nM siRNA pools using Lipofectamine RNAiMAX (Invitrogen) in secondary 1B MEF cells or mESCs upon seeding as previously described (Samavarchi-Tehrani et al. 2010; Golipour et al. 2012). After 24 h, Dox was added and replaced daily. Five days later, cells were fixed and stained for AP activity (Vector red; AP substrate kit, Vector Laboratories) and counterstained for DAPI. All siRNAs are listed in Supplemental Table S7.
Quantitative RT–PCR (qRT–PCR)
Total RNA was isolated using the RNeasy minikit (Qiagen). RNA was converted to cDNA prior to qPCR using SYBR green PCR master mix (Roche). Primer sequences are detailed in Supplemental Table S7.
ChIP-seq
Libraries were prepared using a modified version of the Illumina TruSeq ChIP sample preparation protocol. See the the Supplemental Material for complete details. Each library (10 pM) was sequenced on an Illumina HiSeq 2000. The raw reads in ChIP-seq data sets were mapped to NCBI build 37 (University of California at Santa Cruz [UCSC] mm9) using Bowtie (version 0.12.8) (Langmead et al. 2009). Peaks were called using MACS (version 2.0.10) (Zhang et al. 2008) at a Q-value threshold of 0.05 using input as a control. The enriched regions from each biological replicate of samples were intersected with BEDTools (version 2.13.3) (Quinlan and Hall 2010) to form the final peak set for Gcn5 bioChIPs.
RNA-seq
RNA-seq libraries were prepared according to the Illumina TruSeq standard total RNA sample preparation kit (with Ribo-Zero GoldRS-122-2301). Each library (10 pM) was sequenced on an Illumina HiSeq 2000. RNA-seq reads were aligned to NCBI build 37 (UCSC mm9) using TopHat 2.0.9 (Trapnell et al. 2009). Complete details regarding spike-in normalization for the mESC RNA-seq are described in the Supplemental Material. EdgeR was applied to determine differentially expressed genes (Robinson et al. 2010).
Data for ChIP-seq and RNA-seq have been submitted to Gene Expression Omnibus; accession numbers are pending and will be available on request.
Supplementary Material
Acknowledgments
We thank Yoko Takata, Luis Coletta, Mary Walker, Dr. Louis S. Ramagli, and Kin Chan for RNA-seq and ChIP-seq library preparation as well as next-generation sequencing. We are grateful to Mikhail Bashkurov for quantifying DAPI-positive single cells, and Dr. Kadir Akdemir for his feedback on bioinformatics analysis. We thank Bushra Raj for providing valuable insight on splicing regulators that may directly regulate reprogramming-specific AS events. We acknowledge Malgosia Kownacka from the Embyonic Stem Cell Facility at Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital. We thank Dr. Jianlong Wang (Mount Sinai Hospital, New York) for the kind gift of the pEF1α BirAV5 and pEF1αFLBio plasmids. This work is supported by grants from the National Institutes of Health to S.Y.R.D. (R01067718) and W.L. (R01HG007538), the Ontario Ministry of Resources and Innovation and Stem Cell Network to J.L.W., and the Canadian Institutes for Health Research (CIHR) to B.J.B. Next-generation sequencing was performed in core facilities supported by a National Cancer Institute (NCI) Cancer Center Support Grant (CA016672) (Sequencing and Microarray Facility, Houston) and a Cancer Prevention Research Institute of Texas grant (RP120348) (Smithville).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.255109.114.
References
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. 2011. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. 2013. Chromatin dynamics during cellular reprogramming. Nature 502: 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. 2008. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells 26: 3068–3074. [DOI] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, et al. 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 30: 2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. 2014. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28: 1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Allis CD. 1995. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci 92: 6364–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851. [DOI] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, Dent SY. 2007. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol 27: 3405–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. 2012. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Das S, Jena S, Levasseur DN. 2011. Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. J Biol Chem 286: 42690–42703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Polo JM. 2014. Phases of reprogramming. Stem Cell Res 12: 754–761. [DOI] [PubMed] [Google Scholar]
- Ding X, Wang X, Sontag S, Qin J, Wanek P, Lin Q, Zenke M. 2014. The polycomb protein ezh2 impacts on induced pluripotent stem cell generation. Stem Cells Dev 23: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, et al. 2011. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell 147: 132–146. [DOI] [PubMed] [Google Scholar]
- Ge X, Frank CL, Calderon de Anda F, Tsai LH. 2010. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron 65: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. 2012. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell 11: 769–782. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650. [DOI] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR III, Washburn MP, Abmayr SM, et al. 2006. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol 26: 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, Wu J, Lill JR, Zha J. 2009. The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol Cell Biol 29: 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, et al. 2013. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature 498: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. 2012. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep 2: 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Zhang L, Mao SQ, Li Z, Chen J, Zhang RR, Wu HP, Gao J, Guo F, Liu W, et al. 2014. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14: 512–522. [DOI] [PubMed] [Google Scholar]
- Inoue H, Nagata N, Kurokawa H, Yamanaka S. 2014. iPS cells: a game changer for future medicine. EMBO J 33: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Blencowe BJ. 2012. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol 24: 323–332. [DOI] [PubMed] [Google Scholar]
- Jin Q, Wang C, Kuang X, Feng X, Sartorelli V, Ying H, Ge K, Dent SY. 2014a. Gcn5 and PCAF regulate PPARγ and Prdm16 expression to facilitate brown adipogenesis. Mol Cell Biol 34: 3746–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Zhuang L, Lai B, Wang C, Li W, Dolan B, Lu Y, Wang Z, Zhao K, Peng W, et al. 2014b. Gcn5 and PCAF negatively regulate interferon-β production through HAT-independent inhibition of TBK1. EMBO Rep 15: 1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. 2008. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132: 1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. 2009. Use of in vivo biotinylation to study protein–protein and protein–DNA interactions in mouse embryonic stem cells. Nat Protoc 4: 506–517. [DOI] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. 2006. Myc influences global chromatin structure. EMBO J 25: 2723–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY. 2010. Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, Tora L. 2011. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol Cell 44: 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704. [DOI] [PubMed] [Google Scholar]
- Li S, Shogren-Knaak MA. 2009. The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J Biol Chem 284: 9411–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. 2010. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7: 51–63. [DOI] [PubMed] [Google Scholar]
- Liang G, Taranova O, Xia K, Zhang Y. 2010. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem 285: 25516–25521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, Dent SY. 2007. Developmental potential of Gcn5−/− embryonic stem cells in vivo and in vitro. Dev Dyn 236: 1547–1557. [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. 2012. Revisiting global gene expression analysis. Cell 151: 476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, et al. 2014. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell 15: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Rossi DJ. 2013. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat Protoc 8: 568–582. [DOI] [PubMed] [Google Scholar]
- Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, Krupalnik V, Zerbib M, Amann-Zalcenstein D, Maza I, et al. 2012. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 488: 409–413. [DOI] [PubMed] [Google Scholar]
- Martinez E, Kundu TK, Fu J, Roeder RG. 1998. A human SPT3–TAFII31–GCN5–L acetylase complex distinct from transcription factor IID. J Biol Chem 273: 23781–23785. [DOI] [PubMed] [Google Scholar]
- Martinez-Cerdeno V, Lemen JM, Chan V, Wey A, Lin W, Dent SR, Knoepfler PS. 2012. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS One 7: e39456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout A, Biran A, Meshorer E. 2011. Global epigenetic changes during somatic cell reprogramming to iPS cells. J Mol Cell Biol 3: 341–350. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol 20: 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer DK, Nicol PF, Kimbembe C, Robins SP. 2003. Identification, expression, and tissue distribution of the three rat lysyl hydroxylase isoforms. Biochem Biophys Res Commun 307: 803–809. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller MJ, Soofi A, Braun GS, Li X, Watzl C, Kriz W, Holzman LB. 2004. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. EMBO J 23: 3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu G, Kosaka T, Kawasumi M, Kinoshita T, Takubo K, Akiyama H, Sudo T, Kobayashi T, Oya M, Suda T. 2011. A germ cell-specific gene, Prmt5, works in somatic cell reprogramming. J Biol Chem 286: 10641–10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, Tora L. 2010. The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible _target genes. Cell Mol Life Sci 67: 611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta S, Nishida E, Yamanaka S, Yamamoto T. 2013. Global splicing pattern reversion during somatic cell reprogramming. Cell Rep 5: 357–366. [DOI] [PubMed] [Google Scholar]
- O'Malley J, Skylaki S, Iwabuchi KA, Chantzoura E, Ruetz T, Johnsson A, Tomlinson SR, Linnarsson S, Kaji K. 2013. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature 499: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. 2012. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151: 1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Diaz A, Blouin L, Lebbink RJ, Patena W, Tanbun P, LeProust EM, McManus MT, Song JS, Ramalho-Santos M. 2014. Systematic identification of barriers to human iPSC generation. Cell 158: 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan GC, Orkin SH. 2010. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol 30: 5364–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, et al. 2012. ENCODE whole-genome data in the UCSC genome browser: update 2012. Nucleic Acids Res 40: D912–D917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. 2014. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. 2010. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci 107: 10514–10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara NL, Wolberger C. 2011. A new chapter in the transcription SAGA. Curr Opin Struct Biol 21: 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. 2010. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7: 64–77. [DOI] [PubMed] [Google Scholar]
- Sharma A, Diecke S, Zhang WY, Lan F, He C, Mordwinkin NM, Chua KF, Wu JC. 2013. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J Biol Chem 288: 18439–18447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G, Timmers HT, Pijnappel WW. 2012. ATAC-king the complexity of SAGA during evolution. Genes Dev 26: 527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, et al. 2013. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol 15: 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Slot AJ, Zuurmond AM, Bardoel AF, Wijmenga C, Pruijs HE, Sillence DO, Brinckmann J, Abraham DJ, Black CM, Verzijl N, et al. 2003. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem 278: 40967–40972. [DOI] [PubMed] [Google Scholar]
- van der Vaart B, Manatschal C, Grigoriev I, Olieric V, Gouveia SM, Bjelic S, Demmers J, Vorobjev I, Hoogenraad CC, Steinmetz MO, et al. 2011. SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J Cell Biol 193: 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, et al. 2013. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun 4: 2480. [DOI] [PubMed] [Google Scholar]
- Walker LC, Overstreet MA, Yeowell HN. 2005. Tissue-specific expression and regulation of the alternatively-spliced forms of lysyl hydroxylase 2 (LH2) in human kidney cells and skin fibroblasts. Matrix Biol 23: 515–523. [DOI] [PubMed] [Google Scholar]
- Wang L, Mizzen C, Ying C, Candau R, Barlev N, Brownell J, Allis CD, Berger SL. 1997. Histone acetyltransferase activity is conserved between yeast and human GCN5 and is required for complementation of growth and transcriptional activation. Mol Cell Biol 17: 519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. 2012. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci 109: 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. 2009. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458: 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, et al. 2010. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci 107: 5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. 2000. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet 26: 229–232. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci 97: 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Ichikawa W, Faiola F, Lo SY, Liu X, Martinez E. 2014. MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits. Biochim Biophys Acta 1839: 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.