Abstract
Background
Prostate-specific antigen (PSA) testing has limited accuracy for the early detection of prostate cancer (PCa).
Objective
To assess the value added by percentage of free to total PSA (%fPSA), prostate cancer antigen 3 (PCA3), and a kallikrein panel (4k-panel) to the European Randomised Study of Screening for Prostate Cancer (ERSPC) multivariable prediction models: risk calculator (RC) 4, including transrectal ultrasound, and RC 4 plus digital rectal examination (4+DRE) for prescreened men.
Design, setting, and participants
Participants were invited for rescreening between October 2007 and February 2009 within the Dutch part of the ERSPC study. Biopsies were taken in men with a PSA level ≥3.0 ng/ml or a PCA3 score ≥10. Additional analyses of the 4k-panel were done on serum samples.
Outcome measurements and statistical analysis
Outcome was defined as PCa detectable by sextant biopsy. Receiver operating characteristic curve and decision curve analyses were performed to compare the predictive capabilities of %fPSA, PCA3, 4k-panel, the ERSPC RCs, and their combinations in logistic regression models.
Results and limitations
PCa was detected in 119 of 708 men. The %fPSA did not perform better univariately or added to the RCs compared with the RCs alone. In 202 men with an elevated PSA, the 4k-panel discriminated better than PCA3 when modelled univariately (area under the curve [AUC]: 0.78 vs 0.62; p = 0.01). The multivariable models with PCA3 or the 4k-panel were equivalent (AUC: 0.80 for RC 4+DRE). In the total population, PCA3 discriminated better than the 4k-panel (univariate AUC: 0.63 vs 0.56; p = 0.05). There was no statistically significant difference between the multivariable model with PCA3 (AUC: 0.73) versus the model with the 4k-panel (AUC: 0.71; p = 0.18). The multivariable model with PCA3 performed better than the reference model (0.73 vs 0.70; p = 0.02). Decision curves confirmed these patterns, although numbers were small.
Conclusions
Both PCA3 and, to a lesser extent, a 4k-panel have added value to the DRE-based ERSPC RC in detecting PCa in prescreened men.
Patient summary
We studied the added value of novel biomarkers to previously developed risk prediction models for prostate cancer. We found that inclusion of these biomarkers resulted in an increase in predictive ability.
Keywords: Percentage of free to total PSA, Kallikrein panel (4k-panel), Prostate biopsy, Prostate cancer, Prostate cancer antigen 3 (PCA3), Prostate cancer risk calculator, Validation
1. Introduction
Prostate-specific antigen (PSA) testing is the mainstay of early detection of prostate cancer (PCa) [1]. However, PSA has limited specificity and sensitivity in determining the presence of PCa, leading to unnecessary biopsies and the diagnosis of potentially indolent PCa [2,3]. PSA-based multivariable prediction tools have been developed to improve the prediction of having biopsy-detectable PCa. Well-known externally validated models are the European Randomised Study of Prostate Cancer (ERSPC) risk calculators (RCs) (http://www.prostatecancer-riskcalculator.com/) [4], the Prostate Cancer Prevention Trial calculator (http://deb.uthscsa.edu/URORiskCalc/Pages/calcs.jsp) [5], and the Montreal model [6]. The addition of new biomarkers to an existing prediction tool may increase accuracy. Novel and promising markers in the field of PCa include prostate cancer antigen 3 (PCA3), a noncoding messenger RNA (mRNA) highly overexpressed in PCa tissue [7,8] that can be assessed using urine obtained after digital rectal examination (DRE). A promising serum-based biomarker is the kallikrein panel (4k-panel) that consists of total PSA, free PSA (fPSA), intact PSA, and human kallikrein 2 (hK2) [9,10]. The 4k-panel has been shown to increase predictive capability compared with PSA and DRE alone.
In this study, we aimed to assess the added value of percentage of free to total PSA (%fPSA), PCA3, and the 4k-panel to the ERSPC RCs for prescreened men.
2. Methods
2.1. Participants
Participants were recruited from the Dutch part of the ERSPC study [11,12]. We included 965 men who were invited for rescreening (third, fourth, or fifth time) between October 2007 and February 2009. Serum-based PSA level and PCA3 were measured in all men. The PCA3 score is the ratio of PCA3-to-PSA mRNAs multiplied by 1000 [8]. Men with a PSA level ≥3.0 ng/ml and/or a PCA3 score ≥10 were invited to undergo a DRE, transrectal ultrasound (TRUS), and a lateral sextant biopsy. We set the cut-off for PCA3 as ≥10 to evaluate performance characteristics of the PCA3 in comparison with a biopsy indication driven by PSA values ≥3.0 ng/ml [13]. Assessed prostate volume was categorised with cut points of <30 ml, 30–50 ml, and ≥50 ml [14]. In case of a hypoechogenic lesion, a seventh biopsy was taken. Permission for the present study (ISBN 978-90-5549-653-2) was granted by the medical ethics committee, University Medical Centre Rotterdam, and the Dutch Ministry of Health.
2.2. Tests to predict prostate cancer
The PSA test (Hybritech, Beckman Coulter Inc., Fullerton, CA, USA) was carried out in a standard fashion at the clinical laboratory of the Erasmus University Medical Centre. The PCA3 test (Progensa; Gen-Probe Inc., San Diego, CA, USA) was done at the laboratory of experimental urology at Radboud University Nijmegen Medical Centre. Measurements of the 4k-panel, consisting of four markers (total PSA, fPSA, intact PSA, and hK2), were performed in the Department of Laboratory Medicine at Lund University (Malmö, Sweden) on stored serum samples [15]. Separate marker values as well as an overall 4k-panel predictor were derived using a prespecified formula (ie, the study is an independent validation of a previously specified model [9]). The formula was a mix of linear terms and nonlinear spline transformations of the four markers. A specialised pathologist (G.v.L.) handled the histologic examinations of the biopsy specimens.
2.3. Reference model
Two models from the ERSPC Rotterdam RCs (http://www.prostatecancer-riskcalculator.com/; RC 4+DRE and RC 4 including TRUS) were used as reference models. RC 4+DRE included total PSA (nanograms per millilitre), DRE (normal/abnormal), DRE-assessed volume of the prostate (<30 ml, 30–50 ml, and ≥50 ml), and whether or not there was a previous (negative) biopsy. RC 4 included total PSA (nanograms per millilitre), DRE (normal/abnormal), TRUS (normal/abnormal), TRUS-assessed prostate volume (millilitres), and whether or not there was a previous (negative) biopsy.
Both models are used for men who have previously had PSA screening and a previous biopsy, if indicated, according to the ERSPC Rotterdam screening algorithm [16]. It predicts the chance of a positive sextant biopsy and its degree of aggressiveness; the RC 4+DRE model includes information on prostate volume without the need for a TRUS [17].
2.4. Statistical analyses
The primary outcome measure was any form of PCa versus no cancer, detected by a sextant biopsy, in men with elevated PSA levels (≥3.0 ng/ml). We also assessed the predictive value of %fPSA, PCA3, and the 4k-panel in the total population and in the population with a PSA <3.0 ng/ml.
We assessed the predictive value of %fPSA, PCA3, and the 4k-panel using univariate and multivariable regression models. We refitted the original RCs, RC 4 and RC 4+DRE, to use as the reference. We subsequently refitted the models including %fPSA, PCA3, and/or the 4k-panel. We used the area under the receiver operating characteristic curve (area under the curve [AUC]) to quantify the predictive accuracy of five models: (1) the first reference model (RC 4+DRE), (2) the reference model plus PCA3, (3) the reference model plus the 4k-panel, (4) the reference model plus PCA3 and the 4k-panel, and (5) the reference model plus %fPSA. We used the original RC 4 (ie, including information from TRUS) as the second reference model and used the likelihood ratio test for differences between models.
We applied decision curve analysis [18,19] to evaluate the potential clinical usefulness of making decisions based on the models including the markers. We estimated net benefit (NB) for prediction models by summing the benefits (true-positive biopsies) and subtracting the harms (false-positive biopsies). The harms were weighted by a factor related to the relative harm of a missed cancer versus an unnecessary biopsy. This weighting was derived from the threshold probability (pt) of PCa at which a patient would opt for a biopsy. This threshold can vary between men; we used a pt between 0% and 40% [20]. The interpretation of a decision curve is straightforward; a model with the highest NB at a particular threshold should be chosen over alternative models. The NB was used to calculate the reduction in numbers of biopsies per 100 men with a PSA level ≥3.0 ng/ml [9] and/or a PCA3 score ≥10. We used the following formula: reduction in biopsy per 100 men = (ΔNB/(pt/(1 − pt))*100.
Standard statistical software was used (SPSS v.18.0, IBM Corp, Armonk, NY, USA; R version 2.15.2, R Foundation for Statistical Computing, Vienna, Austria; Stata v.12.0, StataCorp, College Station, TX, USA).
3. Results
Of 965 invited men, 721 (75%) underwent a biopsy. Overall, 163 men (17%) did not meet the PSA or PCA3 inclusion criteria, 39 (4%) could not have a biopsy because of contraindications, and 42 men (4%) refused biopsy. Records of 708 of 721 biopsied participants (98%) were complete including PCA3 and 4k-panel results.
These 708 men were invited for rescreening: 339 originated from the third, 357 originated from the fourth, and 12 originated from the fifth screening round. Participants were aged 64–75 yr at the time of the visit. A previous biopsy was taken from 206 (29%) of all participants. PCa was found in 119 (17%) of the 708 biopsied men, of whom 40 in the group of 202 men had elevated PSA levels (Table 1). A few men had an abnormal TRUS or DRE. Of 708 men, 503 had a PCA3 score ≥10 and a PSA score <3.0 ng/ml. Total PSA and PCA3 levels differed significantly between men with and without PCa (Table 1).
Table 1.
Characteristics of men rescreened in the European Randomised Study of Screening for Prostate Cancer trial
| PSA ≥3.0 ng/ml (n = 202) | Total set (n = 708) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| No cancer n = 162 (80%) | Cancer n = 40 (20%) | p value | No cancer n = 589 (83%) | Cancer n = 119 (17%) | p value | |||||
| Age, yr* | 70.3 | (68.1–72.7) | 70.2 | (68.6–72.4) | 0.98 | 70.3 | (68.1–72.5) | 70.3 | (68.4–72.3) | 0.97 |
|
| ||||||||||
| Previous biopsy | <0.01 | <0.01 | ||||||||
| No | 41 | 25% | 26 | 65% | 403 | 68% | 99 | 83% | ||
| Yes | 121 | 75% | 14 | 35% | 186 | 32% | 20 | 17% | ||
|
| ||||||||||
| Total PSA, ng/ml | 4.6 | (3.7–6.4) | 4.4 | (3.6–6.9) | 0.95 | 1.7 | (0.9–3.2) | 2.1 | (1.4–3.7) | <0.01 |
|
| ||||||||||
| DRE | 0.51 | <0.01 | ||||||||
| Normal | 133 | 82% | 31 | 77.5% | 504 | 86% | 88 | 74% | ||
| Abnormal | 29 | 18% | 9 | 22.5% | 85 | 14% | 31 | 26% | ||
|
| ||||||||||
| Volume classes DRE | 0.03 | 0.53 | ||||||||
| <30 ml | 9 | 6% | 6 | 15% | 115 | 20% | 23 | 19% | ||
| 30–50 ml | 51 | 31% | 17 | 42.5% | 263 | 45% | 60 | 50% | ||
| ≥50 ml | 102 | 63% | 17 | 42.5% | 204 | 35% | 36 | 30% | ||
|
| ||||||||||
| TRUS | 0.85 | 0.38 | ||||||||
| Normal | 155 | 96% | 38 | 95% | 573 | 97% | 114 | 96% | ||
| Abnormal | 7 | 4% | 2 | 5% | 16 | 3% | 5 | 4% | ||
|
| ||||||||||
| 4k-panel | ||||||||||
| Free PSA | 1.14 | (0.86–1.62) | 0.93 | (0.68–1.39) | 0.02 | 0.47 | (0.28–0.84) | 0.56 | (0.39–0.86) | 0.06 |
| Intact PSA | 0.42 | (0.32–0.60) | 0.40 | (0.25–0.58) | 0.40 | 0.20 | (0.12–0.34) | 0.23 | (0.16–0.39) | 0.04 |
| hK2 | 0.05 | (0.04–0.07) | 0.05 | (0.04–0.07) | 1.00 | 0.03 | (0.02–0.05) | 0.04 | (0.03–0.05) | <0.01 |
| 4k-panel score | −2.81 | (−3.37 to −2.18) | −1.69 | (−2.45 to −1.09) | <0.01 | −1.33 | (−2.27 to −0.98) | −1.28 | (−1.76 to −0.97) | 0.04 |
| Probability 4k-panel | 0.06 | (0.03–0.10) | 0.16 | (0.08–0.25) | <0.01 | 0.21 | (0.09–0.27) | 0.22 | (0.15–0.28) | 0.04 |
|
| ||||||||||
| PCA3 score† | 29.5 | (14.0–57.5) | 44.0 | (20.0–118.3) | 0.01 | 31.0 | (18.0–58.5) | 46.0 | (28.0–97.0) | <0.01 |
|
| ||||||||||
| Stage | ||||||||||
| T1C | 31 | 78% | 87 | 73% | ||||||
| T2A | 8 | 20% | 28 | 24% | ||||||
| T2B | 1 | 3% | 2 | 2% | ||||||
| T2C | 0 | 0% | 1 | 1% | ||||||
| T3A | 0 | 0% | 1 | 1% | ||||||
|
| ||||||||||
| Grade | ||||||||||
| Gleason 6 | 31 | 78% | 99 | 83% | ||||||
| Gleason 7 | 5 | 13% | 13 | 11% | ||||||
| Gleason 8 | 3 | 8% | 5 | 4% | ||||||
| Gleason 9 | 1 | 3% | 2 | 2% | ||||||
|
| ||||||||||
| Serious cancer‡ | 9 | 23% | 22 | 18% | ||||||
DRE = digital rectal examination; hK2 = kallikrein protein 2; PSA = prostate-specific antigen; TRUS = transrectal ultrasound.
Continuous variables are noted as median (interquartile range).
PCA3 score = the ratio of PCA3 to PSA messenger RNAs × 1000.
Nominal variables are noted as number and percentage.
In men with PSA levels ≥3.0 ng/ml, the 4k-panel had a higher AUC value compared with PCA3 when studied univariately (AUC: 0.78 vs 0.62; p = 0.01; Table 2; Supplementary Fig. 1–3). The multivariable models with PCA3 or the 4k-panel were equivalent (AUC: 0.80 for RC 4+DRE, 0.78 vs 0.79 for RC 4 with PCA3 and the 4k-panel, respectively).
Table 2.
Incremental enhancement in discrimination for the subgroup of 202 men rescreened in the European Randomised Study of Screening for Prostate Cancer trial with prostate-specific antigen ≥3.0 ng/ml
| Univariate | Added to original risk calculator 4* | Added to original risk calculator 4+DRE† | ||||
|---|---|---|---|---|---|---|
| C‡ | (95% CI) | C | (95% CI) | C | (95% CI) | |
| Reference value§ | 0.53 | (0.44–0.64) | 0.78 | (0.69–0.86) | 0.76 | (0.68–0.83) |
| Kallikrein panel | 0.78 | (0.69–0.85) | 0.80 | (0.71–0.87) | 0.79 | (0.71–0.86) |
| PCA3 | 0.62 | (0.52–0.73) | 0.80 | (0.71–0.87) | 0.78 | (0.70–0.85) |
| Kallikrein panel and PCA3 | 0.75 | (0.65–0.84) | 0.81 | (0.72–0.88) | 0.80 | (0.72–0.87) |
| %fPSA | 0.65 | (0.55–0.75) | 0.80 | (0.71–0.88) | 0.79 | (0.71–0.85) |
%fPSA = percentage of free to total prostate-specific antigen; CI = confidence interval; DRE = digital rectal examination; PCA3 = prostate cancer antigen 3; PSA = prostate-specific antigen.
A model including total PSA (nanograms per millilitre); DRE, normal/abnormal; and assessed DRE volume of the prostate, <30 ml, 30–50 ml, and ≥50 ml.
A model including total PSA (nanograms per millilitre); DRE, normal/abnormal; transrectal ultrasound (TRUS), normal/abnormal; and TRUS-assessed prostate volume (millilitres).
Area under the receiver operator curve.
The reference value for the univariate analysis is total PSA (nanograms per millilitre) and DRE (normal/abnormal); for the multivariate analyses, it is the original risk calculator.
In the total population, PCA3 discriminated better than the 4k-panel (univariate AUC: 0.63 vs 0.56; p = 0.05; Table 3). There was no statistically significant difference between the multivariable model with PCA3 (AUC: 0.73) versus the model with the 4k-panel (AUC: 0.71; p = 0.18). The multivariable model with PCA3 performed better than the reference model (0.73 vs 0.70; p = 0.02). A multivariable model with both markers did not perform better than the multivariable model with PCA3 alone (AUC: 0.73 vs 0.73) in the total data set. The %fPSA did not perform better univariately or added to the RCs compared with the RCs alone in the total population (Table 3).
Table 3.
Incremental enhancement in discrimination in 708 men rescreened in the European Randomised Study of Screening for Prostate Cancer trial
| Univariate | Added to original risk calculator 4* | Added to original risk calculator 4+DRE† | ||||
|---|---|---|---|---|---|---|
| C‡ | (95% CI) | C | (95% CI) | C | (95% CI) | |
| Reference value§ | 0.61 | (0.56–0.67) | 0.70 | (0.64–0.75) | 0.70 | (0.64–0.75) |
| Kallikrein panel | 0.56 | (0.50–0.61) | 0.71 | (0.65–0.76) | 0.71 | (0.65–0.76) |
| PCA3 | 0.63 | (0.58–0.69) | 0.73 | (0.67–0.78) | 0.73 | (0.67–0.77) |
| Kallikrein panel and PCA3 | 0.66 | (0.61–0.70) | 0.73 | (0.68–0.78) | 0.73 | (0.68–0.78) |
| %fPSA | 0.57 | (0.51–0.63) | 0.70 | (0.65–0.76) | 0.70 | (0.64–0.75) |
%fPSA = percentage of free to total PSA; CI = confidence interval; DRE = digital rectal examination; PCA3 = prostate cancer antigen 3; PSA = prostate-specific antigen.
A model including total PSA (nanograms per millilitre); DRE, normal/abnormal; and assessed DRE volume of the prostate, <30 ml, 30–50 ml, and ≥50 ml.
A model including total PSA (nanograms per millilitre); DRE, normal/abnormal; transrectal ultrasound (TRUS), normal/abnormal; and TRUS-assessed prostate volume (millilitres).
Area under the receiver operating curve.
The reference value for the univariate analysis is total PSA (nanograms per millilitre) and DRE, normal/abnormal; for the multivariate analyses, it is the original risk calculator.
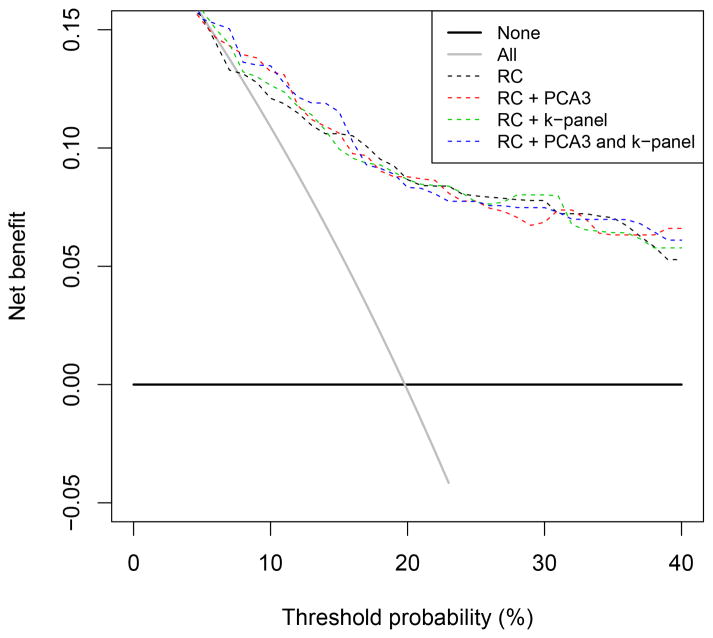
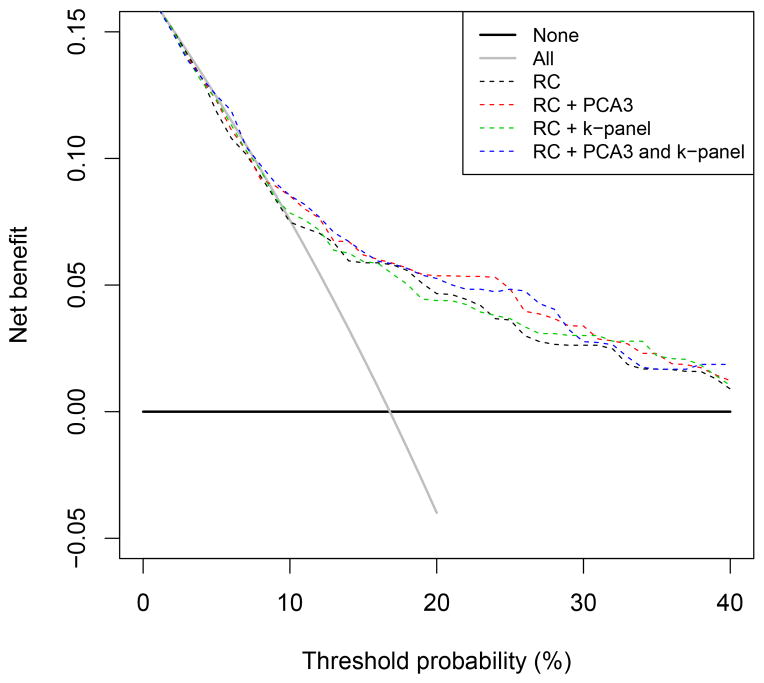
Analyses in men with PSA levels <3.0 ng/ml showed no value for the 4k-panel but some added value of PCA3 (univariate AUC: 0.64 [0.58–0.70], AUC: 0.70 vs 0.66 when added to the reference models, p = 0.01 for RC 4 and p < 0.01 for RC 4+DRE) (see Supplementary Table 1). In men with elevated PSA levels, the NBs of all models were higher than in the total data set (Fig. 1). In this subgroup the use of a model was clinically useful from a threshold of 5%. The reduction in biopsies per 100 men differed between a threshold of 10–30% in the total data set, in favour of the multivariable model with PCA3 and PCA4 plus 4k-panel. In the subgroup of men with elevated PSA, different models were in favour depending on the specific threshold, which also reflected the low number of PCa cases at these thresholds (Fig. 2).
Fig. 1.
Net benefit of prediction models with prostate cancer antigen 3 and/or the kallikrein panel in the subgroup of men with prostate-specific antigen ≥3.0 ng/ml (n = 202).
k-panel = kallikrein panel; PCA3 = prostate cancer antigen 3; RC = risk calculator.
Fig. 2.
Net benefit of prediction models with prostate cancer antigen 3 and/or the kallikrein panel in all men (n = 708).
k-panel = kallikrein panel; PCA3 = prostate cancer antigen 3; RC = risk calculator.
The prediction models had added value over biopsy in all men if the threshold for performing a biopsy was >9% (Fig. 1 and 2). Between thresholds of 9% and 40%, the multivariable model with PCA3 or PCA3 plus 4k-panel had the highest NB and performed better than the reference model at all thresholds. With a cut point of PSA ≥3.0 ng/ml and PCA3 >10, reduction in the number of biopsies per 1000 men at a threshold probability of 12.5% was 89 when PCA3 was added, 50 when the 4k-panel was added, and 124 when both the PCA3 and the 4k-panel marker were added to the original RC. At a threshold probability of 20%, there was a reduction of 11 biopsies per 1000 men when PCA3 was added to the original RC and 7 per 1000 men when both PCA3 and the 4k-panel were added. In contrast, no reduction in the number of biopsies was noted in men with a PSA level ≥3.0 ng/ml.
Results were similar for each of the considered reference models (RC 4+DRE or RC 4 with TRUS) (data not shown).
4. Discussion
In the current study, adding the 4k-panel to a previously developed PCa risk prediction model increased the predictive value in participants with PSA ≥3.0 ng/ml. Adding PCA3 to the previously developed PCa risk prediction model increased the AUC in prescreened men regardless of their total PSA level at time of biopsy. This was equally seen in reference models with and without the inclusion of TRUS and TRUS-assessed volume. Therefore, we advise the model with DRE to estimate prostate volume.
In the past, %fPSA was shown to increase the accuracy of DRE and total PSA significantly [21]. Its limited cost and wide availability in laboratories that run total PSA values are attractive attributes for clinical use. We found a very limited predictive value of %fPSA alone or combined with the RCs.
The usefulness of PCA3 testing for the detection of PCa and possible reduction of unnecessary biopsies has been shown before [22,23]. These studies assessed the added value of PCA3 after selecting men for biopsy solely on the basis of a PSA cut-off level. This implies that PCa in men with PSA values below the threshold will be missed. In addition, assessing the added value of PCA3 in men with a previous negative biopsy, initially selected on the basis of an elevated PSA level, is biased by definition. The benefit from PCA3 as compared with PSA is then overoptimistic. To overcome this attribution bias in the current study, men with a PCA3 score ≥10 were biopsied, even if their PSA level was <3.0 ng/ml [13,24].
Predictions based on the 4k-panel did not differ significantly between cancer and noncancer cases in the total study group while some markers such as intact PSA and Hk2 did differ. In the subgroup analyses of men with PSA level ≥3.0, the PCA3 and 4k-panel scores differed significantly between men with and without PCa, whereas intact PSA and hK2 did not (Table 1). fPSA differed significantly among those in the subgroup men with a PSA level ≥3.0. Hence fPSA may be the most relevant element in the 4k-panel for rescreened men with elevated PSA levels.
The 4k-panel was developed for men with elevated PSA levels and has up to now only been tested in that particular but clinically most relevant setting. Previous studies showed that predictions based on levels of four kallikrein markers in blood distinguish between pathologically insignificant and aggressive PCa with good accuracy [15,25]. We confirmed these results with an increase in predictive capability in addition to a risk prediction model that already had an AUC ≥0.7, albeit in a relatively low number of patients.
With respect to cost effectiveness, data suitable for a direct comparison with our study are scarce. Although data on the cost effectiveness of PCA3 are weak [26], another comparable but cheaper combination of serum-based subforms of PSA, the Prostate Health Index, has been found to be cost effective for screening purposes [27]. For the current study, we assessed cost effectiveness with arbitrarily assumed costs for the PCA3 test and for prostate biopsy (€300 and €249, respectively [28]). The 4k-panel is not commonly available and may be cheaper than a PCA3 test [9]. When adding PCA3 and/or the 4k-panel to previously developed PCa risk prediction model, fewer biopsies are needed to find the same amount of cancers (increased NB; Fig. 1 and 2). However, this did not result in a substantial reduction in prostate biopsies compared with the original RCs alone for pts between 0% and 40%, making it very unlikely that the extended risk model will be cost effective.
One limitation of this study was the prescreened nature of our study cohort. Therefore we compared the performance of models with PCA3 or the 4k-panel with reference models developed for prescreened men, allowing for a fair comparison. This, and the fact that all men were from the Netherlands, may affect external validity. However, elevated PCA3 scores have particularly been demonstrated to increase the probability of a positive repeat biopsy in men with a prior negative biopsy result, independent of PSA [29,30].
Another limitation of this study is the small number of men included, specifically men with a PSA ≥3.0 ng/ml. The relative utility of PCA3 and the 4k-panel need to be confirmed. The number of serious cancers was low (n = 22, of which 9 were in men with PSA levels ≥3.0 ng/ml), limiting separate analyses for this group of patients. In men with PSA ≥3.0 ng/ml (n = 202, of whom 40 had cancer), we used the original RC consisting of four variables and extended this with one or two variables, giving an events per variable (EPV) ratio of 8 or 6.7 that could lead to overfitting of the model. Ideally the EPV would be higher, but EPV values from 5 have been shown to be valid in the context of statistical adjustment for baseline risk factors [31]. We used sextant biopsy in a repeat screening setting and found a 17% cancer detection rate (n = 119), and it is likely that we missed some cases. Even using sextant biopsy for repeat screening, deaths due to PCa occurred at a rate of only 0.03% compared with 0.35% overall [32].
5. Conclusions
Both the PCA3 and, to a lesser extent, a 4k-panel have added value to the DRE-based ERSPC Rotterdam RC in detecting PCa for prescreened men. Further validation is needed, however, and should focus on biomarkers capable of identifying men at elevated risk for potentially aggressive PCa. This is most relevant for men with a previous negative biopsy where such markers may be especially useful.
Supplementary Material
Take-home message.
Both prostate cancer antigen 3 and a kallikrein panel have added some value to the European Randomised study of Screening for Prostate Cancer digital rectal examination–based risk calculator for detecting prostate cancer in prescreened men. Further research should focus on men with a previous negative biopsy, for whom markers may especially be useful.
Acknowledgments
Funding/Support and role of the sponsor: Supported in part by funds from the National Cancer Institute (R01CA160816 and P50-CA92629), the Sidney Kimmel Center for Prostate and Urologic Cancers, David H. Koch through the Prostate Cancer Foundation, the National Institute for Health Research Oxford Biomedical Research Centre Program, Swedish Cancer Society (project no. 11-0624), and Fundación Federico SA. Moniek Vedder and Ewout Steyerberg received funding from the Center for Translational Molecular Medicine (the Prostate Cancer Molecular Medicine project grant). Monique Roobol received funding from the Dutch Cancer Society(KWF94-869, 98-1657, 2002-277, 2006-3518, 2010-4800); The Netherlands Organisation for Health Research and Development (ZonMW-002822820, 22000106, 50-50110-98-311, 62300035), The Dutch Cancer Research Foundation, and an unconditional grant from Beckman-Coulter-Hybritech Inc. Performance of the PSA test (Hybritech, Beckman Coulter Inc., Fullerton, CA, USA), the PCA3 test (PROGENSA, Gen-Probe Inc., San Diego, CA, USA), and the 4k-panel measurements (performed in the Department of Laboratory Medicine [Lund University, Malmo, Sweden]) were sponsored. The funding sources did not have any role in the design or conduct of the study; the collection, management, analysis, or interpretation of the data; or the preparation, review, or approval of the manuscript.
Footnotes
Author contributions: Moniek Vedder had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Roobol, Vickers.
Acquisition of data: Roobol, Lilja.
Analysis and interpretation of data: Vedder, Steyerberg, Roobol, Vickers.
Drafting of the manuscript: Vedder, Roobol.
Critical revision of the manuscript for important intellectual content: Vedder, de Bekker-Grob, Lilja, Vickers, van Leenders, Steyerberg, Roobol
Statistical analysis: Vedder, Steyerberg, Roobol.
Obtaining funding: Steyerberg.
Administrative, technical, or material support: None.
Supervision: Steyerberg, Roobol.
Other (specify): None.
Financial disclosures: Moniek Vedder certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Hans Lilja holds patents for free PSA, hK2, and intact PSA assays and is named, along with Andrew Vickers, on a patent application for a statistical method to detect prostate cancer.
References
- 1.Heidenreich A, Bellmunt J, Bolla M, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol. 2011;59:61–71. doi: 10.1016/j.eururo.2010.10.039. [DOI] [PubMed] [Google Scholar]
- 2.Draisma G, Boer R, Otto SJ, van der Cruijsen IW, et al. Lead times and overdetection due to prostate-specific antigen screening: estimates from the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst. 2003;95:868–78. doi: 10.1093/jnci/95.12.868. [DOI] [PubMed] [Google Scholar]
- 3.Heijnsdijk EA, der Kinderen A, Wever EM, Draisma G, Roobol MJ, de Koning HJ. Overdetection, overtreatment and costs in prostate-specific antigen screening for prostate cancer. Br J Cancer. 2009;101:1833–8. doi: 10.1038/sj.bjc.6605422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steyerberg EW, Roobol MJ, Kattan MW, van der Kwast TH, de Koning HJ, Schroder FH. Prediction of indolent prostate cancer: validation and updating of a prognostic nomogram. J Urol. 2007;177:107–12. doi: 10.1016/j.juro.2006.08.068. discussion 112. [DOI] [PubMed] [Google Scholar]
- 5.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–34. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 6.Karakiewicz PI, Benayoun S, Kattan MW, et al. Development and validation of a nomogram predicting the outcome of prostate biopsy based on patient age, digital rectal examination and serum prostate specific antigen. J Urol. 2005;173:1930–4. doi: 10.1097/01.ju.0000158039.94467.5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–9. [PubMed] [Google Scholar]
- 8.Hessels D, Schalken JA. The use of PCA3 in the diagnosis of prostate cancer. Nat Rev Urol. 2009;6:255–61. doi: 10.1038/nrurol.2009.40. [DOI] [PubMed] [Google Scholar]
- 9.Vickers AJ, Cronin AM, Aus G, et al. A panel of kallikrein markers can reduce unnecessary biopsy for prostate cancer: data from the European Randomized Study of Prostate Cancer Screening in Goteborg, Sweden. BMC Med. 2008;6:19. doi: 10.1186/1741-7015-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A, Roobol MJ, Savage CJ, et al. A four-kallikrein panel for the prediction of repeat prostate biopsy: data from the European Randomized Study of Prostate Cancer screening in Rotterdam, Netherlands. Br J Cancer. 2010;103:708–14. doi: 10.1038/sj.bjc.6605815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroder FH, Denis LJ, Roobol M, et al. The story of the European Randomized Study of Screening for Prostate Cancer. BJU Int. 2003;92(Suppl 2):1–13. doi: 10.1111/j.1464-410x.2003.04389.x. [DOI] [PubMed] [Google Scholar]
- 12.Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 13.Roobol MJ, Schroder FH, van Leeuwen P, et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur Urol. 2010;58:475–81. doi: 10.1016/j.eururo.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 14.Roobol MJ, van Vugt HA, Loeb S, et al. Prediction of prostate cancer risk: the role of prostate volume and digital rectal examination in the ERSPC risk calculators. Eur Urol. 2012;61:577–83. doi: 10.1016/j.eururo.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Vickers A, Cronin A, Roobol M, et al. Reducing unnecessary biopsy during prostate cancer screening using a four-kallikrein panel: an independent replication. J Clin Oncol. 2010;28:2493–8. doi: 10.1200/JCO.2009.24.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roobol MJ, Zhu X, Schroder FH, et al. A calculator for prostate cancer risk 4 years after an initially negative screen: findings from ERSPC Rotterdam. Eur Urol. 2013;63:627–33. doi: 10.1016/j.eururo.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 17.Roobol MJ, Schroder FH, Kranse R ERSPC Rotterdam. A comparison of first and repeat (four years later) prostate cancer screening in a randomized cohort of a symptomatic men aged 55–75 years using a biopsy indication of 3. 0 ng/ml (results of ERSPC, Rotterdam) Prostate. 2006;66:604–12. doi: 10.1002/pros.20352. [DOI] [PubMed] [Google Scholar]
- 18.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–74. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–38. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steuber T, Vickers A, Haese A, et al. Free PSA isoforms and intact and cleaved forms of urokinase plasminogen activator receptor in serum improve selection of patients for prostate cancer biopsy. Int J Cancer. 2007;120:1499–504. doi: 10.1002/ijc.22427. [DOI] [PubMed] [Google Scholar]
- 22.Auprich M, Haese A, Walz J, et al. External validation of urinary PCA3-based nomograms to individually predict prostate biopsy outcome. Eur Urol. 2010;58:727–32. doi: 10.1016/j.eururo.2010.06.038. [DOI] [PubMed] [Google Scholar]
- 23.Auprich M, Chun FK, Ward JF, et al. Critical assessment of preoperative urinary prostate cancer antigen 3 on the accuracy of prostate cancer staging. Eur Urol. 2011;59:96–105. doi: 10.1016/j.eururo.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 24.Roobol MJ, Schroder FH, van Leenders GL, et al. Performance of prostate cancer antigen 3 (PCA3) and prostate-specific antigen in prescreened men: reproducibility and detection characteristics for prostate cancer patients with high PCA3 scores (≥100) Eur Urol. 2010;58:893–9. doi: 10.1016/j.eururo.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 25.Carlsson S, Maschino A, Schroder F, et al. Predictive value of four kallikrein markers for pathologically insignificant compared with aggressive prostate cancer in radical prostatectomy specimens: results from the European Randomized Study of Screening for Prostate Cancer section Rotterdam. Eur Urol. 2013;64:693–9. doi: 10.1016/j.eururo.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malavaud B, Cussenot O, Mottet N, et al. Impact of adoption of a decision algorithm including PCA3 for repeat biopsy on the costs for prostate cancer diagnosis in France. J Med Econ. 2013;16:358–63. doi: 10.3111/13696998.2012.757552. [DOI] [PubMed] [Google Scholar]
- 27.Nichol MB, Wu J, Huang J, Denham D, Frencher SK, Jacobsen SJ. Cost-effectiveness of Prostate Health Index for prostate cancer detection. BJU Int. 2012;110:353–62. doi: 10.1111/j.1464-410X.2011.10751.x. [DOI] [PubMed] [Google Scholar]
- 28.Fandella A. Analysis of costs of transrectal prostate biopsy. Urologia. 2011;78:288–92. doi: 10.5301/RU.2011.8875. [DOI] [PubMed] [Google Scholar]
- 29.Haese A, de la Taille A, van Poppel H, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54:1081–8. doi: 10.1016/j.eururo.2008.06.071. [DOI] [PubMed] [Google Scholar]
- 30.Gittelman M, Hertzman B, Bailen J, et al. PCA3 molecular urine test as a predictor of repeat prostate biopsy outcome in men with previous negative biopsies: a prospective multicenter clinical study. J Urol. 2013;190:64–9. doi: 10.1016/j.juro.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Vittinghoff E, McCulloch CE. Relaxing the rule of ten events per variable in logistic and Cox regression. Am J Epidemiol. 2007;165:710–8. doi: 10.1093/aje/kwk052. [DOI] [PubMed] [Google Scholar]
- 32.Schroder FH, van den Bergh RC, Wolters T, et al. Eleven-year outcome of patients with prostate cancers diagnosed during screening after initial negative sextant biopsies. Eur Urol. 2010;57:256–66. doi: 10.1016/j.eururo.2009.10.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.