Abstract
Recent studies have demonstrated that microRNA-15b (miR-15b) regulates cell cycle progression, proliferationnd apoptosis in glioma cells by _targeting Cyclins. However, the clinical significance of miR-15b in human glioma remains unclear. Therefore, the aim of this study was to investigate the significance of miR-15b expression in diagnosis, prognosis and malignant progression of glioma. Quantitative real-time reverse transcriptive-PCR (qRT-PCR) was performed to examine miR-15b expression levels in 76 glioma tissues (13 grade II, 13 grade III and 50 grade IV gliomas) and seven glioma cell lines, as well as 10 non-neoplastic brain tissues and human astrocyte as control. MiR-15b showed significant increased expression in high-grade gliomas (P ≤ 0.001) and glioma cells (fold change 2.8-7.6) relative to non-neoplastic brains and astrocyte, respectively. Additionally, high miR-15b expression was significantly associated with advanced WHO grade (P ≤ 0.001), advanced patient age (P ≤ 0.001) and low Karnofsky performance score (KPS, P ≤ 0.001). Furthermore, Kaplan-Meier survival analysis and Cox regression analysis showed that patients with high miR-15b expression had significantly poor overall survival rate (P ≤ 0.001) and miR-15b expression was an independent prognosis-predicting factor for glioma patients (P ≤ 0.001; risk ratio = 5.6), respectively. Moreover, miR-15b expression was examined in seven independent patients with primary grade II or III gliomas that spontaneously progressed to grade III or IV gliomas. Statistically significant higher expression (P = 0.01) in the recurrent tumor compared with the corresponding primary tumor was observed in all of the seven patients. Our results suggest that miR-15b may be a prognostic predictor and be involved in malignant progression of glioma.
Keywords: microRNA, miRNA-15b, glioma, up-regulation, prognosis, malignant progression
Introduction
Gliomas are the most frequent and malignant primary brain tumors in human adults. The World Health Organization (WHO) classifies human gliomas into pilocytic astrocytoma (PA, WHO grade I), diffuse astrocytoma (DA, WHO grade II), anaplastic astrocytoma (AA, WHO grade III), and glioblastoma (GBM, WHO grade IV) in the order of increasing malignancy [1]. Despite recent advances in surgery, radiotherapy, and chemotherapy, the prognosis for patients with this tumor remains poor. The grade IV glioma, also known as glioblastoma multiforme (GBM), is the most common and aggressive form of glioma, with a median survival of only 12-15 months as compared to 2-5 years for patients with grade III gliomas and 6-8 years for low grade (I and II) gliomas [1]. Patients with primary GBM have a clinical progression of less than about 3 months. The progression of secondary GBM, however, from grade II or grade III, is slow [2]. Therefore, it is necessary to develop new diagnostic and prognostic tools and effective therapeutics that may be beneficial for improving the clinical management of glioma. Currently, tumor stratifications based on molecular profiles are increasingly prevalent and important. In addition, several recent molecular and genetic profiling studies have identified several markers and unique signatures as prognostic and predictive factors of malignant glioma [3,4].
MicroRNAs (miRNA) are endogenous small noncoding RNA molecules that contribute to the regulation of crucial biological processes by post-transcriptionally regulating expression of their _target genes [5]. Recent studies have proved that miRNAs are aberrantly expressed in various human cancers and exert important regulations on tumor biology by acting as oncogenes or tumor suppressors [6]. In addition, the characterization of miRNA expression patterns in cancer cells is thought to have a substantial value for diagnoses and prognoses as well as for subsequent therapeutic interventions [7-9]. Furthermore, it has been shown that classification of multiple cancers based on miRNA expression profiles is more accurate than that based on mRNA profiles [10]. Several investigations have indicated that expression profiles of miRNAs are associated with patients’ survival and are able to function as prognostic and predictive indicators in glioma [11-15]. Furthermore, several miRNAs that are involved in malignant progression of glioma have also been identified [16]. Thus, the identification of the miRNA expression signature for malignant gliomas, in particular glioblastoma, is of great significance for understanding the molecular mechanisms of tumorigenesis and progression of these malignancies.
MiR-15b, a member of miR-15/16 family, has been reported to be frequently dysregulated and take entirely different roles in various human cancers, by recent studies [17-21]. However, its expression profile in human glioma and involvement in glioma tumorigenesis still remains controversial. Guan et al. in 2010 screened the global expression of miRNAs in human malignant glioma and found that miR-15b had significantly higher expression in primary GBMs than that in AAs, suggesting an oncogenic property of miR-15b in this aggressive brain tumor [15]. Being consistent with their finding, Baraniskin et al. in 2012 showed that miR-15b had remarkably higher expression levels in cerebrospinal fluid (CSF) of glioma patients than in control subjects [22]. In contrast, several studies have demonstrated that miR-15b function as a tumor suppressor by regulating critical biological processes of glioma cells in vitro [23-25]. Xia et al. in 2009 originally showed that overexpression of miR-15b resulted in cell cycle arrest at G0/G1 phase and suppression of miR-15b expression resulted in a decrease of cell populations in G0/G1 and a corresponding increase of cell populations in S phase in glioma cell lines [23]. Zheng et al. found that miR-15b reduced glioma cell invasion and angiogenesis by down-regulating expression of its _target genes NRP-2 and MMP-3 [24]. Sun et al. recently showed that overexpression of miR-15b inhibits proliferation by arrested cell cycle progression and induces apoptosis of glioma cells [25]. Taken together, the role of miR-15b in glioma tumorigenesis and the correlations between its expression and clinicopathological factors of glioma remains unclear. Therefore, the aim of the present study was to investigate the clinical significance of miR-15b expression in human gliomas.
Materials and methods
Glioma specimens and patients
Glioma specimens were obtained from patients during surgery at First Affiliated Hospital of China Medical University. A portion of the tumor tissue was saved and made into paraffin sections for histopathological diagnosis in strict accordance with World Health Organization (WHO) criteria by two established neuropathologists, with differences resolved by careful discussion. And the remaining tissue was snap-frozen in liquid nitrogen then stored at -80°C for RNA extraction and other biological molecular experiments. To analyze the association between miR-15b expression and clinicopathological features of gliomas, a panel of 76 glioma specimens were collected, including 50 primary GBMs (grade IV), 13 AAs (grade III) and 13 diffused astrocytomas (DA, grade II) (see Table 1 for patients’ information in detail). Subsequently, expression level of miR-15b was examined on all of the 76 gliomas and 10 non-neoplastic brain tissues by conventional real-time PCR. These non-neoplastic brain tissues used as controls were obtained by collecting donations with consents from individuals who died in traffic accidents and were confirmed to be free of any prior pathological lesions. On the other hand, to evaluate the association between miR-15b expression and glioma progression, expression level of miR-15b was determined in primary lower-grade (grade II or III) and recurrent higher-grade (grade III or IV) glioma pairs derived from seven independent patients. In addition, a validation step involved analysis of 9 DAs, 8 AAs and 8 secondary GBM from 25 independent patients. For glioma patients, none of them had received chemotherapy or radiotherapy prior to surgery, and all patients were well followed up. Overall survival time was calculated from the date of the initial surgical operation to death. Patients, who died of diseases not directly related to their gliomas or due to unexpected events, were excluded from this study. The present study was approved by the Ethics Committee of China Medical University.
Table 1.
Correlation of miR-15b relative expression level with clinicopathological factors of glioma patients
| Clinicopathological features | No. of cases | miR-15b expression | P | |
|---|---|---|---|---|
|
| ||||
| High (n, %) | Low (n, %) | |||
| WHO grade | ≤ 0.001 | |||
| II | 13 | 0 (0.0%) | 13 (100.0%) | |
| III | 13 | 3 (23.1%) | 10 (76.9%) | |
| IV | 50 | 30 (60.0%) | 20 (40.0%) | |
| Age | ≤ 0.001 | |||
| > 50 | 48 | 29 (60.4%) | 19 (39.6%) | |
| ≤ 50 | 28 | 4 (14.3%) | 24 (85.7%) | |
| Gender | 0.709 | |||
| Male | 41 | 17 (41.5%) | 24 (58.5%) | |
| Female | 35 | 16 (45.7%) | 19 (54.3%) | |
| KPS | ≤ 0.001 | |||
| < 90 | 42 | 26 (61.9%) | 16 (38.1%) | |
| ≥ 90 | 34 | 7 (20.6%) | 27 (79.4%) | |
| Surgery | 0.817 | |||
| GTR | 38 | 17 (44.7%) | 21 (55.3%) | |
| PR | 38 | 16 (42.1%) | 22 (57.9%) | |
Abbreviations: KPS, Karnofsky performance scale; GTR, gross total resection; PR, partial resection.
Glioma cell lines and human astrocyte
The glioma cell lines U87, U251, U373, LN18, LN229, T98G and SF295 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in Dulbecco’s modified eagle’s medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco) and penicillin/streptomycin (100 U/mL). The human astrocyte was a gift from Dr. T. Sasaki (Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan) and maintained in DMEM supplemented with 2% FBS and 1% N-2 supplement (Gibco).
RNA extraction, reverse transcription and real-time PCR quantification for miRNA detection
Total RNA was extracted from frozen tissues of glioma and non-neoplastic brain, glioma cell lines and human astrocyte using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacture’s instruction. RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and RNA quality was measured using a denaturing 15% polyacrylamide gel.
To examine the expression levels of miR-15b in glioma tissues and cell lines, cDNA synthesis and subsequent quantitative real-time PCR were performed using a TaqMan MiRNA Reverse Transcription Kit (Applied Biosystems) and individual TaqMan MiRNA assay (Applied Biosystems), and Applied Biosystems 7500HT Fast Real-Time PCR System (Applied Biosystems), as previously described [26]. RNU6B were used as endogenous controls, non-neoplastic brain tissues and human astrocyte were used for calibrations. Relative quantification of miR-15b expression was calculated with the 2-ΔΔCt method.
Statistical analysis
All computations were carried out using the software of SPSS version19.0 for Windows (SPSS Inc., Chicago, IL, USA). Data were expressed as mean ± standard deviation (SD). Student’s t-test was used to compare the expression levels of miR-15b in different glioma subtypes. The Χ2 test was used to analyze the relationship between miR-15b expression and the clinicopathological characteristics. A life table was calculated according to the Kaplan-Meier method. Risk ratios for the time-to-event endpoint were estimated using the multivariate Cox regression analysis in a forward stepwise method to evaluate the effect of multiple independent prognostic factors on overall survival outcome. Paired samples t-test was used to compare miR-15b expression between primary and recurrent gliomas. Differences were considered statistically significant when P was less than 0.05.
Results
Up-regulation of miR-15b in glioma tissues and cell lines
To evaluate the dysregulation of miR-15b in glioma tissues, we examined miR-15b expression levels in a panel of 76 glioma tissues including 50 primary GBMs (grade IV), 13 AAs (grade III) and 13 DAs (grade II), as well as 10 non-neoplastic brain tissues for control, by qRT-PCR. As shown in Figure 1A, expression of miR-15b was remarkably increased in glioma tissues compared with non-neoplastic brain tissues (Student’s t-test, P ≤ 0.001). High-grade (grade III and IV) gliomas both showed significantly higher expression of miR-15b compared with non-neoplastic brains (P ≤ 0.001, Figure 1A). However, there was no significant difference of miR-15b expression between grade II gliomas and brains. In addition, expression of miR-15b showed a distinctly upward tendency along with the increasing malignancy degree of gliomas (fold changes and P values of grade III vs. II, grade IV vs. III and grade IV vs. II were: 2.27 and 0.002, 2.00 and ≤ 0.001, 4.55 and ≤ 0.001, respectively, Figure 1A). Furthermore, miR-15b expression in glioma cell lines was examined and we found a similarly robust increase in miR-15b expression in seven commonly used model cell lines derived from human malignant gliomas (U87, U251, U373, T98G, LN18, LN229 and SF295, Figure 1B). Its expression was increased about 2.8- to 7.6-fold in these tumor cells compared with human astrocyte (Figure 1B).
Figure 1.
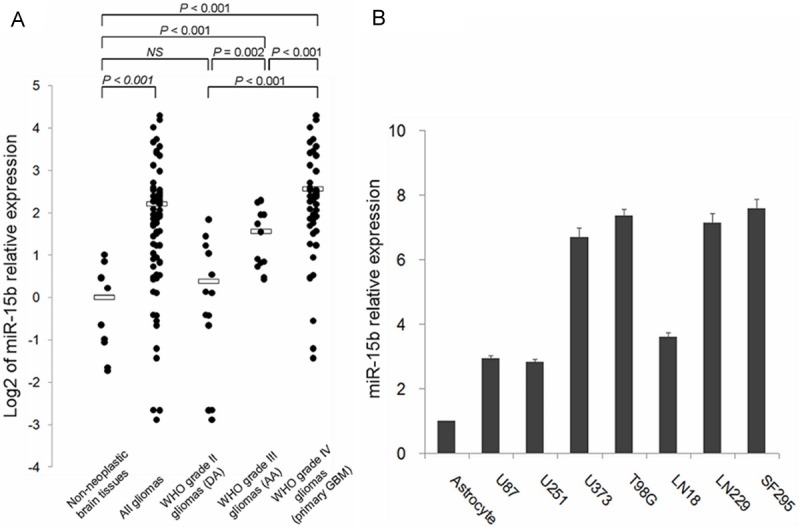
miR-15b expression in 76 glioma tissues (50 primary GBMs, 13 AAs and 13 DAs), 10 non-neoplastic brain tissues, 7 glioma cell lines (U87, U251, U373, T98G, LN18, LN229 and SF295) and normal human astrocyte detected by quantitative reverse transcriptive real-time polymerase chain reaction (qRT-PCR) analysis. A. Dots indicate log2 of the relative quantification (RQ) values of miR-15b expression levels, normalized by RNU6B. Bars indicate log2 of the average RQ values for each group. The expression level of miR-15b was found to be remarkably increased in glioma tissues compared to non-neoplastic tissues (P ≤ 0.001). Both primary GBMs and AAs showed significantly higher expression of miR-15b than non-neoplastic brains tissues (P ≤ 0.001, respectively). Expression of miR-15b showed a distinctly upward tendency along with the increasing malignancy degree of gliomas. B. Columns indicate RQ values of miR-15b expression levels in human astrocyte and glioma cell lines. Glioma cell lines showed about 3- to 7.5-fold higher expression of miR-15b in comparison with normal human astrocyte.
MiR-15b up-regulation correlates with aggressive clinicopathologicl features of gliomas
Subsequently, correlations between miR-15b expression and several clinicopathological factors including malignant degree of tumor, patients’ age at diagnosis, gender, pre-operative Karnofsky performance scale (KPS) and extent of tumor resection in glioma patients were evaluated by Χ2 test. We assigned gliomas to miR-15b low-expression group and high-expression group that were tumors with miRNA expression under and above the median value of expression in all 76 gliomas, respectively (median expression value = 4.62; n = 43 and 33 for low-expression group and high-expression group, respectively). As shown in Table 1, miR-15b high-expression was significantly more frequent in GBMs than AAs and DAs (P ≤ 0.001, Χ2 test). In addition, significant correlations were also found between miR-15b expression and patient age, as well as KPS. The increased expression of miR-15b more frequently occurred in glioma patients with advanced age or low KPS than those with young age or high KPS (P ≤ 0.001, Table 1), respectively. However, no statistically significant correlation was observed between miR-15b expression and other clinicopathological factors (Table 1).
High expression level of miR-15b predicts poor survival in glioma patients
Furthermore, prognostic value of miR-15b expression in glioma patients was evaluated. We performed loglank test and Kaplan-Meier analysis to evaluate the association between miR-15b expression level and clinical information in these glioma patients mentioned above. We observed that miR-15b expression level displayed a significant correlation with glioma patients’ overall survival. As shown in Figure 2A, patients with high-expression level had significantly poor overall survival compared to that with low-expression level of miR-15b (mean overall survivals 386 and 1238 days, respectively; P ≤ 0.001, logrank test). In addition, univariate and multivariate analysis using the Cox proportional hazard regression model was performed to determine whether miR-15b expression level and other clinical parameters are independent factors for prognostic prediction in glioma patients. Our result showed that both miR-15b high expression (P ≤ 0.001; risk ratio 5.6, multivariate Cox regression analysis) and high pathological grade (P ≤ 0.001; risk ratio 11.0, multivariate Cox regression analysis) were independent predictors of poor prognosis in glioma patients (Table 2). Furthermore, the prognostic value of miR-15b expression was also analyzed in patients with high-grade glioma (grade III and IV) and we found that high expression of miR-15b was significantly associated with poor overall survival of patients with these aggressive tumors (mean overall survivals 386 and 713 days for high-expression group and low-expression group, respectively, P ≤ 0.001, Figure 2B).
Figure 2.
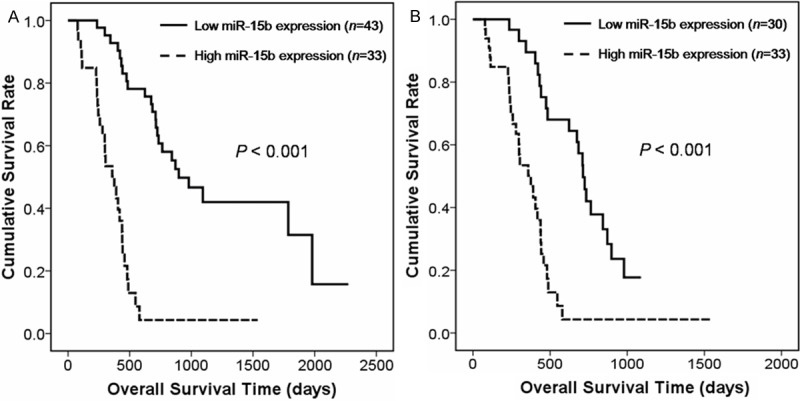
Kaplan-Meier survival curves for glioma patients with high and low expression levels of miR-15b. A. Among 76 glioma patients (50 primary GBMs, 13 AAs and 13 DAs), those with high miR-15b expression (left, dotted line, n = 33) had significantly shorter survival periods than did patients with low miR-15b expression (right, solid line, n = 43; P ≤ 0.001). B. Among 63 high-grade glioma patients (50 primary GBMs and 13 AAs), those with high miR-15b expression (left, dotted line, n = 33) had significantly shorter survival periods than did patients with low miR-15b expression (right, solid line, n = 30; P ≤ 0.001).
Table 2.
Univariate and multivariate Cox regression analysis for overall survival in glioma patients
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Variant | No. of case (%) | Mean OS | 95% CI | P (log-rank) | Variant | RR | 95% CI | P |
| WHO grade | ≤ 0.001 | WHO grade | ≤ 0.001 | |||||
| II | 13 (17.1%) | 1980 | 1687-2273 | IV vs. III vs. II | 11.0 | 4.3-28.1 | ||
| III | 13 (17.1%) | 979 | 643-1315 | |||||
| IV | 50 (65.8%) | 418 | 367-469 | |||||
| Age | ≤ 0.001 | Age | 0.136 | |||||
| > 50 | 48 (63.2%) | 442 | 377-507 | > 50 vs. ≤ 50 | 0.5 | 0.2-1.1 | ||
| ≤ 50 | 28 (36.8%) | 1786 | 651-2921 | |||||
| Gender | 0.905 | Gender | 0.302 | |||||
| Male | 41 (53.9%) | 579 | 263-895 | Male vs. Female | 2.9 | 1.5-5.7 | ||
| Female | 35 (46.1%) | 547 | 331-763 | |||||
| KPS | ≤ 0.001 | KPS | 0.926 | |||||
| < 90 | 42 (55.3%) | 439 | 396-482 | < 90 vs. ≥ 90 | 1.0 | 0.4-2.1 | ||
| ≥ 90 | 34 (44.7%) | 1786 | 405-3167 | |||||
| Surgical resection | 0.141 | Surgery | 0.071 | |||||
| GTR | 38 (50.0%) | 710 | 468-952 | PR vs. GTR | 2.2 | 1.2-4.2 | ||
| PR | 38 (50.0%) | 441 | 325-557 | |||||
| miR-15b expression | ≤ 0.001 | miR-15b expression | ≤ 0.001 | |||||
| Low | 43 (43.4%) | 1238 | 993-1483 | High vs. Low | 5.6 | 2.6-12.0 | ||
| High | 33 (56.6%) | 386 | 280-493 | |||||
Abbreviations: KPS, Karnofsky performance scale; GTR, gross total resection; PR, partial resection; OS, overall survival; RR, risk ratio.
Increased expression of miR-15b is associated with malignant progression of glioma
We also evaluated the correlation between miR-15b expression and glioma progression. MiR-15b expression was examined in seven patients with primary grade II or III gliomas that spontaneously progressed to grade III or IV gliomas by real-time PCR (see Table 3 for patients’ information in detail). As shown in Figure 3 and Table 3, miR-15b exhibited a progression-associated up-regulation, that is, an about 1.3- to 41.6-fold significant higher expression in the recurrent higher-grade glioma as compared to the corresponding primary tumor (P = 0.01, paired samples t test). To validate this result, we analyzed expression of miR-15b in an independent series of 9 DAs, 8 AAs and 8 secondary GBMs from another panel of 25 patients. As shown in Figure 4, median expression of miR-15b was obviously increased along with malignant grade of the tumor. The differences were statistically significant between grade II and III (P = 0.015), grade III and IV (P = 0.016), as well as grade II and IV (P ≤ 0.001) gliomas (Figure 4).
Table 3.
Seven patients with primary grade II or III glioma that recurred as grade III or IV glioma
| Patient No. | Primary tumor | Recurrent tumor | Fold change of miR-15b expression (Recurrent/Primary tumor) |
|---|---|---|---|
| Patient 1 | DA, grade II | AA, grade III | 12.3 |
| Patient 2 | DA, grade II | AA, grade III | 4.4 |
| Patient 3 | DA, grade II | Secondary GBM, grade IV | 13.6 |
| Patient 4 | DA, grade II | Secondary GBM, grade IV | 41.6 |
| Patient 5 | DA, grade II | Secondary GBM, grade IV | 5.7 |
| Patient 6 | AA, grade III | Secondary GBM, grade IV | 2.5 |
| Patient 7 | AA, grade III | Secondary GBM, grade IV | 1.3 |
Abbreviations: DA, diffuse astrocytoma; AA, anaplastic astrocytoma; GBM, glioblastoma.
Figure 3.
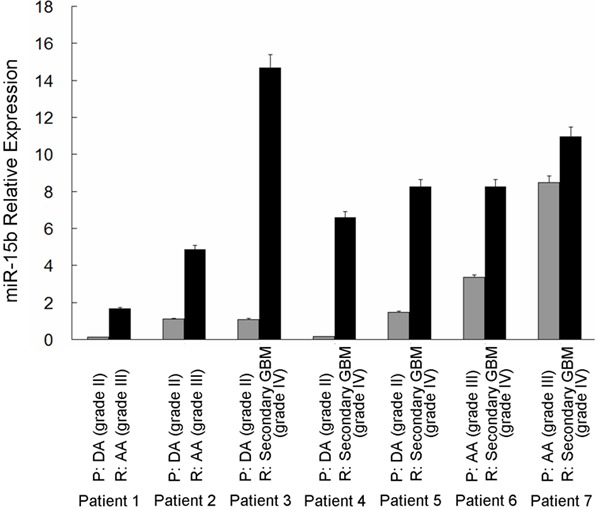
Expression levels of miR-15b in seven patients with primary lower-grade glioma that recurred as higher-grade glioma. MiR-15b showed an about 1.3- to 41.6-fold higher expression in the recurrent glioma as compared to the corresponding primary tumor. Patient numbers 1-7 encode the individual patient; Gray and black columns indicate expression levels of miR-15b in primary and recurrent tumors, respectively. P, primary tumor; R, recurrent tumor.
Figure 4.
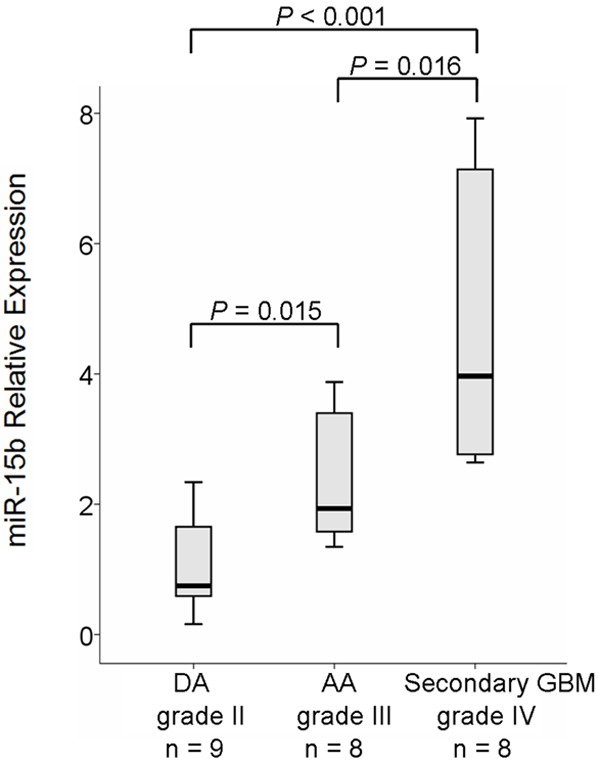
Validation experiment for miR-15b expression in an independent series of 9 DAs, 8 AAs and 8 secondary-GBMs. miR-15b showed a significant progression-associated up-regulation in gliomas with different pathological-grade (P = 0.015, 0.016 and ≤ 0.001 for grade III vs. II, grade IV vs. III and grade IV vs. II, respectively).
Discussion
Recently, several investigations indicate that expressions of miRNAs are associated with patients’ survival and involved in the malignant progression of human glioma [11-16]. In the present study, we explored the expression profile of miR-15b in various histological subtypes of glioma and analyzed the associations between miR-15b expression and patients’ survival, as well as the malignant progression of the tumor. As results of our analysis, there are four points of findings. Firstly, miR-15b was remarkably up-regulated in human glioma tissues and glioma cell lines compared with non-neoplastic brain tissues and normal astrocyte, respectively; Secondly, the increased miR-15b expression in glioma tissues was significantly correlated with advanced tumor progression and aggressive clinicopathological features; Thirdly, glioma patients with high miR-15b expression level in tumor tissues had significantly poorer overall survival and high expression of miR-15b was a statistically significant risk factor of poor survival in glioma patients; Finally, miR-15b was significantly up-regulated in the recurrent higher-grade gliomas compared with the corresponding primary lower-grade tumors. Our data suggest that miR-15b expression could be a valuable marker of prognosis and malignant progression of glioma. To our knowledge, this is the first study to analyze the expression profile and clinical significance of miR-15b in large panel of glioma patients.
Recent studies have indicated that miR15b was frequently deregulated in human cancer. Xi et al. in 2006 originally reported that miR-15b was significantly overexpressed in colorectal cancer compared to the paired normal colorectal sample [17]. Up-regulation of miR-15b was also revealed in the cervical cancer tissues comparing with the matched normal cervix by Wang et al. in 2008 [18]. Similarly, Satzger et al. in 2010 showed that miR-15b was overexpressed in human melanomas and high expression of miR-15b was significantly associated with poor recurrence free survival and overall survival in melanoma patients [19]. In addition, they found that down-regulation of miR-15b could reduce cell proliferation and increase apoptosis in melanoma cell lines, suggesting the oncogenic role of this miRNA in tumorigenesis of melanoma. In contrast, several studies have shown evidences that miR-15b was statistically reduced in tumor tissues, such as gastric cancer and hepatocellular carcinoma [20,21]. These findings illustrate that miR-15b is differently expressed in tumors originate from diverse organic tissues. However, the expression level and the role of miR-15b in human gliomas remain controversial. In, 2009, Xia et al. firstly reported that the expression of miR-15b was elevated in human glioma cell lines [23]. Of interest, they performed function analysis and found that miR-15b could induce cell cycle arrest at G0/G1 phase and decrease the cell populations in S phase in U87 glioma cells, by _targeting cell cycle-related molecule, CCNE1 (encoding cyclin E1), suggesting a tumor suppressive role of this miRNA. Similarly, a recent studies by Sun et al. reported that overexpression of miR-15b inhibited proliferation by arrested cell cycle progression and induced apoptosis, by directly _targeting Cyclin D1 [25]. In contrast with the data of Xia et al., they observed markedly decreased expression of miR-15b in glioma tissues in comparison to the normal brain tissues and an obviously downward trend of miR-15b expression with ascending tumor pathological grades. In addition, Zheng et al. in 2014 also showed evidences that miR-15b exists as an anti-tumor factor in gliomas. By employing an in vivo 9L homograft glioma tumor animal model and an in vitro hypoxic cell culture model, they revealed that miR-15b could reduce glioma cell invasion and angiogenesis through down-regulating its _target, NRP-2 [24]. From above discussion, miR-15b might function as a tumor suppressive miRNA in glioma. On the contrary, Guan et al. in 2010 screened the global expression of 365 miRNAs in human malignant glioma by microRNA qPCR-array method and found that miR-15b expression was significantly increased in primary GBMs as compared with AAs, suggesting an oncogenic property of miR-15b in this aggressive brain tumor [15]. In addition, a recent report by Baraniskin et al. demonstrated that miR-15b was the most abundant miRNA in cerebrospinal fluid (CSF) samples from patients with glioma [22]. According to their data, miR-15b showed a 25-fold higher mean expression level in CSF samples from patients with glioma than that in control subjects, 5-fold and 17-fold higher mean expression levels than that in patients with primary CNS lymphoma (PCNSL) and brain metastases or leptomeningeal carcinoma, respectively. In consistent with both of the results from Guan et al. and Baraniskin et al., we in the present study showed the up-regulated expression of miR-15b in glioma tissues and cell lines and the significant correlation of high miR-15b expression and the poor outcome of glioma patients, suggesting the oncogenic potential of this miRNA in glioma. Taken together, unlike most of the dysregulated miRNAs in gliomas, miR-15b expression was reported to be down- or up-regulated by different research groups. Such discrepancy might partly due to the differences in methodology, platform, and control samples between various studies mentioned above. Therefore, investigation on a much larger scale of glioma specimens should be necessary to determine the aberrant expression of this specific miRNA.
On the other hand, the molecular basis of glioma malignant progression has been investigated in previous studies by analyzing chromosomal and genetic aberrations, as well as changes in mRNA expression levels [27]. However, the dysregulation and potential roles of miRNAs in glioma progression remains unclear. Malzkorn et al. in 2010 investigated the expression profiles of 157 miRNAs in patients with primary WHO grade II gliomas that spontaneously progressed to WHO grade IV secondary GBMs and identified 12 miRNAs showing increased expression and two miRNAs showing reduced expression upon glioma progression [16]. Among these candidate miRNAs, miR-15a, and the family member of miR-15b we studied in the present study, showed a progression-associated up-regulation in recurrent high-grade glioma in comparison with the corresponding primary low-grade glioma [16]. Similarly, we found that expression of miR-15b was significantly up-regulated during progression from low-grade to AAs and secondary GBMs in our patients, suggesting that this miRNA be involved in malignant progression of glioma.
In conclusion, we in the current study showed that expression of miR-15b was remarkably increased in glioma tissues and cell lines. In addition, the up-regulation of miR-15b was significantly correlated with the poor overall survival of glioma patients and the progression from low-grade to high-grade gliomas. Our data suggest that miR-15b might take an important oncogenic role in glioma tumorigenesis and malignant progression. Such findings would lead to new approaches of diagnostic and therapeutic for glioma patients. Hence, the detailed biological mechanism of miR-15b up-regulation in glioma tumorigenesis and progression deserves further study.
Acknowledgements
This study was supported by Natural Science Foundation of China (Grant No: 81302190) and the Scientific Research Foundation for the Returned Overseas Chinese Scholars of China (Grant No: 2013-1792).
Disclosure of conflict of interest
None.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumors of central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayers DN Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houshmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K Cancer Genome Atlas Research Network. Identification of a CpG island methlator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–63. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 6.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–76. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic out-come in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J. Clin. Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancer. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Hermansen S, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW. MiR-21 expression in the tumour cell compartment holds unfavorable prognostic value in gliomas. J Neurooncol. 2013;111:71–81. doi: 10.1007/s11060-012-0992-3. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Lu S, Geng S, Ma S, Liang Z, Jiao B. Expression and clinical significance of microRNA-326 in human glioma miR-326 expression in glioma. Med Oncol. 2013;30:373. doi: 10.1007/s12032-012-0373-y. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Zhang J, Yan W, You G, Bao Z, Li S, Kang C, Jiang C, You Y, Zhang Y, Chen CC, Song SW, Jiang T. Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer. 2013;119:814–24. doi: 10.1002/cncr.27826. [DOI] [PubMed] [Google Scholar]
- 14.Zhi F, Chen X, Wang S, Xia X, Shi Y, Guan W, Shao N, Qu H, Yang C, Zhang Y, Wang R, Zen K, Zhang CY, Zhang J, Yang Y. The use of has-miR-21, has-miR-181b and has-miR-106a as prognostic indicator of astrocytoma. Eur J Cancer. 2010;46:1640–9. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, Ma X, Hayashi K, Sasaki T. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16:4289–97. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 16.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–50. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M, Ju J. Prognostic values of microRNAs in colorectal cancer. Biomark Insights. 2006;2:113–21. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLos One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stazger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2010;126:2553–62. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 20.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. MiR-15b and miR-16 modulate multidrug resistance by _targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 21.Chung GE, Yoon JH, Myung SJ, Lee JH, Lee SH, Kim SJ, Hwang SY, Lee HS, Kim CY. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23:113–9. [PubMed] [Google Scholar]
- 22.Baraniskin A, Kuhnhenn J, Schlegel U, Maghnouj A, Zöllner H, Schmiegel W, Hahn S, Schroers R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012;14:29–33. doi: 10.1093/neuonc/nor169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M, Li D, Zhao Y, Ge R, Li G, Chen Y, He ML, Kung HF, Lai L, Lin MC. MicroRNA-15b regulates cell cycle progression by _targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380:205–10. doi: 10.1016/j.bbrc.2008.12.169. [DOI] [PubMed] [Google Scholar]
- 24.Zheng X, Chopp M, Liu Y, Buller B, Jiang F. MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett. 2013;329:146–54. doi: 10.1016/j.canlet.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun G, Shi L, Yan S, Wan Z, Jiang N, Fu L, Li M, Guo J. MiR-15b _targets cyclin D1 to regulate prolifeation and apoptosis in glioma cells. Biomed Res Int. 2014;2014:687826. doi: 10.1155/2014/687826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time PCR of microRNA by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riemenschneider MJ, Reifenberger G. Astrocytic tumors. In: von Deimling A, editor. Gliomas: Recent Results in Cancer Research. Berlin: Springer; 2009. pp. 3–24. [DOI] [PubMed] [Google Scholar]


