Abstract
The bacterial CRISPR–Cas9 system has emerged as a multifunctional platform for sequence-specific regulation of gene expression. This Review describes the development of technologies based on nuclease-deactivated Cas9, termed dCas9, for RNA-guided genomic transcription regulation, both by repression through CRISPR interference (CRISPRi) and by activation through CRISPR activation (CRISPRa). We highlight different uses in diverse organisms, including bacterial and eukaryotic cells, and summarize current applications of harnessing CRISPR–dCas9 for multiplexed, inducible gene regulation, genome-wide screens and cell fate engineering. We also provide a perspective on future developments of the technology and its applications in biomedical research and clinical studies.
Complex and dynamic transcription regulation of multiple genes and their pathways drives many essential cellular activities, including genome replication and repair, cell division and differentiation, and disease progression and inheritance. Understanding the complex functions of a gene network requires the ability to precisely manipulate and perturb expression of the desired genes by repression or activation. However, until recently, we lacked such simple, robust technologies. RNA-mediated interference (RNAi), which uses small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs), has been one major approach for sequence-specific gene suppression in eukaryotic organisms1. Although RNAi is a convenient tool for studying gene function, allowing transcript-specific degradation through Watson–Crick base-pairing between mRNAs and siRNAs or shRNAs, its effects can be inefficient and nonspecific2. In addition to RNAi, customized DNA-binding proteins such as zinc-finger proteins or transcription activator-like effectors (TALEs) have been used as tools for sequence-specific DNA _targeting and gene regulation3. These proteins robustly _target DNA through programmable DNA-binding domains and can recruit effectors for transcription repression or activation in a modular way4–9. However, because each DNA-binding protein needs to be individually designed, their construction and delivery for the purpose of simultaneously regulating multiple loci is technically challenging10. Methods for gene overexpression include the use of cDNA overexpression vectors or vector libraries, but cloning large cDNA sequences into viral vectors and manipulating several gene isoforms simultaneously is difficult, and synthesizing large-scale libraries is costly. An ideal technology for genome regulation would therefore combine the convenience and scalability of RNAi with the robustness and modularity of DNA-binding proteins.
The discovery of the bacterial CRISPR–Cas system has inspired the development of a new approach for nucleotide base-pairing-mediated DNA _targeting. The type II CRISPR system uses an endonuclease, Cas9, which is guided by a single guide RNA (sgRNA) that specifically hybridizes and induces a double-stranded break (DSB) at complementary genomic sequences11–14. Using an engineered nuclease-deficient Cas9, termed dCas9, enables the repurposing of the system for _targeting genomic DNA without cleaving it15. As detailed below, recent work has suggested that dCas9 is a flexible, RNA-guided DNA recognition platform, which enables precise, scalable and robust RNA-guided transcription regulation.
In this Review, we first provide a very brief overview of the CRISPR–Cas9 technology for genome editing, before focusing on the development of CRISPR–dCas9 tools for transcription activation and repression in diverse organisms. We highlight the advantages and limitations of the current dCas9 technology, and also present a sampling of current applications of the technology in biological research and potential future clinical studies.
From editing to transcription control
CRISPR–Cas is an RNA-mediated adaptive immune system found in bacteria and archaea, in which it protects host cells from invasion by foreign DNA elements11. CRISPR–Cas is currently divided into two major classes and five types, of which type II is the most widely used for genome-engineering applications16. Discovery of key components of the type II CRISPR system and elucidation of its mechanism were integral to its use as a genome-engineering tool. These include the demonstration that Streptococcus thermophilus could specifically cleave double-stranded DNA, mediated by Cas9 (REFS 11,12); the discovery of a short DNA sequence adjacent to the RNA-binding site, later termed the protospacer-adjacent motif (PAM), as the CRISPR–Cas mechanism for discriminating self from non-self17; the discovery of a small transactivating CRISPR RNA (tracrRNA), which directs the post-transcriptional processing and maturation of the CRISPR RNA (crRNA) through sequence complementarity18; and, lastly, the demonstration that the CRISPR–Cas9 system from S. thermophilus could function in Escherichia coli and provide resistance against foreign plasmids19. On the basis of these findings about CRISPR–Cas9 biology, it was demonstrated that the Streptococcus pyogenes Cas9 protein can bind to a tracrRNA–crRNA complex or to a designed, chimeric sgRNA to generate a double-strand break (DSB) at a specific site of the _target DNA in vitro13,14. Another report similarly showed that S. thermophilus Cas9 could interact with the tracrRNA–crRNA complex to cut DNA14. Demonstrations of the use of Cas9 and RNAs for genome editing in vivo rapidly followed this seminal observation20–25 (FIG. 1a). Further information on the genome-editing applications of CRISPR–Cas9 can be found in other reviews26–29.
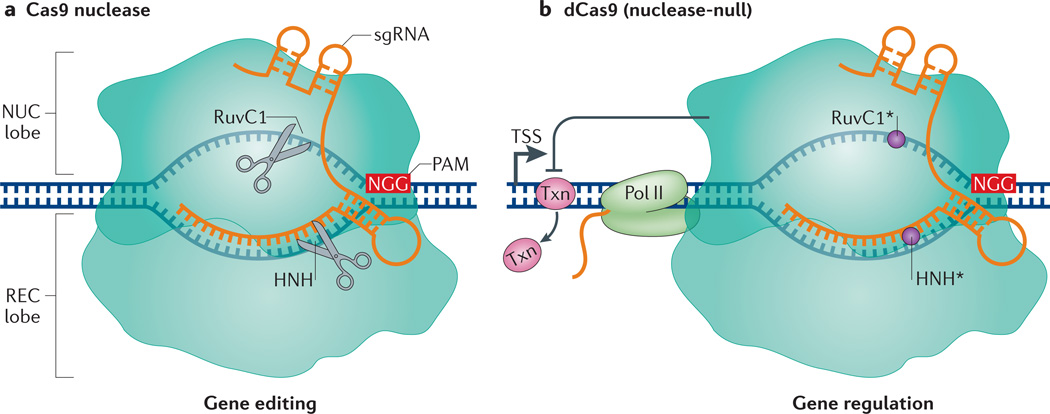
Figure 1. Gene editing versus gene regulation using Streptococcus pyogenes Cas9 and dCas9.
a | The S. pyogenes Cas9 endonuclease consists of a nuclease (NUC) lobe and a recognition (REC) lobe. Cas9 is _targeted to specific DNA sequences by direct pairing of the chimeric single guide RNA (sgRNA) with the _target DNA. This _targeting relies on the presence of a 5′ protospacer-adjacent motif (PAM) in the DNA, which in S. pyogenes is usually NGG. Binding mediates cleavage of the _target sequence by two nuclease domains, RuvC1 and HNH. b | The S. pyogenes dCas9 protein contains mutations in its RuvC1 (D10A) and HNH (H841A) domains, which inactivate its nuclease function (circles). dCas9 retains the ability to _target specific sequences through the sgRNA and PAM. dCas9 binding downstream of the transcription start site (TSS) can block transcription elongation by blocking RNA polymerase II (Pol II) or the binding of important transcription factors (Txn).
In addition to using the nuclease Cas9 for editing genomic sequences, the CRISPR–Cas9 technology can be used as a sequence-specific, non-mutagenic gene regulation tool. This repurposing was first demonstrated by introducing mutations into the S. pyogenes Cas9 in its two nuclease domains, HNH and RuvC15,30 (FIG. 1b). The resulting nuclease-deficient dCas9 is unable to cleave DNA but retains the ability to specifically bind to DNA when guided by a sgRNA. As discussed below, dCas9 allows for direct manipulation of the transcription process without genetically altering the DNA sequence. Furthermore, it allows the recruitment of diverse effector proteins for gene regulation at the transcription level. Other uses of the dCas9 protein include chromosome imaging in live cells and dissection of long-range chromatin interactions31–35 (BOX 1).
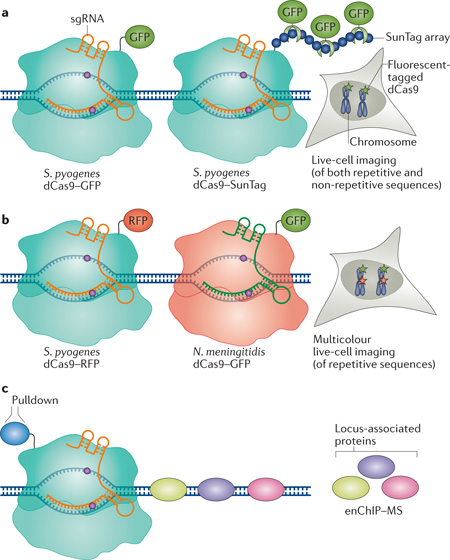
Box 1. Live-cell imaging and probing chromatin interactions using dCas9.
In addition to its use in transcription regulation, endonuclease-deficient Cas9 (dCas9) has been utilized as a tool for chromosome imaging and for identifying chromatin interactions. Using dCas9 tagged with enhanced GFP (EGFP) and one single guide RNA (sgRNA) _targeting telomeric repetitive elements, researchers were able to image telomere dynamics in live retinal pigment epithelium cells or HeLa cells31. The SunTag method was used to improve genomic imaging by amplifying the fluorescent signal35 (see the figure, part a). This approach has been extended to non-repetitive sequences; however, it requires the use of multiple sgRNAs tiling the genomic locus of interest31. The use of orthogonal dCas9 proteins (of Streptococcus pyogenes, Neisseria meningitidis and Streptococcus thermophilus), each tagged with a different fluorescent protein, has been demonstrated for multicolour genomic locus imaging in live cells32 (see the figure, part b). This enabled the imaging of multiple genomic loci simultaneously and the determination of the distance between different loci. In addition to imaging, dCas9 was used to probe molecular interactions in vivo at specific genomic regions33. Immunoprecipitation with an antibody against tagged dCas9 _targeted to a specific genomic locus by a sgRNA (known as engineered DNA-binding molecule-mediated chromatin immunoprecipitation) followed by mass spectrometry (enChIP–MS), allowed the identification of _target-specific interacting proteins (see the figure, part c).
Transcription repression by CRISPRi
Bacteria lack the machinery for RNAi, and simple platforms for _targeted gene regulation in bacteria have been limited. The utility of dCas9 for sequence-specific gene repression was first demonstrated in E. coli as a technology called CRISPR interference (CRISPRi). By pairing dCas9 with a sequence-specific sgRNA, the dCas9–sgRNA complex can interfere with transcription elongation by blocking RNA polymerase (Pol). It can also impede transcription initiation by disrupting transcription factor binding15,30,36,37 (FIG. 1b). In bacteria, the CRISPRi method using dCas9 is highly efficient in suppressing genes; is specific, with minimal off-_target effects; and is multiplexable, such that several genes can be simultaneously controlled using multiple sgRNAs. Unlike the permanent genetic modifications induced by the nuclease Cas9, gene repression using CRISPRi is reversible15. A disadvantage is that dCas9 may repress downstream genes within an operon (polar effects) instead of an individual gene. The CRISPRi platform thus provides a robust RNA-guided approach for gene repression in bacteria; however, further studies are needed to expand the method to selectively perturb gene expression on a genome-wide scale. Efficient dCas9-mediated transcription repression in bacteria demonstrated the possibility of using RNA-guided mechanisms for transcription repression and activation in diverse organisms15.
The introduction of CRISPRi into mammalian cells using dCas9 alone achieved only modest repression of enhanced GFP (egfp) in the human HEK293T reporter cell line15. When _targeting endogenous genes such as the transferrin receptor CD71, C-X-C chemokine receptor type 4 (CXCR4) and tumour protein 53 (TP53), up to 80% repression was observed37,38. To achieve enhanced repression, the Krüppel-associated box (KRAB) or four concatenated mSin3 interaction domains (SID4X) was fused to the carboxyl terminus of dCas9. Together with a _target-specific sgRNA, the dCas9–KRAB or dCas9–SID4X fusion proteins can efficiently repress endogenous genes (CXCR4, CD71, Kruppel-like factor 4 (KLF4) or SRY-box 2 (SOX2)) in mammalian cells38–40 (FIG. 2a). This repression was further enhanced by fusing KRAB to the amino terminus of dCas9, leading to strong repression of endogenous genes41. The level of dCas9- or KRAB–dCas9-mediated knockdown of endogenous genes was highly dependent on the sgRNA _targeting site, suggesting that the chromatin structure or the presence of regulatory elements may limit the level of repression. In yeast, a different mammalian transcription repressor domain, Max-interacting protein 1 (Mxi1), was used for effective repression38. CRISPRi has been used in genome-wide screens and for the manipulation of cell fate, which are discussed below.
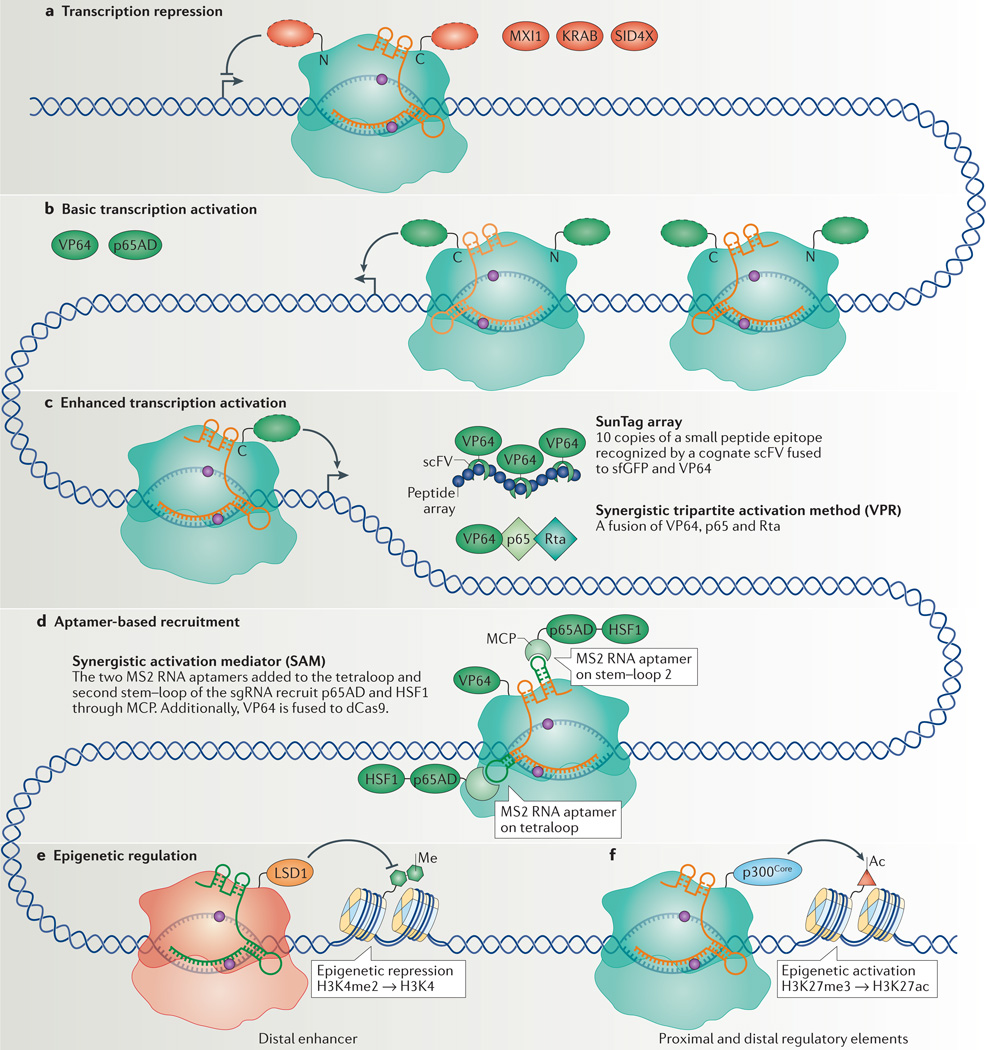
Figure 2. CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) for transcription repression and activation.
a | Transcription repression by nuclease-deficient Cas 9 (dCas9) can be improved by fusing dCas9 with different repressor domains (red dashed ovals), including MAX-interacting protein 1 (MXI1), Krüppel-associated box (KRAB) domain or four concatenated mSin3 domains (SID4X), to either amino or carboxyl termini. b | Initial strategies for transcription activation included fusing dCas9 with different activation domains (green dashed ovals), including multiple repeats of the herpes simplex VP16 activation domain (VP64 or VP160) or the nuclear factor-κB (NF-κB) transactivating subunit activation domain (p65AD). Improved levels of transcription activation were achieved with VP64 fused at both N and C termini. These methods require the use of multiple single guide RNAs (sgRNAs; different shades of orange) to recruit multiple dCas9 fusion proteins to achieve efficient transcription activation. c | In methods for enhanced transcription activation, it is sufficient to use only one sgRNA to recruit one dCas9 per _target gene. The SunTag activation method uses an array of small peptide epitopes (blue circles) fused to the C terminus of dCas9 to recruit multiple copies of single-chain variable fragment (scFV) fused to super folder GFP (sfGFP; for improving protein folding), fused to VP64. The synergistic tripartite activation method (VPR) uses a tandem fusion of three transcription activators, VP64, p65 and the Epstein–Barr virus R transactivator (Rta), to achieve enhanced transcription activation. d | The aptamer-based recruitment system (synergistic activation mediator (SAM)) utilizes dCas9 with a sgRNA encoding MS2 RNA aptamers at the tetraloop and the second stem–loop (shown in dark green) to recruit the MS2 coat protein (MCP) that is fused to two activators, p65 and heat shock factor 1 (HSF1). Additionally, VP64 is fused to dCas9. e | Epigenetic regulation can be carried out by fusion of epigenetic regulators to dCas9. Fusion of the histone demethylase LSD1 to Neisseria meningitidis Cas9 removes the histone 3 Lys4 dimethylation (H3K4me2) mark from _targeted distal enhancers, leading to transcription repression. f | The fusion of the catalytic core of the histone acetyltransferase p300 (p300Core) to dCas9 can acetylate H3K27 (H3K27ac) at _targeted proximal and distal enhancers, which leads to transcription activation.
Transcription activation by CRISPRa
CRISPR-mediated gene activation, termed CRISPRa, uses dCas9 fusion proteins to recruit transcription activators. A fusion of dCas9 with the ω-subunit of the E. coli Pol allowed assembly of the holoenzyme at a _target promoter for gene activation in E. coli36. There are currently limited reports on CRISPRa in bacteria, and more work is needed to achieve robust and consistent gene activation in bacteria.
The fusion of VP64 or of the p65 activation domain (p65AD) to dCas9 in mammalian cells could activate both reporter genes and endogenous genes, with a single sgRNA38,42–44 (FIG. 2b). However, the use of multiple sgRNAs was necessary to achieve significant activation of the endogenous genes tested (interleukin 1 receptor antagonist (IL1RN), achaete-scute family bHLH transcription factor 1 (ASCL1), Nanog homeobox (NANOG), myogenic differentiation 1 (MYOD1), vascular endothelial growth factor A (VEGFA) and neurotrophin 3 (NTF3))42,43. Protein engineering approaches were adopted to optimize the efficiency of activation. For example, it was determined that stronger activation could be achieved with VP64 fused simultaneously at both amino and carboxyl termini45. The addition of multiple copies of VP16 (for example, dCas9–VP160) was reported; however, the efficient activation of endogenous IL1RN, octamer-binding 4 (OCT4; also known as POU5F1) and SOX2 still required multiple sgRNAs46.
The complexity of genome-wide activation screens or cell fate reprogramming experiments necessitate the use of one efficient sgRNA per gene. The enhancement of gene activation observed with multiple sgRNAs suggested that recruitment of many activators could increase activation efficiency. One study used dCas9 fused with a carboxy-terminal SunTag array, which consisted of 10 copies of a small peptide epitope35. A cognate single-chain variable fragment (scFV) fused to a superfolder GFP (sfGFP; for improving protein folding) and to VP64 (scFV–sfGFP–VP64) recognized these peptides and recruited multiple copies of VP64 to a single dCas9. Using dCas9–SunTag, significant activation of CXCR4 was achieved with a single sgRNA, leading to the modulation of cell migration35 (FIG. 2c). An additional study screened different activator domains and members of the Mediator complex and Pol II complex for highly efficient activation of endogenous genes. The screen led to the development of a tripartite activator domain that consisted of VP64, p65AD and the Epstein–Barr virus R transactivator Rta47 (VPR) (FIG. 2c). The dCas9–VPR fusion showed improved activation of endogenous coding and non-coding genes using multiple sgRNAs when compared with dCas9–VP64. The system was also tested in Saccharomyces cerevisiae, Drosophila melanogaster and Mus musculus cells for activating endogenous loci47.
In addition to dCas9 engineering, sgRNA engineering was also shown to enhance the efficiency of gene activation. The recruitment of VP64 using protein-interacting RNA aptamers incorporated into the sgRNA has achieved activation of the gene encoding endogenous zinc-finger protein 42, using multiple sgRNAs48. An improvement, termed the synergistic activation mediator (SAM) system, was achieved by adding MS2 aptamers to the sgRNA; MS2 recruits its cognate MS2 coat protein (MCP) fused to p65AD and heat shock factor 1 (HSF1) (FIG. 2d). The SAM technology, together with dCas9–VP64, further increased endogenous gene activation compared with dCas9–VP64 alone and was shown to activate 10 genes simultaneously49. Although each of these improvements expanded the CRISPRa toolbox, it will be necessary in the future to compare activation by these methods across many endogenous genes, and in a variety of cell types, to determine which tool is best suited for specific genes and in different cells.
dCas9-mediated epigenetic modifications
The ability to manipulate epigenetic modifications, such as histone acetylation and methylation and DNA methylation, would allow for the interrogation of epigenetic regulation of cellular function. The histone demethylase LSD1 (Lys-specific histone demethylase 1) fused to Neisseria meningitidis dCas9 was recently used for gene repression50. Using dCas9–LSD1 and a sgRNA in mouse embryonic stem cells (ES cells) to _target the distal enhancer region of the endogenous transcription factor gene Oct4, the authors demonstrated the repression of Oct4 and loss of pluripotency. However, downregulation of Oct4 expression was not seen when the complex _targeted the proximal enhancer region, which is known to regulate Oct4 expression in epiblast cells51. This indicates that this epigenetic regulatory system can allow delineation between cell type-specific enhancers (FIG. 2e). Additionally, rather than catalysing a specific histone modification, it was recently demonstrated that _targeting dCas9–KRAB with a sgRNA to the HS2 enhancer in the globin locus control region led to H3K9 trimethylation at the enhancer, thus silencing the expression of multiple globin genes52. The use of these tools to silence transcription by _targeting regulatory regions, instead of the _target gene itself, further expands the capacity of dCas9 as a versatile transcription manipulation tool.
In addition to utilizing activation domains to achieve endogenous gene activation, the catalytic core of the human acetyltransferase p300 was recently fused to dCas9 (Cas9–p300Core) for _targeted epigenetic regulation. _target genes were activated by catalysing the acetylation of histone H3 Lys27 (H3K27ac) at both promoters and enhancers53. Although potential off-_target binding may lead to spurious activation, owing to the possibility of activating distant enhancers, dCas9–p300Core was found to be specific and robust, only activating the _targeted gene (FIG. 2f). Taken together, these studies demonstrate that dCas9 fused to epigenetic modifiers can modulate chromatin states and gene expression, thereby providing powerful tools for probing the interactions between the epigenome, regulatory elements and gene expression. The ability to _target epigenetic modifications to a gene in a combinatorial fashion may allow the temporal and spatial regulation of genes that are natively regulated by a complex set of interacting transcription factors54.
Simultaneous activation and repression
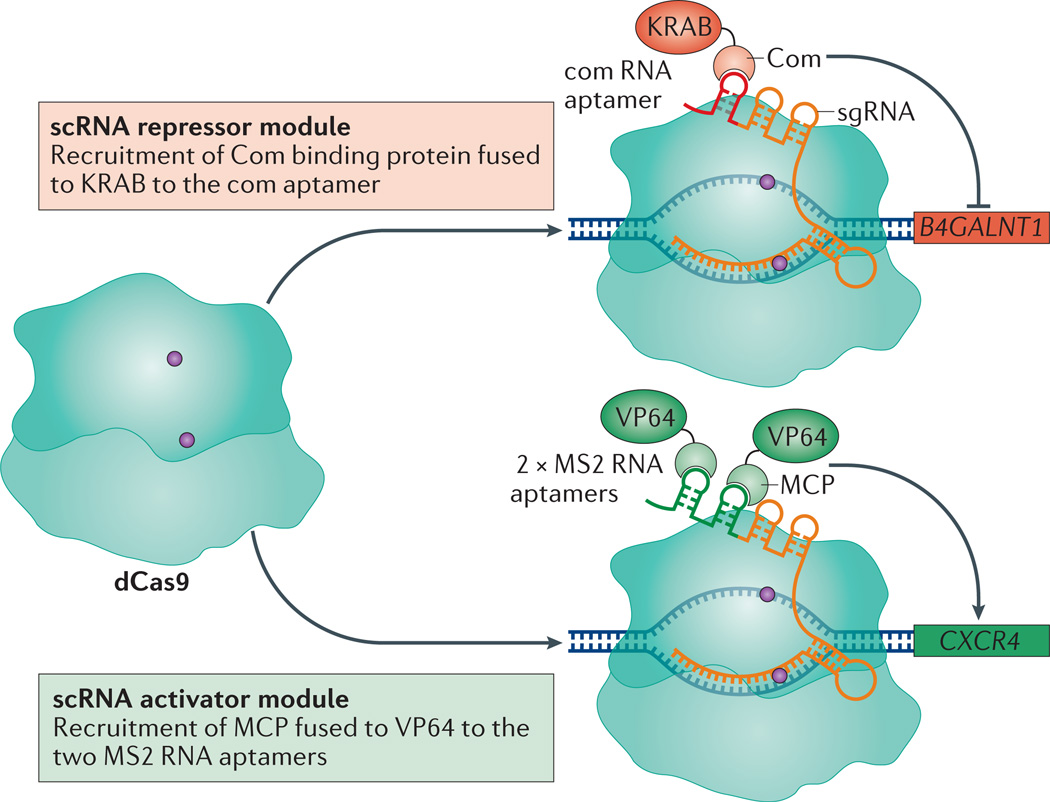
CRISPR–dCas9 can _target several genes simultaneously by using multiple sgRNAs. Recently, a method for simultaneous repression and activation of genes was established using scaffold RNAs (scRNAs)55. The scRNAs are designed by extending the sgRNA sequence with orthogonally acting protein-binding RNA aptamers (MS2, PP7 or com)55. Each scRNA can encode information both for DNA _target recognition and for recruiting a specific repressor or activator protein. By changing the DNA _targeting sequence or the RNA aptamers in a modular fashion, multiple dCas9–scRNAs can simultaneously activate or repress multiple genes in the same cell (FIG. 3). This functionality could facilitate the study of regulatory networks and genetic interactions. For example, a scRNA-based strategy was developed to modulate a branched metabolic pathway in yeast cells55, wherein different combinations of scRNAs were used to activate and repress alternative sets of enzymes for the production of distinct metabolites. In mammalian cells, two scRNAs were used to simultaneously activate CXCR4 with two MS2 scRNAs recruiting VP64, and repress β-1,4-N-acetyl-galactosaminyl transferase 1 (B4GALNT1) with a com scRNA recruiting KRAB (FIG. 3).
Figure 3. Simultaneous transcription activation and repression.
Multiplexed transcription activation and repression is carried out using single guide RNAs (sgRNAs) modified with RNA aptamers, termed scaffold RNAs (scRNAs). The com aptamer recruits the aptamer-binding protein, Com, which was fused to a Krüppel-associated box (KRAB) domain for transcription repression of β-1,4-N-acetyl-galactosaminyl transferase 1 (B4GALNT1). In the same cells, two MS2 aptamers were used to recruit the MS2 coat protein (MCP) fused to VP64 for transcription activation of C-X-C chemokine receptor type 4 (CXCR4).
The diversity of CRISPR–Cas systems highlights the potential of using orthogonal dCas9 proteins for parallel gene regulation56. Two limiting factors for this approach are the design of functional cognate sgRNAs and the characterization of the full PAM landscape. In addition to the S. pyogenes Cas9, CRISPR–Cas systems from several other bacteria (such as S. thermophilus, N. meningitidis, Treponema denticola and Staphylococcus aureus) have been examined and characterized with functional sgRNAs and PAMs20,57–59. However, as the recognition of PAM sequences in human cells may not always be the same as when characterized in vitro or inferred bioinformatically, it is necessary to test the full set of PAM sequences for each Cas9 in mammalian cells57. Nevertheless, the possibility of using scRNAs and/or orthogonal dCas9 proteins for parallel gene regulation would facilitate the manipulation and the study of complex gene networks.
The specificity of Cas9 and dCas9
The specificity of the nuclease Cas9 in mammalian cells remains a major concern for the use of the technology, in particular for clinical purposes. Compared to the bacterial cells in which CRISPR–Cas9 has evolved, the several-hundred-fold larger mammalian genomes might present many more off-_target binding sites to the system. Thus, the off-_target effects of CRISPR–Cas in genome-wide binding, editing and regulation have been examined extensively. It is important to distinguish the difference between the three cases (binding, editing and regulation), as off-_target binding may not necessarily have editing or regulatory effects. To examine binding specificity, two studies mapped the genome-wide binding sites of dCas9 with multiple different sgRNAs in mouse ES cells and HEK293T cells. Chromatin immunoprecipitation followed by deep DNA sequencing (ChIP–seq) analysis revealed that dCas9 had bound to many off-_target genomic sites60–62. However, through _targeted sequencing of the dCas9 binding sites, these studies demonstrated that cleavage by Cas9 at off-_target sites was substantially lower than at on-_target sites. These results suggested that although the high level of off-_target binding of Cas9 is a concern, only a small subset of off-_target binding sites were cleaved efficiently59,60,63. In addition to binding, various approaches were used to characterize editing specificity63–67. These studies suggest that off-_target effects are a concern for gene editing in mammalian cells, but that they may be highly dependent on the _target gene, the sequence of the designed sgRNA, the cell type, and the context of the genomic sequence and its epigenetic state28.
Gene repression was found to be quite specific when the transcriptome of HEK293T cells expressing dCas9–KRAB with a _targeting or a non-_targeting sgRNA was assayed by RNA sequencing (RNA-seq)38,52. Similar results were reported when using dCas9–VP64 for _targeted gene activation42,68. In another study, it was observed that, on average, more than two sequence mismatches between the sgRNA and the _target gene abolished CRISPRi regulatory activity in a set of genes, and therefore that dCas9 used for gene regulation can tolerate fewer mismatches compared with Cas9 used for gene editing41. To test the concern that the fused effector domains might contribute additional off-_target binding of dCas9, ChIP–seq was conducted using dCas9–KRAB or dCas9–VP64 with multiple sgRNAs. The results showed that genome-wide binding profiles were similar to those of dCas9 without an effector domain and were also highly specific52,62,68.
It is thought that the specificity of dCas9 for gene regulation comes from the fact that effective transcription regulation requires dCas9–sgRNA to bind within a small ‘window’ of sequence around the transcription start site (TSS) and to interact with local transcription factors or Pol complexes. A set of rules was determined to optimize the efficacy of sgRNAs in modulating gene expression by testing a library of sgRNAs _targeting the region surrounding the TSSs of 49 genes that had previously been shown to make cells susceptible to ricin41,69. For CRISPRi, strong repression was achieved when KRAB–dCas9 was _targeted to a window from −50 to +300 bp relative to the TSS of a specific gene, with maximum repression detected at +50 to +100 downstream of the TSS. In addition, sgRNAs with protospacer lengths of 18–21 bp were more active than those with longer protospacers, whereas a sequence of identical bases (such as TTTT or CCCC) in the sgRNAs had a negative effect on repression. Neither the choice of _targeting strand nor the GC content of the sgRNAs correlated with repression levels. For CRISPRa using dCas9–SunTag, optimal sgRNA-mediated gene activation was found when _targeting a window between −50 and −400 bp upstream of the TSS; for activation using the SAM system, the optimal window was determined to be between −200 and +1 bp relative to the TSS49. Thus, most off-_target binding events of dCas9, which occur outside these sequences, may not lead to changes in transcription68. Furthermore, many off-_target binding events may be transient and therefore insufficient for modulating transcription of nearby genes. Although the current data in mammalian cells demonstrate that the CRISPRi and CRISPRa systems are specific, further studies are needed to fully understand the causes of off-_target binding and to develop more strategies to minimize it.
CRISPR–dCas9 applications
The CRISPR–dCas9 system is a broadly applicable tool for genome-scale screening, manipulation of dynamic gene programmes and modulation of cell fates. Here, we describe these applications and compare them to alternative approaches.
Genome-wide screens
The ability to regulate essentially any genomic locus enables the study of gene function on a global scale. RNAi has been used for genome-wide screens; however, concerns about its efficiency and specificity still remain1,2. Overexpression screening methods have relied on the construction and delivery of cDNA vector libraries; however, difficulties exist in manipulating multiple gene isoforms simultaneously, in addition to the high cost and difficulty of cloning such cDNA libraries. The ability to easily design and clone sgRNAs makes the Cas9 system a powerful approach for genome-wide screens using oligonucleotide synthesis. Several studies have used Cas9 to conduct genetic knockouts for genome-scale loss-of-function screens70–73.
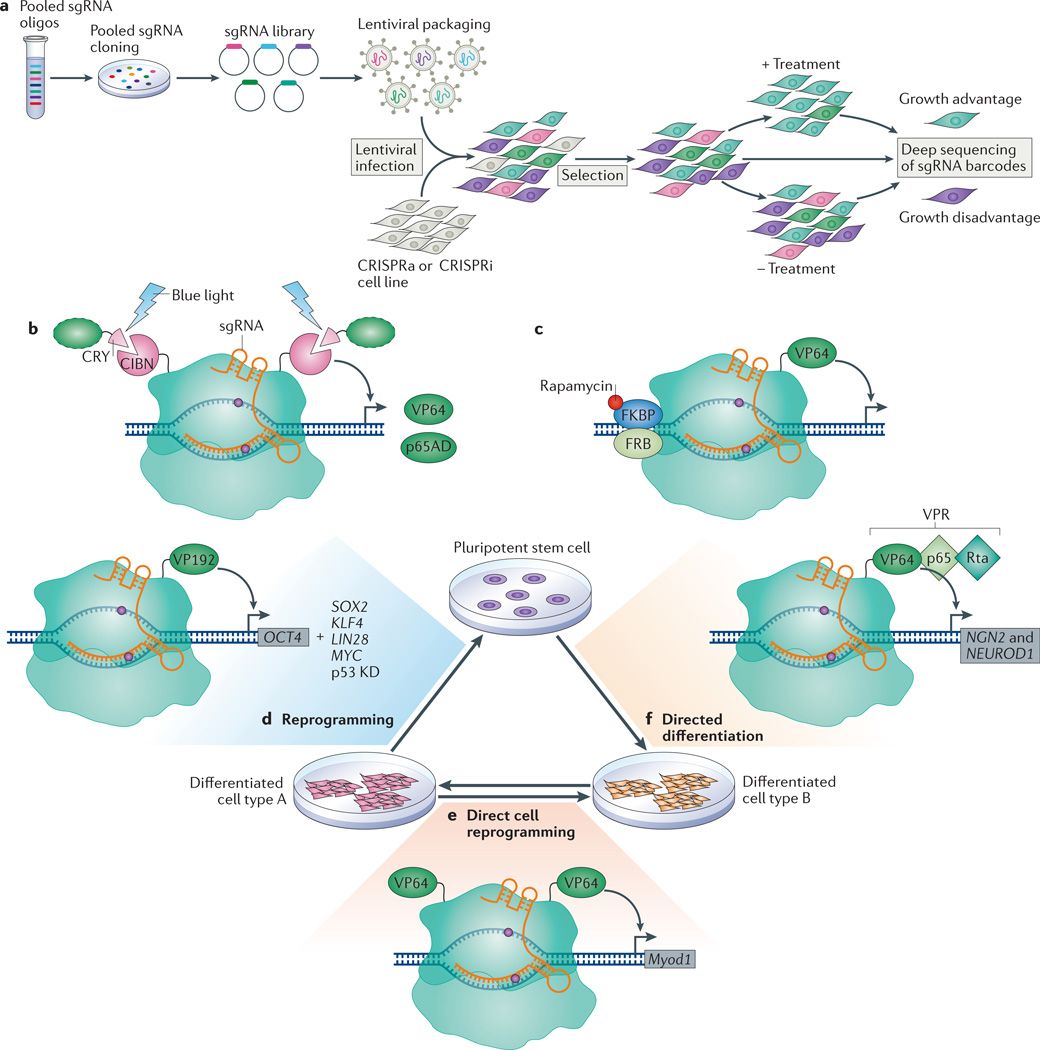
Distinct from Cas9-mediated screens, the dCas9 systems allow for both genome-wide loss-of-function (using CRISPRi) and gain-of-function (using CRISPRa) screens. The CRISPRi and CRISPRa screens are based on the pooled approach, in which sgRNAs are synthesized as a mixture of oligonucleotides and then cloned in mixture to generate an sgRNA vector library (FIG. 4a). This library is packaged into viral particles that are used to transduce mammalian cells at a low multiplicity of infection, achieving genomic integration rates of one sgRNA per cell. The different sgRNAs are barcoded so that their identity can be assayed by deep sequencing to infer which gene is _targeted, and thus activated or repressed, in any particular cell. The relative abundance of each sgRNA at the end of the screen is indicative of the effect of silencing or activating the _targeted gene under the specific experimental conditions (FIG. 4a).
Figure 4. Applications of the CRISPR–dCas9 technology.
a | Overview of pooled screening using CRISPR interference (CRISPRi) or CRISPR activation (CRISPRa). Growth-based screens identify _targeting single guide RNAs (sgRNAs) that confer growth advantage or disadvantage on the basis of sgRNA enrichment or depletion in the final population, assessed using deep sequencing. b | Optogenetics-based transcription control using light-dependent peptide heterodimerization. The amino-terminal fragment of CIB1 (CIBN) is fused to both the amino and carboxyl termini of dCas9. The blue light-sensitive cryptochrome 2 protein CRY undergoes a conformational change in the presence of blue light that enables its heterodimerization with CIBN, thus recruiting activation domains (green dashed oval) such as the herpes simplex virus VP16 activation domain (VP64) or the nuclear factor-κB (NF-κB) transactivating subunit activation domain (p65AD). c | In this chemically inducible system for transcription activation, rapamycin induces the dimerization of FK506-binding protein (FKBP) and the FKBP–rapamycin-binding domain (FRB). Thus, a dCas9 protein that is split into dCas9 (N terminus)–FRB and FKBP–dCas9 (C terminus)–VP64 can be reassembled by the introduction of rapamycin. d | The _targeted activation of octamer-binding 4 (OCT4) with dCas9–VP192 (12 repeats of VP16) can replace transgenic expression of OCT4 to achieve reprogramming from a differentiated cell to a pluripotent stem cell, but this also requires the transgenic expression of SRY-box 2 (SOX2), Krüppel-like factor 4 (KLF4), LIN-28 homologue A (LIN28) and MYC, and the knockdown (KD) of p53. e | Fusion of two VP64 domains to dCas9 induced the expression of myogenic differentiation 1 (Myod1) in mouse embryonic fibroblasts, which caused direct cell reprogramming into skeletal myocytes. f | Activation of neurogenin 2 (NGN2) and neuronal differentiation 1 (NEUROD1), using dCas9–VPR (where VPR is a complex of VP64, p65AD and Epstein–Barr virus R transactivator Rta) and a mixed pool of sgRNAs, directed the differentiation of pluripotent stem cells into neuron-like cells.
A recent study used a genome-wide sgRNA library _targeting each gene with ten sgRNAs per gene for CRISPRi screening. Myelogenous leukaemia K562 cells expressing KRAB–dCas9 and the sgRNA library were cultured with or without a chimeric toxin composed of the diphtheria toxin catalytic subunit linked to cholera toxin (CTx–DTA)41. The screen revealed both known and unanticipated genes that control sensitivity to CTx–DTA41. Additionally, using a large library of non-_targeting sgRNAs, the researchers found that 99.5% of control sgRNAs had no activity, thus demonstrating the high specificity of the CRISPRi system. Robust repression (80–99% knockdown of genes) was demonstrated by validating the top hits individually. The strong repression and low off-_target activity are clear advantages of CRISPRi; however, there are still important uses for RNAi-based screens. For example, CRISPRi modulates transcription at the TSSs of endogenous genes; therefore, it is difficult to _target specific splice isoforms. By contrast, RNAi can be _targeted to specific mature transcripts74. Therefore, the use of CRISPRi and RNAi in conjunction may hold the potential for more complete analysis of gene function.
Two reports have demonstrated the use of CRISPRa for genome-wide screens. As a complementary approach to the CRISPRi screen, one CRISPRa screen utilized the dCas9–SunTag system to probe genes that modulate sensitivity to CTx–DTA41. Interestingly, this gain-of-function screen provided both new and complementary information to the results of the CTx–DTA CRISPRi screen. In another study, the SAM system was used to activate all human transcript isoforms in a malignant melanoma cell line and screen for genes that confer resistance to an inhibitor of the proto-oncogene Ser/Thr kinase B-RAF (BRAF)49. They discovered novel resistance-conferring candidates, in addition to validating known resistance genes. The advantages of CRISPRa screens over the cDNA overexpression approach include the ability to assay the consequences of activating an endogenous gene locus, and the ability to drive the expression of multiple splicing isoforms with one _targeting sgRNA49. The possibility of manipulating multiple genes in single cells may enable large-scale screens that will help to elucidate genetic interactions and uncover networks of proteins that are important for cell fate and function.
Gene regulation by inducible CRISPR–dCas9 tools
CRISPR–dCas9 can be combined with other tools to control gene expression in a spatial and temporal manner, which is useful for understanding dynamic gene networks. Several transcription control strategies based on optogenetics have been developed that utilize light-inducible peptide heterodimerization, in which one peptide is fused to a DNA-binding protein and another to a transcription activator. Two groups recently created such light-activated dCas9-effectors using the cryptochrome-based blue light-sensing system CRY–CIB heterodimerizing domains to recruit VP64 or p65AD to dCas9 (REFS 75,76) (FIG. 4b). By illuminating cells with blue light, both studies demonstrated the activation of endogenous genes using a mixture of four sgRNAs; the highest activation levels were comparable to those obtained with the dCas9–VP64 system in HEK293 cells. Although using single sgRNAs with these systems resulted in poor gene activation, these studies demonstrated that _targeted gene regulation could be spatially and temporally controlled in a reversible manner using light. In addition to optical induction, a chemically-inducible system for activating endogenous loci has also been developed on the basis of rapamycin-dependent dimerization of a split dCas9–VP64 (REF. 77) (FIG. 4c). In the future, discovery of other optogenetically inducible (for example, the recently reported nMag system78) or chemically inducible dimerization systems may expand and optimize inducible dCas9-based regulation tools, which would then ideally be introduced into whole organisms to drive activation or repression of precise spatial and temporal gene expression programmes in vivo.
Cell fate engineering
By specifically controlling gene expression, CRISPRi and CRISPRa can be used to modulate cell identity, reprogramming and differentiation. To reprogramme HEK293T cells into induced pluripotent stem cells (iPSCs), the human OCT4 promoter was activated by _targeting of multiple sgRNAs with dCas9–VP64; only modest gene activation was achieved79, which was considerably enhanced by expression of the epigenetic modifier p300, albeit to levels that were still not sufficient to drive reprogramming. Although somatic cell reprogramming into human iPSCs solely using CRISPRa has not been achieved, dCas9–VP192 (12 copies of VP16) in combination with multiple sgRNAs _targeting the OCT4 promoter was able to replace transgenic OCT4 expression, but reprogramming still required the overexpression of the additional reprogramming factors80 (FIG. 4d). In addition to somatic cell reprogramming into iPSCs, direct lineage reprogramming has been attempted using CRISPRa tools. A fusion of two VP64 domains flanking dCas9 was used to induce the transcription of Myod1 in mouse embryonic fibroblasts, which caused them to differentiate into skeletal myocytes81 (FIG. 4e). Activation of human MYOD1 was also achieved; however, levels were much lower than for mouse Myod1 and were not sufficient to reprogramme human fibroblasts to skeletal myocytes81. These results are promising for the use of CRISPRa in reprogramming; however, it is clear that current levels of activation are insufficient to drive reprogramming or direct lineage reprogramming of most human cell types.
Achieving robust and homogenous differentiation of pluripotent cells will be essential for disease modelling, and CRISPRa or CRISPRi have potential to direct such differentiation. The activation of a key marker of endoderm, SOX17, was achieved using dCas9–VP64 with multiple sgRNAs in human ES cells82. The same group then tested the ability of dCas9–KRAB to repress OCT4 in human ES cells. They achieved significant repression of OCT4, as well as downregulation of NANOG, influencing the pluripotency expression network82. Additionally, enhanced activation systems such as dCas9–VPR have been utilized for the differentiation of human iPSCs into neuronal cells by activating neurogenin 2 (NGN2) and neuronal differentiation 1 (NEUROD1) with a mixed pool of 30 _targeting sgRNAs47 (FIG. 4f). These studies provide promising evidence of the ability to use CRISPRi or CRISPR for direct reprogramming and differentiation. This would provide a new, CRISPR-based approach for cell fate modulation, improving our ability to use pluripotent stem cells for disease studies and for future therapies.
Conclusions and future perspectives
The CRISPR–Cas9 nuclease system offers a powerful approach for precisely modifying genomic sequences, allowing the study of gene function at nucleotide resolution. The ability to correct genetic mutations in a permanent manner will be an important aspect of this tool for future therapeutics. Another advantage of the CRISPR–Cas9 nuclease is that it enables complete genetic loss of function. However, loss of function often results in bimodality, wherein a cell population exhibits loss of function, while other cells acquire in-frame mutations and may retain gene function. This can be partially alleviated by the use of homology-directed repair with CRISPR–Cas9 but, to date, this process remains inefficient. We believe that CRISPRi and CRISPRa are useful tools to use in concordance with gene-editing strategies. The nuclease-deactivated dCas9 offers the ability to transiently or stably control gene expression without altering the genomic sequence. Partial loss-of-function studies are important for our understanding of gene function, in particular when studying essential genes. In addition, this technology offers a relatively simple method for manipulating the expression of multiple genes and thus is also important for the study of polygenic diseases. Furthermore, CRISPRi and CRISPRa may allow quantitative tuning of gene expression and thus understanding of how gene dosage drives processes such as cell proliferation and differentiation or disease progression. Studies to enhance the efficiency of repression or activation, the development of inducible tools, and the creation of improved orthogonal systems for parallel activation and repression in the same cell will further broaden the use of CRISPRi and CRISPRa.
The strength of the Cas9 nuclease system in studying human disease has been demonstrated by the correction of genetic mutations in animal models83–87. Genome-wide association studies have identified many disease- and trait-associated genetic variants, with up to 93% of these found outside the protein-coding sequence. This implies that the aberrant regulation of gene expression and non-coding RNAs is important in the aetiology of diseases88. Thus, methods for manipulating gene expression could be vital for disease research. The use of CRISPRi for in vivo gene regulation is likely to offer an alternative to RNAi for studying gene function, and for modelling and therapeutics. Perhaps even more significantly, in vivo activation studies are likely to benefit from CRISPRa, as activation of multiple genes can be achieved simply by expressing several small sgRNAs.
The CRISPRi and CRISPRa technologies will benefit from the discovery and use of other Cas9 orthologues, in addition to the creation of dCas9-knock-in animal models. The most commonly used S. pyogenes Cas9 protein is encoded by a 4.2 kb gene, which is just within the packaging limit of adeno-associated virus (AAV) vectors. Recently, a smaller Cas9 orthologue, from S. aureus, was shown to have similar editing capabilities to the S. pyogenes Cas9, but its gene is 25% shorter. The smaller size facilitates its packaging with a sgRNA cassette into a single AAV vector for in vivo delivery59. Other Cas9-like proteins, such as Cpf1 (CRISPR from Prevotella and Francisella 1), have been shown to exhibit different mechanisms for DNA cleavage89. It would be interesting to look at the nuclease-deactivated versions of these proteins and explore their potential for sequence-specific gene regulation. For example, they may show different binding affinities and/ or interact with local transcription factors differently. In summary, there is much to be explored before we develop fully comprehensive CRISPR–dCas9 or dCas9-like toolkits for transcription regulation and related biomedical research and clinical applications.
Acknowledgments
The authors thank the members of the Qi lab for advice and helpful discussions. L.S.Q. acknowledges support from the U.S. National Institutes of Health (NIH) Office of the Director (OD) and National Institute of Dental & Craniofacial Research (NIDCR). A.A.D. acknowledges support through the Milton Safenowitz Post Doctoral Fellowship for ALS Research. This work was supported by NIH R01 DA036858 (to W.A.L. and L.S.Q.), the Howard Hughes Medical Institute (grant to W.A.L.) and NIH DP5 OD017887 (to A.A.D. and L.S.Q.).
Glossary
- CRISPR–Cas
(Clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins). CRISPR are bacterial DNA loci containing short repeat segments that match foreign DNA elements. Together with Cas proteins, they form an adaptive immune system in bacteria and archaea, which can acquire sequence segments from foreign DNA and use these sequences to recognize and destroy the foreign _target DNA.
- Type II CRISPR system
CRISPR–Cas system that encodes cas9, cas1 and cas2 within the CRISPR–cas loci, in addition to a tracrRNA, which is partially complementary to repeats in the CRISPR array.
- Single guide RNA
(sgRNA). A synthetic RNA chimera containing a hairpin that links the transactivating CRISPR RNA (tracrRNA) to the crRNA and functions similarly to the native crRNA–tracrRNA duplex, directing Cas9 to a specific genomic locus.
- Protospacer-adjacent motif
(PAM). A short sequence in the _target DNA (not in the guide RNA) that is necessary for the successful _targeting of Cas9. The PAM sequence varies between bacterial species. In Streptococcus pyogenes, it is NGG (which is more effective) or NAG (less effective).
- Transactivating CRISPR RNA
(tracrRNA). A small RNA encoded upstream of the CRISPR locus in type II CRISPR systems, with a 24-nucleotide sequence complementary to repeats of the CRISPR RNA (crRNA) precursor transcripts. Essential for the processing of pre-crRNA to mature crRNA.
- CRISPR RNA
(crRNA). Small RNAs transcribed from the protospacer within the CRISPR array. Together with the transactivating crRNA (tracrRNA), crRNA guides Cas9 to a specific genomic locus.
- Krüppel-associated box
(KRAB). A conserved domain of a transcription repressor that can be fused to DNA-binding proteins for _targeted transcription repression.
- mSin3 interaction domains
Interaction domains that are present on multiple transcriptional repressor proteins.
- VP64
A transcription activator composed of four tandem copies of the herpes simplex virus VP16 activation domain connected by Gly-Ser linkers. VP64 is often fused to DNA-binding proteins for _targeted transcription activation.
- p65 activation domain
(p65AD). The principal transactivation domain of the 65 kDa polypeptide of the nuclear form of the NF-κB transcription factor.
- Single-chain variable fragment
(scFV). A fusion protein in which the epitope-binding regions of the heavy and light chains of an antibody are connected by a short linker peptide and are expressed in soluble form in cells.
- Mediator complex
A multi-subunit complex that is required for the transcription of most RNA polymerase II transcripts.
- RNA aptamers
RNA molecules that have high affinity and specificity for _target molecules.
- Optogenetics
A technique that utilizes optics for achieving spatiotemporal gene regulation of cells in living tissues.
- CRY–CIB heterodimerizing domains
A light-inducible protein interaction between the blue light-sensitive cryptochrome 2 protein (CRY2) and its interacting partner CIB1 from Arabidopsis thaliana.
- Direct lineage reprogramming
The conversion of fully differentiated cells of a certain type into another cell type, while bypassing the intermediate pluripotent state.
- Adeno-associated virus (AAV) vectors
Viral vectors with small packaging capacity, commonly used in gene therapy, which can infect both dividing and non-dividing cells and do not integrate into the host genome. AAV vectors have been approved for clinical use.
Footnotes
Competing interests statement
The authors declare no competing interests.
FURTHER INFORMATION
CRISPRi protocol by Nature Protocols: http://www.nature.com/nprot/journal/v8/n11/full/nprot.2013.132.html
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 2014;15:591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson AL, et al. Expression profiling reveals off-_target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 3.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene _targeting in human cells. Science. 2003;300:763–763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 5.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 6.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christian M, et al. _targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabadi AM, Gersbach CA. Engineering synthetic TALE and CRISPR/Cas9 transcription factors for regulating gene expression. Methods. 2014;69:188–197. doi: 10.1016/j.ymeth.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 11.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 12.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 13. Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. This paper shows that Cas9, when paired with a tracrRNA–crRNA complex or a chimeric sgRNA, can recognize and cut specific DNA sequences.
- 14.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. This paper describes the nuclease-deficient Cas9 (dCas9) and its use as an RNA-guided DNA-binding platform for gene repression in bacteria and mammalian cells.
- 16.Makarova KS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015;13:1–15. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the _targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 18.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. Along with references 21 and 22, this paper demonstrates the use of the nuclease Cas9 for genome editing in mammalian cells.
- 24.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR–Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SW, Kim S, Kim JM, Kim JS. _targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 26.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR–Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 27.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR–Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sander JD, Joung JK. CRISPR–Cas systems for editing, regulating and _targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson MH, et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma H, et al. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc. Natl Acad. Sci. USA. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem. Biophys. Res. Commun. 2013;439:132–136. doi: 10.1016/j.bbrc.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Fujita T, et al. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) Sci. Rep. 2013;3:3171. doi: 10.1038/srep03171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. This paper demonstrates an optimized dCas9 system fused with a tandem peptide array (SunTag) for enhanced transcription activation.
- 36.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR–Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji W, et al. Specific gene repression by CRISPRi system transferred through bacterial conjugation. ACS Synth. Biol. 2014;3:929–931. doi: 10.1021/sb500036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawhorn IE, Ferreira JP, Wang CL. Evaluation of sgRNA _target sites for CRISPR-mediated repression of TP53. PLoS ONE. 2014;9:e113232. doi: 10.1371/journal.pone.0113232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. This paper describes the use of dCas9 fusion proteins for gain-of-function and loss-of-function screens.
- 42.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao X, et al. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by _targeting enhancers. Nucleic Acids Res. 2014;42:e155–e155. doi: 10.1093/nar/gku836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. This paper, together with references 38 and 42–45, describes the use of dCas9 fused with the repressor KRAB or the activator VP64 for manipulating gene expression in mammalian and yeast cells.
- 47. Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. This paper demonstrates the use of a tripartite system (VPR) for efficient transcription activation.
- 48.Mali P, et al. CAS9 transcriptional activators for _target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature. 2014;517:583–588. doi: 10.1038/nature14136. This paper describes the use of an engineered activator system (SAM) for efficient transcription activation and for genome-wide activation screening.
- 50.Kearns NA, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeom YI, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 52.Thakore PI, et al. Highly specific epigenome editing by CRISPR–Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hilton IB, et al. Epigenome editing by a CRISPR–Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. Along with reference 50, this paper describes the fusion of dCas9 with a histone-modifying enzyme for _targeted epigenetic modifications.
- 54.Keung AJ, Bashor CJ, Kiriakov S, Collins JJ, Khalil AS. Using _targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell. 2014;158:110–120. doi: 10.1016/j.cell.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zalatan JG, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. This paper describes the development of scRNAs with dCas9 for multiplexed and parallel transcription activation and repression in mammalian cells.
- 56.Briner AE, et al. Guide RNA functional modules direct cas9 activity and orthogonality. Mol. Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 57.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-_target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 62.O’Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ. A genome-wide analysis of Cas9 binding specificity using ChIP–seq and _targeted sequence capture. Nucleic Acids Res. 2015;43:3389–3404. doi: 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-_target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu Y, et al. High-frequency off-_target mutagenesis induced by CRISPR–Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu PD, et al. DNA _targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems _targeting β-globin and CCR5 genes have substantial off-_target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polstein LR, et al. Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res. 2015;25:1158–1169. doi: 10.1101/gr.179044.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–922. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 71.Shalem O, et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR–Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 74.Kampmann M, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc. Natl Acad. Sci. USA. 2015;112:E3384–E3391. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Polstein LR, Gersbach CA. A light-inducible CRISPR–Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR–Cas9-based photoactivatable transcription system. Chem. Biol. 2015;22:169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 77.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR–Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 79.Hu J, et al. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res. 2014;42:4375–4390. doi: 10.1093/nar/gku109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balboa D, et al. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Rep. 2015;5:448–594. doi: 10.1016/j.stemcr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chakraborty S, et al. A CRISPR/Cas9-Based system for reprogramming cell lineage specification. Stem Cell Rep. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Platt RJ, et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swiech L, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR–Cas9. Nat. Biotechnol. 2014;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Niu Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene _targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 88.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]