Abstract
The opportunistic fungal pathogen Candida albicans is an increasingly common threat to human health. Candida albicans grows in several morphologies and mutant strains locked in yeast or filamentous forms have attenuated virulence in the murine model of disseminated candidiasis. Thus, the ability to change shape is important for virulence. The transcriptional repressors Nrg1p and Tup1p are required for normal regulation of C. albicans morphology. Strains lacking either NRG1 or TUP1 are constitutively pseudohyphal under yeast growth conditions, and display attenuated virulence in the disseminated model. To dissect the relative importance of hyphae and pseudohyphae during an infection, we used strains in which the morphological transition could be externally manipulated through controlled expression of NRG1 or TUP1. Remarkably, hyphal form inocula retain the capacity to cause disease. Whilst induction of a pseudohyphal morphology through depletion of TUP1 did result in attenuated virulence, this was not due to a defect in the ability to escape the bloodstream. Instead, we observed that pseudohyphal cells are cleared from tissues much more efficiently than either hyphal (virulent) or yeast form (avirulent) cells, indicating that different C. albicans morphologies have distinct interactions with host cells during an infection.
Keywords: Candida albicans, filamentation, virulence, pseudohyphae, NRG1, TUP1
Different morphological forms of Candida albicans are similar in the ability to escape the bloodstream but pseudohyphae are cleared more efficiently.
Graphical Abstract Figure.
Different morphological forms of Candida albicans are similar in the ability to escape the bloodstream but pseudohyphae are cleared more efficiently.
INTRODUCTION
Candida albicans is part of the normal human flora, but this opportunistic pathogen remains the main causative agent of invasive fungal infections and is associated with high morbidity and mortality rates (Wey et al. 1988; Viudes et al. 2002). Candida albicans pathogenesis depends on complex interactions between fungal cells and the host immune system (Casadevall and Pirofski 1999; 2001; 2002; 2003). Moreover, the ability of C. albicans to grow in different morphologies, including yeast cells, pseudohyphae and hyphae, is a pivotal aspect of its capacity to move from the commensal to the disease-causing state. We have previously demonstrated that C. albicans virulence can be altered by manipulating the morphology of the fungus in vivo (Saville et al. 2003; 2006). Mixtures of different cell types are found in infected tissues and in biofilms, suggesting a role for each during infection, although there is variation in the degree of filamentation and biofilm formation in clinical isolates (Hirakawa et al. 2015). Yeast cells are important for establishment of biofilms (Finkel and Mitchell 2011) and for dissemination: they adhere better than filaments to endothelial cells under conditions of flow (Grubb et al. 2009) and strains kept in the yeast form are able to escape the bloodstream and invade tissues (Saville et al. 2003; Chen et al. 2006). However, C. albicans mutant strains that grow only in the yeast (Lo et al. 1997; Stoldt et al. 1997; Saville et al. 2003) or filamentous (Braun and Johnson 1997; Murad et al. 2001) forms are either avirulent or have attenuated virulence in the murine model of systemic candidiasis. Typically a yeast cell culture is used as the starting point for in vitro biofilm formation or to produce infecting inocula in animal models, mainly due to ease of handling and quantification. The handful of previous studies that have examined the virulence potential of filamentous inocula performed only pseudohyphal infections (Braun and Johnson 1997; Murad et al. 2001). The underlying cause of the attenuation observed with these filamentous inocula remains unclear.
Two transcriptional repressors, Nrg1p and Tup1p, control the expression of many genes involved in a variety of processes and are required for normal regulation of C. albicans morphology. Tup1p is a general transcriptional repressor that must be brought to its site of action through interactions with various co-repressors, specifically DNA binding proteins such as Nrg1p. TUP1 is expressed constitutively (Bassilana, Hopkins and Arkowitz 2005) and is known to play a role in regulating cellular morphology because strains lacking TUP1 grow as pseudohyphae under both yeast and hypha-inducing conditions (Braun and Johnson 1997). In contrast, NRG1 transcript levels are high in yeast conditions, but fall rapidly upon hyphal induction (Braun, Kadosh and Johnson 2001; Murad et al. 2001; Cleary et al. 2012). In strains where NRG1 has been deleted, cells grow exclusively as pseudohyphae under yeast conditions, but are able to respond to hypha inducing signals (Braun, Kadosh and Johnson 2001; Murad et al. 2001). In the disseminated model the tup1Δ strain is avirulent and the nrg1Δ strain has attenuated virulence even though the latter strain is still able to form hyphae under inducing conditions (Braun, Kadosh and Johnson 2001; Murad et al. 2001). When mutant strains lacking either TUP1 or NRG1 are grown in vitro under yeast conditions the resulting pseudohyphae form large clumps of filaments (Braun and Johnson 1997; Braun et al. 2000; Braun, Kadosh and Johnson 2001; Murad et al. 2001), making them technically challenging to work with. Unfortunately, since wild-type C. albicans strains form hyphae in vivo, mutant strains such as these currently remain the only available means to study the contribution of the pseudohyphal form to virulence. Whether the decreased virulence of these strains reflected an inability of the pseudohyphal cells to leave the bloodstream and enter the deep organs, or was due to mis-regulation of _target genes in the absence of these transcriptional regulators was left unanswered. To more closely examine the role of morphology at different stages of the infectious process, we used C. albicans strains in which we could control expression of either NRG1 or TUP1, and therefore manipulate the morphology of the fungal cells, both in vitro and in vivo. These strains enabled us to examine the virulence of both hyphal, and pseudohyphal, infecting inocula and also to determine whether the type of filament formed before or after injection could influence the outcome of the infection.
MATERIALS AND METHODS
Strains and media
The yeast strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Strains were routinely maintained as −80°C frozen stocks and grown on yeast extract–peptone–dextrose (YPD). Expression from the tetO promoter was abolished by the addition of 20 μg ml–1 doxycycline to the growth medium or, for the in vivo studies, at 2 mg ml−1 in a 5% sucrose solution as the drinking water for the +DOX animal groups. All plasmid manipulations were performed with Escherichia coli strain DH5α with selection on Luria-Bertani plates containing 100 μg ml−1 ampicillin when necessary.
Table 1.
Strains used in this study.
| Strain | Parent | Genotypea | Reference |
|---|---|---|---|
| CAF2-1 | wild-type | Gillum et al. (1984) | |
| nrg1Δ (BCa23-3) | CAI4 | nrg1::hisG-URA3-hisG/nrg1::hisG | Braun, Kadosh and Johnson (2001) |
| THE1 | ade2::hisG/ade2::hisG ura3::λ imm434/ura3:: λ imm434 ENO1/ eno1::ENO1-tetR-ScHAP4AD-3xHA-ADE2 | Nakayama et al. (2000) | |
| tet-NRG1 (SSY50-B) | THE1 | NRG1/nrg1::URA3-tetO-NRG1 | Saville et al. (2003) |
| tup1Δ (BCa2-10) | CAI4 | ura3::λimm434/ura3::λimm434, tup1::hisG/tup1::hisG-URA3-hisG | |
| Braun, Kadosh and Johnson (2001) | |||
| tet-TUP1 (4T3-6) | THE1 | RPS1 /RPS1::URA3-tetO-TUP1, tup1Δ::FRT /tup1Δ::SAT1-FLIPb | |
| This study. |
The full genotype is that of the parental strain with additional modifications as indicated.
SAT1-FLIP denotes the SAT1 flipper cassette.
Table 2.
Plasmids used in this study.
Fluorescence microscopy
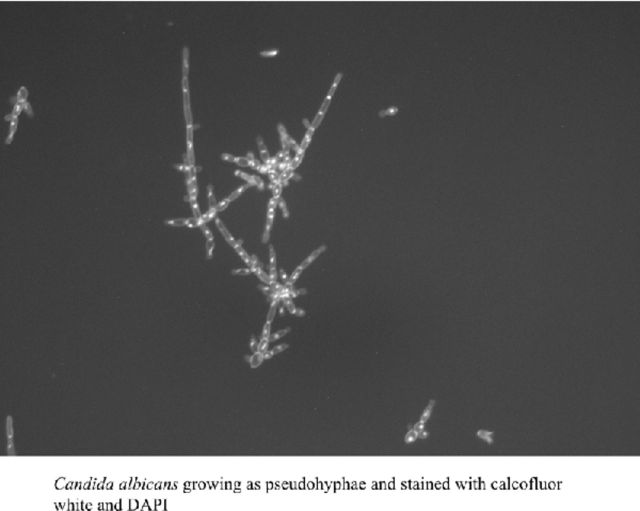
Candida albicans cells were stained using calcofluor white (Sigma), and nuclei were revealed through the use of Vectashield mounting medium containing DAPI (4′,6-diamidino-2-phenylindole; Vector Labs). Cell images were captured using a fluorescence microscope (Axiovert 200M with X-cite 120 fluorescence illumination unit, Carl Zeiss Inc., Germany)) equipped with a digital camera.
Strain construction
Both alleles of TUP1 were sequentially deleted from the transactivator-containing strain THE1 using the SAT1 flipper (Reuss et al. 2004). To construct over-expression strains, the vector CIp10tetO was constructed by liberating the tetO sequence from CIpSATtetO (Cleary et al. 2012) with KpnI and XhoI, then ligating the product between the KpnI and XhoI sites of CIp10. A regulatable allele of TUP1 was constructed by amplifying the coding sequence of TUP1 from C. albicans (strain SC5314) genomic DNA using PfuTurbo® (Stratagene) and primers TUP1_FOR with TUP1_REV. The PCR product was ligated into the SmaI site of pMT3000 to produce plasmid pMTTUP1. Sequence analysis showed no changes in DNA sequence in our clone compared to the reference found in the Candida Genome Database (Arnaud et al. 2005) (retrieved May 20, 2010). The TUP1 coding sequence was liberated from pMTTUP1 by digestion with XhoI and MluI and ligated between the XhoI and MluI sites of CIp10tetO to form CIp10tetOTUP1.
Candida albicans strains were transformed using a modified electroporation transformation method (Kohler, White and Agabian 1997). Nourseothricin-resistant transformants were selected on YPD agar plates containing 100 μg ml–1 nourseothricin (Werner Bioagents, Jena, Germany) as described previously (Reuss et al. 2004). When using the URA3 marker, prototrophic transformants were selected on YNB plates without amino acids (Sherman, Fink and Hicks 1986). Using a CIp10-based vector ensures that URA3 is integrated at a highly transcribed genomic location (Murad et al. 2000) thus eliminating position effects on virulence. Transformants were screened by Southern blot analysis as previously described (Church and Gilbert 1984; Saville et al. 2003; Cleary et al. 2010). Strains in which the TUP1 replacement cassette had integrated correctly were grown in yeast extract–peptone–maltose (YPM) to activate the FLP recombinase and excise the deletion cassette. Resulting nourseothricin-sensitive colonies were selected on YPD agar plates containing 20 μg ml–1 nourseothricin (Werner Bioagents, Jena, Germany) as described previously (Reuss et al. 2004). Gene expression was measured either using northern blot analysis or quantitative real-time PCR as described previously (Cleary and Saville 2010; Cleary et al. 2010). Integration of CIp10-based constructs at RPS1 was confirmed by PCR. For each C. albicans transformation, several independent transformants were isolated and examined to ensure that the different isolates behaved identically. Representative results from single isolates are shown.
Filamentation assays
For filamentation assays in liquid media, strains were grown overnight at 28°C, washed in sterile PBS and diluted 1:20 into fresh RPMI-1640 supplemented with L-glutamine and buffered with MOPS (Angus Buffers and Chemicals) and incubated with shaking at 37ºC.
Murine virulence assays
For injection, liquid cultures were grown overnight at 28°C in YPD. Some cells were then subcultured: for 3 h with and without DOX (for TUP1 depletion); for 90 minutes in YPD plus serum at 37°C to induce hypha formation in the wild-type strain (CAF2-1); for 90 minutes in YPD with doxycycline at 37°C to induce hypha formation in the tet-NRG1 strain (SSY50-B). Cultures containing hyphae were rested briefly to allow very large clumps to settle out. The remaining suspended material containing individual hyphae and small clumps were harvested via pipette, centrifuged and washed three times in sterile pyrogen-free saline. Cells were counted using a haemocytometer, and appropriate dilutions made so that the required dosage of cells could be injected in a final volume of 200 μl into the lateral tail veins of 6- to 8-week-old female BALB/c mice. Confirmation of the number and viability of cells present in the infecting inocula was performed by plate count.
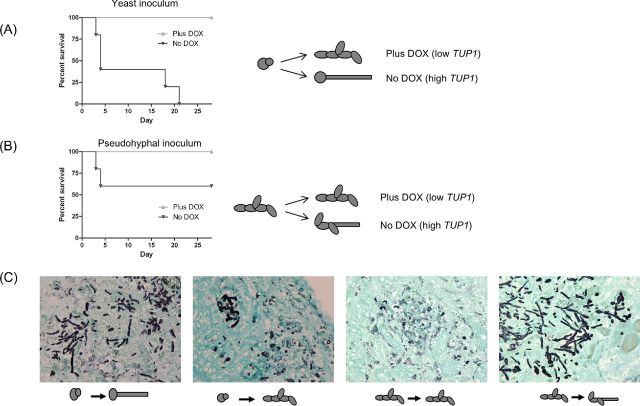
The following doses were used for the virulence studies: for the tet-NRG1 strain (Fig. 1), approximately 9.5 × 105 CFU; for the wild-type strain CAF2-1 (Fig. 1), approximately 5.7 × 105 CFU; for the tet-TUP1 studies (No DOX, Fig 3A), approximately 5 × 105 CFU and for infections with TUP1-depleted inocula (Plus DOX, Fig. 3B), approximately 5.3 × 105 CFU.
Figure 1.
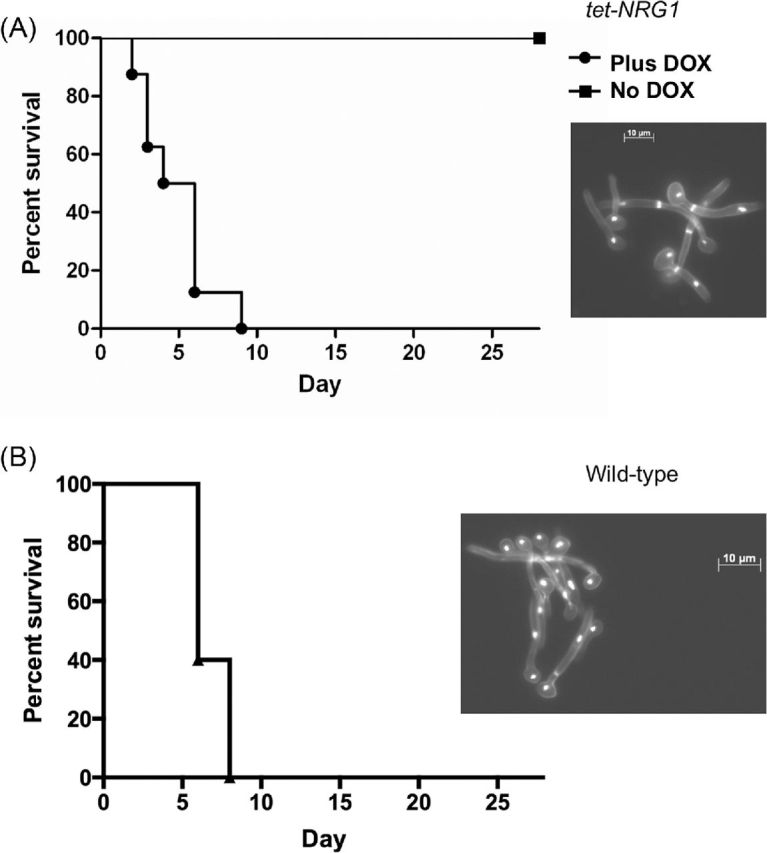
Infections with hyphae are virulent. (A) The tet-NRG1 strain (SSY50-B) was induced to form hyphae (90 minutes, 37°C, YPD+DOX) and approximately 9.5 × 105 CFU injected into mice. Cell morphology of the infecting inoculum is illustrated in the inset panel. Virulence was attenuated when the infecting cells were subsequently directed to yeast form growth (No DOX group). (B) Approximately 5.7 × 105 CFU preformed hyphae (90 minutes, 37°C, YPD + serum) of a wild-type strain (CAF2-1) were injected into mice. Cell morphology of the inoculum is again illustrated in the inset panel. In both cases, preformed hyphae were able to cause disease in mice.
Figure 3.
TUP1 transcript depletion attenuates virulence. (A) Mice succumbed to infection with the tet-TUP1 strain injected as yeast (approximately 5 × 105 CFU) and allowed to form hyphae like a wild-type strain (No DOX, dark lines). However, abolition of TUP1 transcription upon infection (Plus DOX, light lines) resulted in all of the mice surviving. (B) When cells were injected in the pseudohyphal form (after TUP1 depletion, approximately 5.3 × 105 CFU), virulence was attenuated, even when cells were able to subsequently form hyphae (No DOX, dark lines). (C) Histopathological examination of kidneys retrieved from infected mice confirmed that we were able to manipulate C. albicans/fungal morphology in vivo as well as in vitro. Cartoons represent the in vitro morphology prior to injection and predicted outcomes. Note the evidence of fungal element degradation in the DOX treated mice (pseudohyphal infection samples, third panel from left). Sections are from kidneys retrieved from mice sacrificed on day 3 post-infection (see Fig. 5B).
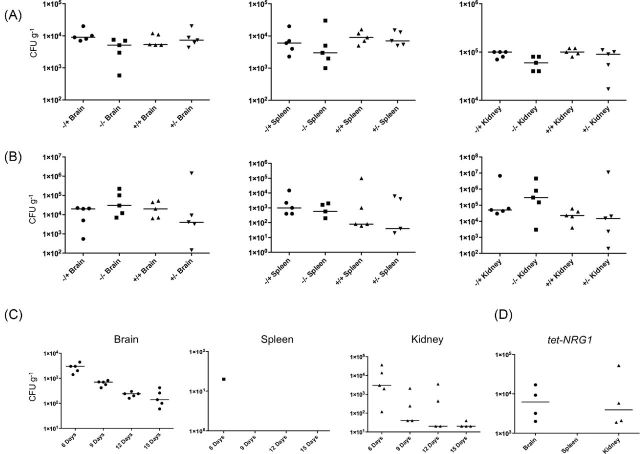
The following doses were used for the fungal burden determination experiments: for 6 h sacrifice (Fig. 4A), approximately 1 × 106 CFU (No DOX) or 1.2 × 106 CFU (Plus DOX, TUP1-depleted) of the tet-TUP1 strain; 3 day sacrifice (Fig. 4B), approximately 5.2 × 105 CFU (No DOX) or 5.8 × 105 (Plus DOX) CFU of the tet-TUP1 strain and for later time points (Fig. 4C), approximately 5.4 × 105 CFU. For the tet-NRG1 strain infections (Fig. 4D), approximately 1.9 × 106 CFU were used.
Figure 4.
Examination of organ fungal burdens at various time points post-infection. Organs were removed from infected mice, homogenized, and samples plated to obtain fungal burdens (CFU g−1). Median burdens are indicated by a horizontal line. Differences in median burdens between pairs of samples were assessed using the Mann–Whitney Test. Some data points included in the analysis result in a score of zero on a logarithmic scale and are not shown. (A) There were only two statistically significant differences 6 h after infection: between the brain burdens of the Plus DOX and No DOX treated groups following yeast form infection (P = 0.0278) and between the kidney burdens of the Plus DOX treated, pseudohyphal (+/+) and the No DOX treated, yeast form (−/−) infections (P = 0.0196). (B) After 3 days, there were no statistically significant differences between any of the groups (P > 0.5). (C) Fungal burdens were determined in the organs retrieved from doxycycline-treated mice sacrificed at various intervals after infection with yeast form cells. By day 6 post-infection there were much fewer fungal organisms recovered from the tissues than at day 3 and by day 15, no fungal organisms were detectable in the spleen and only very few remained in some of the brain samples. (D) Fungal burdens were examined in the organs retrieved from mice infected with the tet-NRG1 strain maintained in the yeast form (No DOX) and sacrificed after 27 days. Even after this extended period, there are numerous fungal organisms present in the brain and kidney.
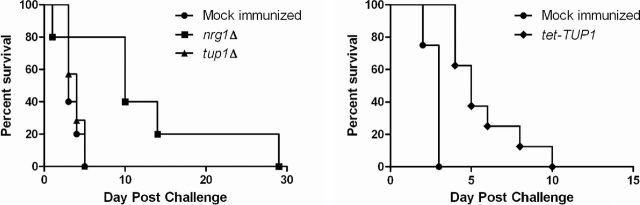
For the immunization experiments (Fig. 5), mice injected with the nrg1Δ strain (approximately 4.7 × 105 CFU), the tup1Δ strain (approximately 4.3 × 105 CFU), or a saline control were subsequently challenged with approximately 5.2 × 105 CFU of wild-type strain CAF2-1. Mice injected with the tet-TUP1 strain (approximately 4.3 × 105 CFU) or saline (control) were subsequently challenged with approximately 2.8 × 105 CFU of wild-type strain CAF2-1.
Figure 5.
Immunization assays. Mice were infected with one of the three indicated strains (or saline) and subsequently challenged with a wild-type strain (CAF2-1). Inoculation with the tup1Δ strain (approximately 4.3 × 105 CFU) did not provide protection from the subsequent wild-type strain challenge (approximately 5.2 × 105 CFU), with no statistically significant difference between the survival curves of this strain and the saline control (P = 0.6475). Immunization with the nrg1Δ mutant strain (approximately 4.7 × 105 CFU) did offer some protection and significantly altered the survival kinetics (P = 0.0348). Immunization with our regulatable tet-TUP1 strain (approximately 4.3 × 105 CFU, mice treated with DOX) also caused a significant alteration in the survival times (P = 0.0002) compared to the mock-immunized control when challenged with a wild-type strain (approximately 2.8 × 105 CFU).
Groups of five to eight mice were used for each condition. Days on which the animals died were recorded; severely moribund animals were humanely sacrificed to minimize suffering and recorded as having died the following day. In all experiments, one kidney was processed for histopathology, whereas the other kidney, the brain and the spleen were homogenized and fungal loads determined by plating dilutions onto Sabouraud agar plates. All experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of The University of Texas Health Science Center at San Antonio or by the Institutional Animal Care and Use Committee (IACUC) of The University of Texas at San Antonio (both AAALAC accredited institutions) and performed in accordance with institutional regulations. Mice were allowed a one week acclimatization period before experiments were started.
Statistical analyses
For statistical analyses of survival curves, the Log-rank (Mantel–Cox) test was used to detect statistically significant differences between groups. For statistical analyses of fungal burdens (CFU), the Mann–Whitney test was used to detect statistically significant differences between pairs of groups. Analyses were performed using Prism 6 (GraphPad Software Inc).
Histopathology
Kidneys excised from deceased or sacrificed mice were fixed in 10% buffered formalin and stored at 4°C until required. Kidneys were embedded in paraffin, tissue slices were cut and stained with Grocott-Gomori methenamine-silver (GMS) (Grocott 1955) prior to microscopic evaluation.
RNA isolation and quantitative PCR
RNA was isolated using the MasterPure Yeast RNA Extraction Kit (Epicentre Biotechnologies) and treated with amplification grade DNase I (Invitrogen) to remove any genomic DNA. For quantification of transcripts, cDNA was synthesized with random hexamers (Applied Biosystems) using the MasterScript cDNA synthesis kit (5prime). Primer pairs (Table 3) were used in conjunction with GoTaq® qPCR Master Mix (Promega) and twin.tec real-time 96 well PCR plates (Eppendorf) in an ABI 7300 Real Time PCR System (Applied Biosystems). Dissociation curves were analyzed for all reactions to verify the presence of single peaks/products. Expression levels were analyzed using ABI 7300 System SDS Software (Applied Biosystems). All quantitative real-time PCR experiments were performed in biological triplicate.
Table 3.
Oligonucleotides used in this study.
| Name | Sequencea | Reference |
|---|---|---|
| TUP1_LHF _UPS | 5′- TTGGGCAGTGAGCTCTTTACTACTC -3′ | This study. |
| TUP1_LHF _DS | 5′- ATACATGGACATCCGCGGTGGGGATGG -3′ | This study. |
| TUP1_RHF_UPS | 5′- GGCGATTGTAACTCGAGAATTTGGAA -3′ | This study. |
| TUP1_RHF_UPS | 5′- GGGCCCTTCTCAAGTCTGCTAAACACG-3′ | This study. |
| TUP1_For | 5′- CTCGAGATGTCCATGTATCCCCAACG-3′ | This study. |
| TUP1_Rev | 5′- CAATTGCACTTATTTTTTGGTCCATTTCC-3′ | This study. |
| ACT1-S | 5′-ATGTGTAAAGCCGGTTTTGCCG-3′ | (Toyoda et al. 2004) |
| ACT1-A | 5′-CCATATCGTCCCAGTTGGAAAC-3′ | (Toyoda et al. 2004) |
| TUP1qFOR | 5′- GCCCATAGAAAAATCAAAGAGG-3′ | This Study. |
| TUP1qREV | 5′- GCCATTCTTCATTTGCCTGT-3′ | This Study. |
Underlined sequences indicate restriction enzyme recognition sites engineered into the oligonucleotides.
RESULTS AND DISCUSSION
Hyphal inocula are virulent
Animal infection studies using various C. albicans mutant strains that are defective in the ability to transition between the different morphotypes have demonstrated that the capacity to change morphology is pivotal for C. albicans to cause disseminated disease. Infected tissues and biofilms contain a mixture of yeast and filamentous cells, suggesting that each form provides some contribution to virulence. Indeed yeast cells appear to be particularly important for dissemination (Saville et al. 2003), whilst hyphae are essential for cell-cell adhesion and biofilm formation (Finkel and Mitchell 2011). All of the previous studies that have examined the virulence of filamentous inocula have used mutant strains that are unable to grow as yeast and the injected cells were in the pseudohyphal form (Braun et al. 2000; Braun, Kadosh and Johnson 2001; Murad et al. 2001). Whilst all of these reported that the filamentous inocula displayed reduced virulence, the nature of the defect remained unexplored. Moreover, despite the fact that all of these prior studies used wild-type strain cells grown in the yeast form for the control group infections, they are widely used as evidence to suggest that pre-formed C. albicans filaments are attenuated for virulence in the mouse model of haematogenously disseminated candidiasis. In contrast, we set out to test the virulence of both hyphal and pseudohyphal inocula and to examine whether the type of filament could influence the outcome of the infection.
To evaluate the infectious potential of hyphal cells, we used our regulatable tet-NRG1 strain (SSY50-B), in which one allele of NRG1 has been placed under the control of the tetO promoter and the other allele remains under the control of the native promoter, in the haematogenously disseminated murine model of candidiasis. We have previously shown that when NRG1 over-expression is induced at various time points after a yeast form infection that virulence becomes attenuated and several animals ultimately survive the infection (Saville et al. 2006). In the present study, hypha formation was induced by growth in YPD Plus DOX at 37ºC for 90 minutes. These hyphae were injected into two groups of mice, half treated with DOX and half untreated. Consistent with our previous observations, in conditions where NRG1 transcription was elevated and the tet-NRG1 strain reverts to, and is kept in, the yeast form (No DOX group), all of the mice survived. In contrast, when tet-NRG1 expression remained off (Plus DOX), thus permitting continued hypha formation in vivo, all of the mice succumbed to the infection (Fig. 1A). To further verify this observation and confirm that it was not a peculiarity of our tet-NRG1 strain, we repeated this experiment using hyphae (induced by growth in YPD plus serum at 37ºC for 90 minutes) of the wild-type strain CAF2-1; this also resulted in mice succumbing to the infection. Our observation that preformed hyphal inocula of a wild-type strain or the tet-NRG1 strain have the capacity to cause disease is in contrast to the previous studies which reported that infections with filamentous forms (pseudohyphae grown in yeast conditions) were either avirulent or highly attenuated for virulence in this animal model (Braun et al. 2000; Murad et al. 2001).
Regulatable tet-TUP1 expression
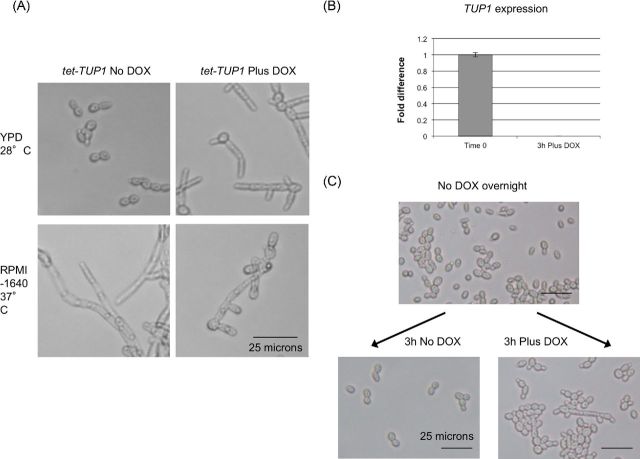
The tup1Δ null strain is constitutively pseudohyphal and unable to form hyphae even under inducing conditions (Braun and Johnson 1997). In liquid media the cells form large aggregates, making the strain difficult to manipulate in vitro and adversely affecting the accuracy of cell counts and administration of infecting inocula. It was possible that the previously observed avirulent phenotype of the tup1Δ null strain could result from either a failure to disseminate related to the altered morphology, or to inappropriate expression of a gene or genes involved in virulence in the animal model. To more precisely study the contribution of the pseudohyphal morphology to virulence, we constructed a strain in which we could manipulate expression of TUP1. Both alleles of TUP1 were deleted from the transactivator-containing strain THE1 and an ectopic tet-regulatable TUP1 allele introduced at the RPS1 locus to produce the tet-TUP1 strain. In the absence of doxycycline TUP1 is expressed from the tet-regulated allele and the strain behaves similar to the wild-type, growing as yeast cells under non-inducing conditions and forming hyphae in response to appropriate stimuli such as growth at 37ºC (Fig. 2A, left panels). In the presence of DOX, TUP1 expression ceases (Fig. 2B, right panels) and the strain grows exclusively as pseudohyphae (Fig. 2A). The manipulability of this strain would allow us to infect with either yeast or pre-formed pseudohyphae, and to more easily isolate individual cells or small clumps of cells than when using the tup1Δ null mutant. The results of infection experiments using this strain are described below and summarized in Table S1 (Supporting Information).
Figure 2.
Abolishing TUP1 transcription from a regulatable promoter induces pseudohyphae. (A) When TUP1 is expressed (No DOX) our tet-TUP1 strain forms yeast under yeast growth conditions (YPD, 28°C) and hyphae under inducing conditions (RPMI-1640, 37°C). When grown in the presence of DOX, the strain forms pseudohyphae in both yeast and hypha inducing conditions. (B) Quantitative real-time PCR analysis was used to verify that expression of TUP1 from the tet-regulated allele was modulated by addition of doxycycline to the medium and that 3 h growth (YPD, 28°C) was sufficient to reduce TUP1 expression below the detectable level. Levels were normalized by ACT1 and expressed relative to the time zero sample (therefore those results are 1.0). Error bars represent the standard error of the mean. (C) After growth in the presence of doxycycline (YPD, 28°C) for 3 h chains and clumps of elongated pseudohyphal cells form. Cells grown in the absence of doxycycline continued to grow as isolated yeast form cells under the same conditions.
TUP1 transcript depletion attenuates virulence in a disseminated model
The tet-TUP1 strain was cultured in the absence of DOX to produce yeast form cells, which were injected into mice. After injection of these yeast cells into mice, expression of TUP1 was modulated through the omission or addition of doxycycline to the drinking water. The injected yeast form cells were permitted to behave like a wild-type strain and form hyphae (No DOX) or induced to form pseudohyphae via TUP1 transcript depletion (Plus DOX). The tup1Δ strain is avirulent (data not shown and (Murad et al. 2001)) and we observed that modulation of TUP1 expression post-infection was sufficient to significantly attenuate virulence (P = 0.0018) (Fig. 3A).
To generate a pseudohyphal inoculum, we depleted the TUP1 transcript prior to injection by culturing the tet-TUP1 strain in the presence of DOX for 3 h. As a pseudohypha or small clump of cells is made up of more than a single fungal element, the plate count method of estimating CFUs used during infection must therefore underestimate the dose of a pseudohyphal inoculum compared to one in the yeast form. We attempted to minimize this variance by collecting cells at a time point where TUP1 has been depleted, but before large aggregates form (Fig. 2B and C). After injection of these pseudohyphae into mice, TUP1 expression was modulated through the omission or addition of doxycycline to the drinking water; cells would either continue to grow as pseudohyphae in the absence of TUP1 expression (Plus DOX) or be permitted to form hyphae as in a wild-type strain when TUP1 expression was restored (No DOX). Cells kept in the pseudohyphal form (Plus DOX) were avirulent whilst those permitted to form hyphae (No DOX) demonstrated attenuated virulence with a little over half (or 60%) of the mice surviving the infection (Fig. 3B).
Infections with pseudohyphal cells show varying degrees of attenuation compared to yeast form infections. Previously reported results, and our own experiments using pseudohyphal inocula of the constitutively filamentous nrg1Δ or tup1Δ strains (not shown) or preformed pseudohyphae of our regulatable TUP1 strain, reveal differing virulence properties; the tup1Δ mutant is completely avirulent whilst the other two strains are still capable of killing at least some of the mice. Interestingly, this difference in virulence correlates to the nrg1Δ and our tet-TUP1 strain (in the absence of DOX) retaining the ability to form hyphae upon proper inducing signals, whereas strains lacking TUP1 expression cannot.
TUP1 transcript depletion has little effect on dissemination and proliferation in vivo
It was not known whether the previously reported avirulence of the tup1Δ mutant was due to a defect in dissemination due to its pseudohyphal form, or perhaps to a change in the capacity of these organisms to replicate within the host due to changes in TUP1-dependent gene expression. Since differences in morphology can influence adhesion to host cells under conditions of flow (Grubb et al. 2009) and could conceivably affect the ability of cells of different morphologies to escape the bloodstream, we used our newly constructed tet-TUP1 strain to explore the role of morphology in this process. To that end, we examined the fungal burden in different tissues from animals infected with yeast or pseudohyphal (TUP1-depleted) inocula and sacrificed 6h or 3 days post infection.
Our overall observation was that there was no marked difference in organ fungal burdens between the different infectious inocula or post-infection treatment in Pairwise comparisons using the Mann–Whitney test to compare median burdens. In the organs removed after 6 h, most comparisons showed no significant differences between the groups (Fig. 4A). In two exceptions, between the brain burdens of the Plus DOX (−/+) and No DOX (−/−) treated groups following yeast form infection (P = 0.0278) and between the kidney burdens of the Plus DOX treated, pseudohyphal (+/+) and the No DOX treated, yeast form (−/−) infections (P = 0.0196), there were higher burdens in the TUP1-depleted (pseudohyphal growth in vivo) sample. No significant differences were noted (P > 0.5) in the fungal burdens in organs removed 3 days after infection (Fig. 4B). The similarity in fungal burdens, irrespective of infecting cell morphology or of post-infection treatment, lead us to conclude that the observed defect in virulence resulting from the absence of TUP1 is not due to these pseudohyphal cells being unable to disseminate into the deep organs during the early stages of infection. Histopathological examination of the kidneys retrieved from infected animals indicated that the DOX treatment regulated the morphology of the fungal cells in vivo as it had in vitro (Fig. 3C; GMS stained sections). Perhaps most interesting, however, was the observation that there appeared to be greater evidence of fungal element degradation in kidneys retrieved from DOX treated animals (Fig. 3C, TUP1 depletion, infecting cells are pseudohyphal).
When cells are maintained in the yeast form, through NRG1 over-expression, they are able to persist for a considerable time without causing disease (Fig. 4D, Saville et al. 2003), before eventually being cleared from the organs by the host immune system. Since we noted an apparent degradation of the TUP1 transcript-depleted fungal elements at early time points during our histological analysis, we performed a more extended time course infection and sacrifice experiment to examine how long these pseudohyphal cells were able to persist in the tissues. Yeast form cells of the tet-TUP1 strain were injected into DOX treated mice (where the cells would form pseudohyphae as a result of reduced TUP1 expression) and fungal burdens in various organs determined 6, 9, 12 and 15 days post-injection. We observed a rapid decline in the number of viable fungal organisms recovered from the organs and, remarkably, by day 15 the organs were virtually sterile, except for a very few cells remaining in the brain (Fig. 4C). Taken together with our other results, it seems that TUP1 depletion attenuates virulence without strongly influencing dissemination or proliferation during the early stages of infection. Perhaps this attenuation is a consequence of differences in interactions with the host immune system between the TUP1-depleted cells and wild-type, virulent organisms. TUP1 might be required for expression of virulence factors required to overcome host defences, and thus TUP1-depleted cells, whilst able to disseminate throughout the body, are unable to cause disease. The rapid clearance of these cells from the host suggests additional changes in the relationship between the immune system and the pathogen. Perhaps TUP1 depletion results in the de-repression of genes encoding surface proteins not normally expressed during an infection. Such a change might help to recruit immune cells to the site of infection, alert the immune system, or stimulate specific immune mechanisms such as phagocytosis.
Infections with different cell morphologies provide varied protection to a subsequent challenge
Given that the TUP1-depleted cells apparently stimulate an efficient immune response, we sought to examine whether exposure to these pseudohyphal cells might confer protection against a subsequent wild-type challenge, as we have previously demonstrated for cells kept in the yeast form (Saville et al. 2009). We therefore infected mice with three different strains (nrg1Δ, tup1Δ or TUP1-depleted tet-TUP1) along with a mock-immunized saline control group. After 14 days, the mice were challenged with a wild-type C. albicans strain (CAF2-1).
Interestingly, infection with a sub-lethal dose of the nrg1Δ strain offered some protection from a subsequent wild-type challenge (Fig. 5). However, infection with the tup1Δ strain did not confer any protection (Fig. 5) with no significant difference in survival kinetics between the mock-immunized and the tup1Δ-immunized groups (P = 0.6475). To assess the protective potential of the tet-TUP1 strain, yeast form cells were injected into mice treated with DOX, where TUP1 would be depleted and the infecting cells would grow as pseudohyphae in vivo. Unlike the tup1Δ strain, this treatment did offer modest protection against a subsequent wild-type strain challenge, with significant differences in the survival times between the mock-immunized, control group and the tet-TUP1-immunized groups (P = 0.0002). Although both of these strains (tup1Δ and tet-TUP1 + DOX) lack TUP1 during the infection, the morphology of the infecting cells was different (yeast versus pseudohyphae) and this, intriguingly, resulted in altered virulence kinetics following the wild-type strain challenge (Fig. 5). It should be noted, however, that all of the mice still succumbed to the infection, which is markedly different to the almost complete protection offered by our tet-NRG1 strain maintained in the yeast form (Saville et al. 2009).
SUMMARY
In this study we have explored the contributions of the differing filamentous C. albicans morphologies to virulence. Previous studies suggested that filamentous inocula were avirulent, but by growing cells in hypha-inducing conditions prior to infection we observed that pre-formed hyphal inocula possess the capacity to cause disease in the disseminated model. The influence of pseudohyphae and the role of the transcriptional regulator TUP1 in virulence are complex, and can be difficult to study due to the necessity of using genetically altered strains. Whilst induction of a pseudohyphal morphology through depletion of TUP1 did result in attenuated virulence, this was not due to a defect in the ability to escape from the bloodstream and enter the deep organs. Rather, TUP1-depleted, pseudohyphal cells are cleared from tissues much more efficiently than yeast (which are avirulent) or cells that are able to form hyphae and make the transition between different morphologies (which are virulent). Our results further demonstrate the differing contributions each of the various cell types makes towards C. albicans virulence, the complex nature of host immune interactions with infecting cells, and suggest that manipulating fungal gene expression and/or cell morphology in a clinical setting could influence survival outcomes.
Supplementary Material
Acknowledgments
We would like to thank Joachim Morschhaüser for providing plasmid pSFS2, Mark Paget for providing plasmid pMT3000, Alistair Brown for providing plasmid CIp10, Hironobu Nakayama for providing strain THE1 and Alexander Johnson for providing the tup1Δ (Bca2-10) and nrg1Δ (Bca23-3) strains. We would also like to thank Dr Karen Wozniak for helpful discussions.
SUPPLEMENTARY DATA
FUNDING
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health [Grants RO1 AI064562 to JLLR and RO1 AI063256-01 to SPS].
Conflict of interest. None declared.
REFERENCES
- Arnaud MB, Costanzo MC, Skrzypek MS, et al. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 2005;33:D358–63. doi: 10.1093/nar/gki003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M, Hopkins J, Arkowitz RA. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot Cell. 2005;4:588–603. doi: 10.1128/EC.4.3.588-603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Head WS, Wang MX, et al. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics. 2000;156:31–44. doi: 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–9. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–61. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski L. Host-pathogen interactions: the attributes of virulence. J Infect Dis. 2001;184:337–44. doi: 10.1086/322044. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun. 1999;67:3703–13. doi: 10.1128/iai.67.8.3703-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. What is a pathogen? Ann Med. 2002;34:2–4. doi: 10.1080/078538902317338580. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Pirofski LA. “Anti-virulence” genes–further muddling the lexicon? Response from Arturo Casadevall and Liise-anne Pirofski. Trends Microbiol. 2003;11:413–4. doi: 10.1016/s0966-842x(03)00209-9. [DOI] [PubMed] [Google Scholar]
- Chen CG, Yang YL, Cheng HH, et al. Non-lethal Candida albicans cph1/cph1 efg1/efg1 transcription factor mutant establishing restricted zone of infection in a mouse model of systemic infection. Int J Immunopath Ph. 2006;19:561–5. doi: 10.1177/039463200601900312. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. P Natl Acad Sci USA. 1984;81:1991–5. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary IA, Lazzell AL, Monteagudo C, et al. BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol Microbiol. 2012;85:557–73. doi: 10.1111/j.1365-2958.2012.08127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary IA, Mulabagal P, Reinhard SM, et al. Pseudohyphal regulation by the transcription factor Rfg1p in Candida albicans. Eukaryot Cell. 2010;9:1363–73. doi: 10.1128/EC.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary IA, Saville SP. An analysis of the impact of NRG1 overexpression on the Candida albicans response to specific environmental stimuli. Mycopathologia. 2010;170:1–10. doi: 10.1007/s11046-010-9297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–18. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–82. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- Grocott RG. A stain for fungi in tissue sections and smears using Gomori's methenamine-silver nitrate technic. Am J Clin Pathol. 1955;25:975–9. doi: 10.1093/ajcp/25.8_ts.0975. [DOI] [PubMed] [Google Scholar]
- Grubb SE, Murdoch C, Sudbery PE, et al. Adhesion of Candida albicans to endothelial cells under physiological conditions of flow. Infect Immun. 2009;77:3872–8. doi: 10.1128/IAI.00518-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa MP, Martinez DA, Sakthikumar S, et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 2015;25:413–25. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler GA, White TC, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol. 1997;179:2331–8. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–49. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Murad AM, Lee PR, Broadbent ID, et al. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast. 2000;16:325–7. doi: 10.1002/1097-0061(20000315)16:4<325::AID-YEA538>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–52. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Mio T, Nagahashi S, et al. Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans. Infection and immunity. 2000;68:6712–19. doi: 10.1128/iai.68.12.6712-6719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Hintermann G, Smith CP. Construction and application of streptomycete promoter probe vectors which employ the Streptomyces glaucescens tyrosinase-encoding gene as reporter. Gene. 1994;146:105–10. doi: 10.1016/0378-1119(94)90842-7. [DOI] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, et al. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–27. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Bryant AP, et al. Inhibition of filamentation can be used to treat disseminated candidiasis. Antimicrob Agents Ch. 2006;50:3312–6. doi: 10.1128/AAC.00628-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Chaturvedi AK, et al. Efficacy of a genetically engineered Candida albicans tet-NRG1 strain as an experimental live attenuated vaccine against hematogenously disseminated candidiasis. Clin Vaccine Immunol. 2009;16:430–2. doi: 10.1128/CVI.00480-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, et al. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–60. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks J. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, et al. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–91. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda M, Cho T, Kaminishi H, et al. Transcriptional profiling of the early stages of germination in Candida albicans by real-time RT-PCR. FEMS Yeast Res. 2004;5:287–96. doi: 10.1016/j.femsyr.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Viudes A, Peman J, Canton E, et al. Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur J Clin Microbiol Infect Dis. 2002;21:767–74. doi: 10.1007/s10096-002-0822-1. [DOI] [PubMed] [Google Scholar]
- Wey SB, Mori M, Pfaller MA, et al. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch Intern Med. 1988;148:2642–5. doi: 10.1001/archinte.148.12.2642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.