Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the 4th leading cause of cancer-related mortality in the United States. Aggressive treatment regimens have not changed the disease course, and the median survival has just recently reached a year. Several mechanisms are proposed to play a role in PDAC therapeutic resistance, including hypoxia, which creates a more aggressive phenotype with increased metastatic potential and impaired therapeutic efficacy. AP Endonuclease-1/Redox Effector Factor 1 (APE1/Ref-1) is a multi-functional protein possessing a DNA repair function in base excision repair and the ability to reduce oxidized transcription factors, enabling them to bind to their DNA _target sequences. APE1/Ref-1 regulates several transcription factors involved in survival mechanisms, tumor growth, and hypoxia signaling. Here, we explore the mechanisms underlying PDAC cell responses to hypoxia and modulation of APE1/Ref-1 redox signaling activity, which regulates the transcriptional activation of hypoxia inducible factor 1 alpha (HIF1α). Carbonic anhydrase IX (CA9) is regulated by HIF1α and functions as part of the cellular response to hypoxia to regulate intracellular pH, thereby promoting cell survival. We hypothesized that modulating APE1/Ref-1 function will block activation of downstream transcription factors, STAT3 and HIF1α, interfering with hypoxia-induced gene expression. We demonstrate APE1/Ref-1 inhibition in patient-derived and established PDAC cells results in decreased HIF1α–mediated induction of CA9. Furthermore, an ex vivo 3D tumor co-culture model demonstrates dramatic enhancement of APE1/Ref-1-induced cell killing upon dual-_targeting of APE1/Ref-1 and CA9. Both APE1/Ref-1 and CA9 are under clinical development, therefore these studies have the potential to direct novel PDAC therapeutic treatment.
Keywords: APE1, CA9, HIF-1, redox, hypoxia, synthetic lethality
Introduction
Pancreatic ductal adenocarcinoma (PDAC) continues to be the 4th leading cause of cancer related mortality in the United States in both genders with a five-year survival rate of 5–7% (1). Approximately, 80% of patients present with advanced disease due to local invasion or metastatic disease (1, 2). Treatment options for this patient population are limited to palliative chemotherapy, and even the most aggressive treatment strategies fail to extend life beyond one year for most patients (1, 3). The disappointing result in pancreatic cancer may be explained by the complexity of this disease. Several mechanisms of resistance have been proposed to play a role in resistance to therapy. The presence of cancer-associated fibroblasts (CAFs) as well as other cell types within a desmoplastic stroma, hypoperfusion of the tumor, hypoxia, multidrug resistance and other mechanisms have been reported (4–6). Lack of clinical efficacy is due, at least in part, to the fibrosis that accompanies the disease. In the studies presented here, tumor-stroma effects were monitored as the importance of the stroma in PDAC is well-established (7, 8). _targeting one pathway is very unlikely to alter the natural course of this disease. However affecting critical survival mechanisms of this cancer is crucial to produce any positive results (9, 10).
Hypoxic conditions in pancreatic tumors are associated with poor prognosis. Oxygen deprivation leads to stabilization of hypoxia inducible factor 1 alpha (HIF1α), a transcription factor that upregulates a variety of factors that contribute to increased drug resistance, proliferation, and migration/invasion in tumor cells (11–13). HIF-1 transcriptional activity depends on stabilization of its α subunit, which is _targeted for degradation under normoxic conditions by proline hydroxylation and subsequent von Hippel-Lindau protein (VHL)-mediated ubiquitination. Stable HIF1α dimerizes with the constitutively expressed b subunit to activate genes with hypoxia-response elements (HREs) in their promoters (14, 15). No HIF-1-specific inhibitors currently exist, so _targeting its vital transcriptional _targets and the enzymes that regulate HIF-1 activity are promising ways to modulate hypoxia signaling in cancer cells (15, 16).
Apurinic/Apyrimidinic Endonuclease/Redox Factor-1 (APE1/Ref-1) is a multifunctional protein that is involved in responses to oxidative stress, acting on both oxidative and alkylative DNA damage (via its endonuclease activity in base excision repair) (17–19) and augmenting activity of various transcription factors (via its redox signaling) (20–23), as well as contributing to clearance of RNA with damaged bases (24). APE1/Ref-1 expression and redox activity are increased in PDAC tissue, and its upregulation increases tumor cell migration, proliferation, and survival (17, 23). We previously demonstrated that APE1/Ref-1 redox activity regulates several transcription factors involved in pancreatic cancer signaling, including HIF-1α, as well as STAT3, and NFκB (20, 21).
Carbonic Anhydrase IX (CA9) is a transmembrane protein that regulates pH in tumor cells under low oxygen conditions and contributes to cell proliferation and transformation (25, 26). CA9 was reported to be an endogenous sensor to HIF-1 activity (27) and CA9 inhibitors were more potent in 3D culture due to the regions of hypoxia (28). One report demonstrated a strong correlation in vivo between pSTAT3 and CA9 expression and postulated that the regulation of CA9 by STAT3 could be important in invasion through IL-6 signaling (29). Both of these transcription factors are regulated by APE1/Ref-1, so the impact of APE1/Ref-1 redox activity on HIF1 activity as well as CA9 expression was evaluated in PDAC following exposure to hypoxia.
Here we describe the mechanisms by which APE1/Ref-1 regulates hypoxia signaling through HIF1α-mediated transcription. Inhibiting APE1/Ref-1 redox signaling with the selective inhibitor APX3330 (also called E3330) resulted in decreased STAT3 activity (20) and HIF1α activity (21) leading to decreased expression of CA9, a major HIF-1 _target within cells (27). We also demonstrate that hypoxia stimulates interactions between APE1/Ref-1 and its redox _targets, HIF1α and STAT3, but not NFκB in both PDAC tumor and stromal cells supporting our approach.
We also show for the first time evidence using patient-derived tumor cells in the presence of appropriate stromal components, CAFs, 3D spheroid size and proliferation are dramatically reduced upon combination treatment with APE1/Ref-1 inhibitor, APX3330 and CA9 inhibitor, SLC-0111 (Clinical Trial NCT02215850) (30, 31). Upon blockade of multiple hypoxia signaling pathways with APX3330 and SLC-0111, we observe a dramatic effect on 3D tumor spheroid growth even in the presence of the protective environment of the CAFs. APE1/Ref-1 redox inhibitor APX3330 (20, 22, 32, 33) is slated for Phase 1 clinical trials in mid-2016, and the CA9 inhibitor is currently in clinical trials. By decreasing APE1/Ref-1 redox activity we can impair the tumor cells’ response to hypoxia (21) and potentially improve the response to therapy. Therefore the combination of these agents may have proximate clinical applications.
Methods and Materials
Cell Culture
Cells were maintained in culture as previously described (20–22). Patient-derived tumor cells, Pa03C, Pa02C, and Panc10.05 and CAF19 cells were a kind gift from Dr. Anirban Maitra (The Johns Hopkins University) (34). Upon receipt of the cells in 2011 as well as in June of 2015, we used STR analysis (CellCheck with IDEXX BioResearch) to confirm that we indeed received the aforementioned cells from Dr. Maitra and that they were mycoplasma-free. MIA-PaCa2 cells were purchased from and authenticated by ATCC (Manassas, VA). Cancer-associated fibroblasts, UH1303-02 were isolated using the outgrowth method from patient tumor tissue as previously described (35). All patient-derived lines were passaged only 10 times before new stocks frozen prior to authentication were resuscitated. Hypoxia exposure was achieved using a Ruskinn Invivo2 200 hypoxia work station. CMV-EGFP-WT APE1/Ref-1 lentiviral construct was used to overexpress APE1/Ref-1 as previously described (36). To detect the cells for imaging, a CMV-EGFP lentiviral construct was used as previously described (36). Additionally, 150 pfu/cell of the pCL7TdTOMwo lentiviral vector was incubated with Pa03C and Panc10.05 cells for 48 hours to make cells stably express TdTomato.
Western Blot Analysis
Western blots were performed as previously described (20–23) with antibodies for APE1/Ref-1 (Novus Biologicals; Littleton, CO), HIF1α (GeneTex; Irvine, CA), STAT1, STAT3, (Cell Signaling; Danvers, MA), NFκB (abcam; Cambridge, MA), CA9 (Santa Cruz; Dallas, Texas), Vinculin (Sigma; St. Louis, MO), and Actin (NeoMarkers; Fremont, CA).
Co-immunoprecipitation
Samples were co-immunoprecipitated using the Pierce Co-IP kit (Thermo Scientific) with modifications as previously described (22).
Transfection
PDAC and CAF cells were transfected with APE1/Ref -1 siRNA as previously described (20, 22, 23). siRNAs used were: #1 or scrambled control (previously reported) and two LifeTech validated siRNAs (#2: s1445 and #4: s1447) (22). APE1/Ref-1 siRNA #1 was used as the standard siRNA unless otherwise specified.
Transient Luciferase Reporter Assays
MIA-PaCa-2 cells were co-transfected with constructs containing luciferase driven by HIF1α and a Renilla luciferase reporter vector as previously described (22) using X-tremeGENE 9 DNA transfection reagent (Roche, Indianapolis, IN) along with siRNA as described above. Firefly and Renilla luciferase activities were assayed by using the Dual Luciferase Reporter Assay System (Promega Corp.) as before (20, 22).
qRT-PCR Reactions
qRT-PCR was used to measure the mRNA expression levels of CA9 as previously described (21). Cells were treated with APE1/Ref-1 siRNA or increasing amounts of APX3330 in the presence or absence of hypoxia (1% and 0.2 % O2) for 24 h, then total RNA was extracted from cells using the QiagenRNeasy Mini kit (Valencia, CA) (37). First-strand cDNA synthesis and quantitative PCR were performed as previously described (21). The relative quantitative mRNA level was determined using the comparative Ct method using 18S rRNA, RPLP0, and B2M as reference genes (37, 38). The primers for CA9, 18S, RPLP0, and B2M are commercially available (Applied Biosystems).
Inhibitors
Compounds were prepared and used as previously described: APX3330 (32, 33) and SLC-0111 (30, 31). The concentrations of APX3330 used are within clinically tolerated levels established previously by Eisai pharmaceutical company through a prior development program that evaluated the toxicology and phase I/II safety and clinical profile in non-cancer patients. Additionally, the SLC-0111 analog FC13-555A was synthesized as described in Supplemental Methods. The structure of each inhibitor can be seen in Supplemental Figure 1.
Cell Proliferation
PDAC cell proliferation in monolayer was measured using the Alamar Blue assay as previously described (22). Cells treated with APX3330 and SLC-0111 were exposed to hypoxia for six days followed by addition of Alamar Blue reagent (Invitrogen) and subsequent fluorescence analysis. Fold change refers to the fluorescence reading for cells treated with indicated inhibitors compared to cells growing in normal media.
pH Assay
Intracellular pH was evaluated using the pHrodo Red AM Intracellular pH Indicator (LifeTech). PDAC cells treated with APX3330 and SLC-0111 were exposed to hypoxia for 48 hours followed by analysis with pHrodo Red AM dye. Intracellular pH Calibration Buffers (LifeTech) were used to create a standard curve of fluorescence intensity for determination of pH values. Results were normalized to MTS analysis to account for changes in proliferation as before (20–22). Fluorescent images were acquired using a confocal/two-photon Olympus Fluoview FV-1000 MPE system (Olympus American) at the Indiana Center for Biological Microscopy facility (Indianapolis, IN).
Statistical Analysis
qPCR data points for scrambled, siAPE1/Ref-1, and hypoxia treatments were analyzed using the 2−ΔΔCT method and analysis of covariance (ANCOVA) models as previously described (22, 39). Data points in tests with multiple treatment groups were analyzed using post-hoc Multiple Comparisons Tests (Tukey, Dunnett, or Sidak as appropriate). For evaluation of data curves using multiple drugs, an extra-sum-of-squares F test was used to compare the goodness-of-fit of a nonlinear regression curve shared between groups with that of separate curves for each group. Differences between the treatment groups and control group were considered significant if p<0.05 following Bonferroni corrections as appropriate. Statistical analyses were performed using SAS (Version 9.3, Copyright ©2010 SAS Institute Inc. Cary, NC) and Prism (Version 6.0f, Copyright ©2014 GraphPad Software Inc. La Jolla, CA).
HIF-1 −/− MEF Generation
HIF-1-floxed mouse embryonic fibroblast (MEF) cells were generated as previously reported (40) and transduced with Ad-CMV-Cre (Cre adenovirus) or Ad-GFP (control) vector (Vector BioLabs; Malvern, PA) for 24 hours using 5 ng/mL polybrene to produce HIF-1-deficient cells (40). PCR was used to verify the deletion of HIF (Supplemental Figure 2).
3D Co-Culture
Ultra low attachment 96-well plates (Corning Inc., Life Sciences) were used to generate 3-dimensional tumor spheroids in the presence and absence of CAFs (75 μL/well) as described previously (41, 42). Cells were stably transduced with EGFP (green) or TdTomato (red) as indicated to preserve the genetic characteristics of the low passage patient cells (34, 42). Cells were re-suspended in colorless DMEM growth media containing 3% Reduced Growth Factor Matrigel (RGF, BD Biosciences) and 5% FBS. Following plating, cells were treated on Days 4 and 8 with media containing 5% serum, 3% RGF Matrigel, and inhibitors as indicated. On Day 12, spheroids were analyzed using Thermo ArrayScan high-content imaging system (43). Images of 3D structures were captured by ArrayScan using a 2.5× objective for TdTomato and EGFP; then 2D projections were processed to quantify differences in total intensity and total area of both CAFs and tumor.
Results
APE1/Ref-1 interactions with HIF1α and STAT3 are stimulated by hypoxia
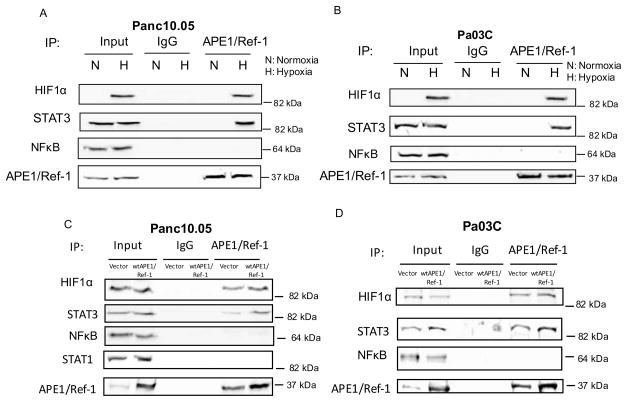
We previously published data demonstrating decreased STAT3, HIF1α, and NFκB activity following knockdown of APE1/Ref-1 and/or inhibition of APE1/Ref-1 redox signaling with the selective inhibitor APX3330 (also called E3330) (20, 21). Similarly, we found that inhibition of APE1/Ref-1 led to a decrease in a major HIF-1 _target within cells, Carbonic Anhydrase IX (CA9) (21, 28). To further dissect the role of APE1/Ref-1 in hypoxia signaling and more clearly determine whether hypoxia stimulates interactions between APE1/Ref-1 and its redox _targets, endogenous APE1/Ref-1 was immunoprecipitated from lysates of two low passage PDAC cell lines (Panc10.05 and Pa03C) under normoxic and hypoxic (0.2% O2) conditions. These cell lines are representative of ductal adenocarcinoma, were previously sequenced (34), and have the common G12D mutation in KRAS as well as missense mutations in p53. Panc10.05 was derived from a primary PDAC tumor, is wt for p16(INK4A), but has a SMAD4 deletion; however Pa03C was isolated from a metastatic lesion in the liver and is wt for both p16(INK4A) and SMAD4. IPs were probed for HIF1α, STAT3, and NFκB. HIF1α and STAT3, but not NFκB were detected in the pull-down fractions under hypoxic conditions, but these interactions were not detected under normoxic conditions (Figure 1A–B). Controls of TNFα (NFκB) and IL-6 (STAT3) were performed to show that interactions between APE1/Ref-1 and the transcription factors it regulates do indeed occur under normoxic conditions with appropriate stimulation (Supplemental Figure 3). Interactions of APE1/Ref-1 with HIF1α and STAT3 were obvious under hypoxic conditions.
Figure 1. APE1/Ref-1 interactions with HIF1α and STAT3 are stimulated by hypoxia in PDAC cells.
Cell extracts were prepared from Panc10.05 (A) and Pa03C (B) cells and also from cells that overexpress APE1/Ref-1 (C, D) following exposure to hypoxia (0.2%) for 24 hr. Extracts were immunoprecipitated with anti-APE1/Ref-1 antibody or IgG. The immunoprecipitated complexes were then probed for HIF1α, STAT3, NFkB, STAT1, or APE1/Ref-1. n = 3–4 per experiment. Typical results shown.
With overexpression of APE1/Ref-1, the interaction with HIF1α and STAT3 remained intact while NFκB was still not detected in IPs from cells overexpressing APE1/Ref-1 following exposure to hypoxia indicating that the amount of APE1/Ref-1 was not limiting in the above panels (Figure 1C–D). To demonstrate that APE1/Ref-1’s interactions with the transcription factors are specific to signaling in hypoxia, we probed for another STAT family member, STAT1. IPs from 10.05 cells were probed for STAT1, which was not detected regardless of the levels of APE1/Ref-1 or oxygen conditions (Figure 1C). Due to the complexity of the disease and the signaling between various cell types in the pancreatic tumor microenvironment, we investigated APE1/Ref-1 interactions with HIF1α, STAT3, and NFκB in CAFs. These CAFs are non-tumorigenic and although they have activated signaling pathways due to their association with the tumor, they are non-transformed. The results were identical to the result with PDAC cells: APE1/Ref-1 interacts with HIF1α and STAT3 under hypoxia, but not NFκB (Supplemental Figure 4). In light of CA9 inhibitors beginning clinical trials and our previous data demonstrating transcriptional regulation of CA9 following APE1/Ref-1 blockade (21), we focused these studies on HIF1α signaling and the regulation of the downstream molecule CA9 through APE1/Ref-1.
APE1/Ref-1 protein expression contributes to hypoxia-induced HIF1α-mediated transcription
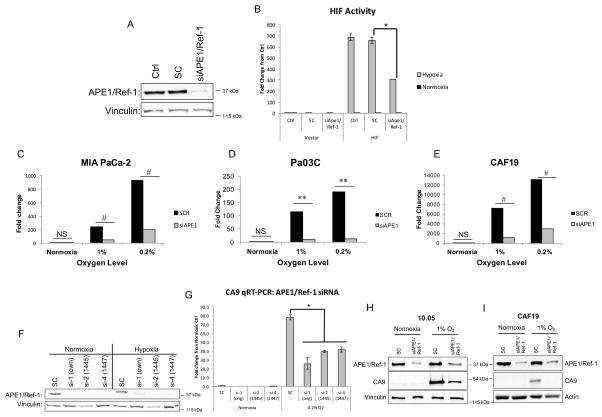
To show that the interactions between APE1/Ref-1 and HIF1α are functionally important, we evaluated the contribution of APE1/Ref-1 to HIF-1 transcriptional activity by co-transfecting MIA PaCa-2 cells with HIF1α-driven firefly luciferase or pLuc-MCS (vector control) alongside APE1/Ref-1 siRNA or scrambled control and exposing cells to hypoxia for 24 hrs. APE1/Ref-1 knock-down resulted in a significant reduction (~47%) in hypoxia-induced HIF1α activity (Figure 2A–B).
Figure 2. APE1/Ref-1 protein expression contributes to hypoxia-induced HIF1α-mediated transcription.
MIA-PaCa2 cells were assayed for HIF1 activity using a luciferase and Renilla reporter assay following APE1/Ref-1 knock-down. A. Knock-down of APE1/Ref-1 was confirmed via western blot. B. HIF1α-driven luciferase expression was evaluated following hypoxia (24 hr, 0.2% oxygen vs. normoxia controls; n=3). C–E. Following APE1/Ref-1 knock-down and 24 hrs in hypoxia, CA9 mRNA levels were evaluated via qPCR in the cell lines described (n=3). F. APE1/Ref-1 knock-down in MIA-PaCa2 cells with three different siRNAs was confirmed via western blot. G. CA9 mRNA levels were evaluated via qPCR in SC and knocked-down samples from three siRNAs following hypoxic conditions (24 hrs, 0.2% oxygen vs. normoxia controls; representative experiment of n=3). H–I. CA9 protein levels were evaluated via western blot in 10.05 and CAF19 cells following transfection with SC or APE1/Ref-1 siRNA and hypoxia (24 hr, 1% oxygen, representative blots of n=3). *p<0.001 (Tukey’s Multiple Comparisons Test); **p<0.01 & #p<0.001 (ANCOVA). For CA9 western blots (H–I), p<0.05 for SC vs. siAPE under hypoxia (Tukey’s Multiple Comparisons Test).
Effects of APE1/Ref-1 on HIF transcriptional activity were further evaluated by examining hypoxia-mediated transcription of HIF-1 _target, CA9. We compared CA9 mRNA levels in two PDAC cell lines and one pancreatic CAF cell line following APE1/Ref-1 knock-down and exposure to hypoxia. Bar graph represents the fold change of mRNA expression level of CA9. Hypoxia-induced CA9 mRNA levels were attenuated by APE1/Ref-1 knock-down in all cell lines at both levels of hypoxia (Figure 2C–E). Variability in the amount of induction in different cell lines may be partially attributable to the extremely low baseline CA9 expression under normoxic conditions. APE1/Ref-1 knock-down similarly attenuated CA9 mRNA levels under hypoxic conditions in two additional primary PDAC cell lines (Supplemental Figure 5). These results were validated in MIA-PaCa-2 cells exposed to hypoxia using two additional APE1/Ref-1-_targeting siRNAs, and similar results were obtained (Figure 2F–G). To verify the reduction in CA9 also occurred at the protein level, hypoxia-induced CA9 protein levels were evaluated via western blot following APE1/Ref-1 knock-down in PDAC cells and pancreatic CAF cells. APE1/Ref-1 knock-down resulted in a ~70% reduction in hypoxia-induced CA9 protein levels (Figure 2H–I).
Hypoxia-induced CA9 transcription is HIF-1-dependent
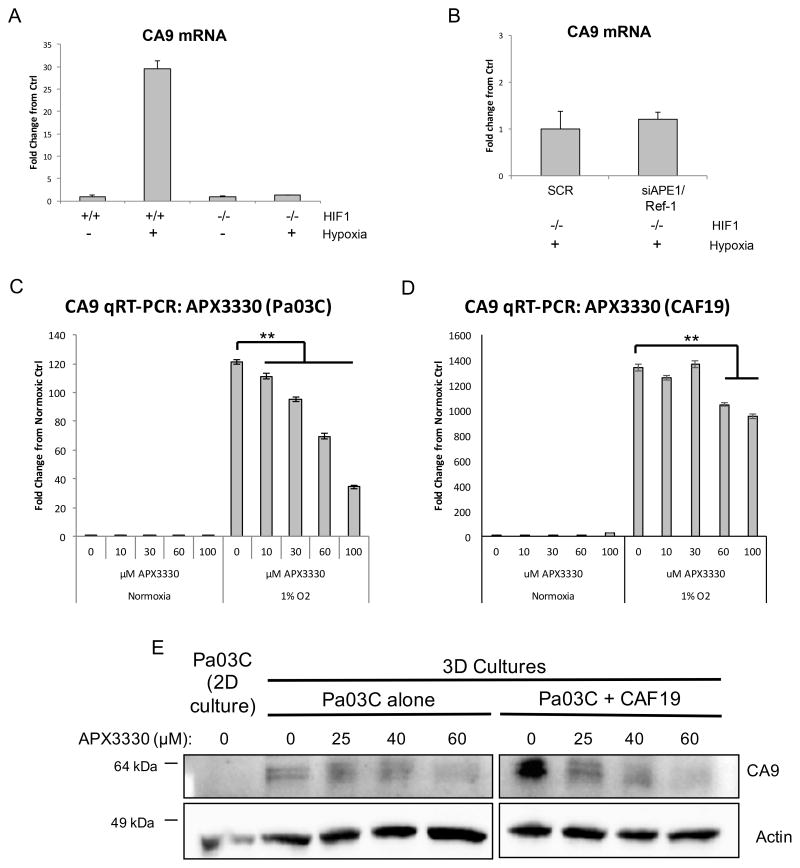
To confirm that the effects of APE1/Ref-1 and hypoxia on CA9 transcription were mediated by HIF-1 activity, we evaluated hypoxia-induced CA9 mRNA levels in HIF-1-deficient (−/−) MEFs following APE1/Ref-1 knock-down. As expected, in HIF-1 proficient MEFs, CA9 is induced 30-fold compared to normal oxygen controls. In HIF-1 −/− MEFs CA9 mRNA levels were not induced by exposure to hypoxia (Figure 3A), or affected by APE1/Ref-1 knock-down (Figure 3B), indicating that CA9 transcription is HIF-1-dependent, regardless of APE1/Ref-1 expression or oxygen levels. HIF-1 depletion in these cells was confirmed by PCR (Supplemental Figure 2).
Figure 3. APE1/Ref-1 redox signaling affects CA9 transcription in a HIF-1-dependent manner.
A. HIF-1-proficient (+/+) and HIF-1-deficient (−/−) mouse embryonic fibroblasts (MEFs) were exposed to 0.2% oxygen for 24 hrs, and CA9 mRNA levels were evaluated by qPCR, representative experiment of n=3. B. HIF-1 −/− MEFs were transfected with SC or APE1/Ref-1-directed siRNA and incubated at 0.2% oxygen for 24 hrs and CA9 mRNA levels were evaluated via qPCR, representative experiment of n=3. C–D. CA9 mRNA levels were evaluated via qPCR following APX3330 treatment and hypoxia (24 hr, 1% oxygen; representative graph of n=3). E. Pa03C cells were collected from monolayer (2D) cultures and 3D tumor spheroid cultures grown in the presence or absence of CAFs following treatment with APX3330, and CA9 protein levels were evaluated via western blot (representative blot of n=2). *p<0.01 & **p<0.001 (Tukey’s Multiple Comparisons Test).
Inhibition of APE1/Ref-1 redox signaling affects CA9 transcription
As a multi-functional protein, APE1/Ref-1 is also involved in base excision repair (BER) of DNA lesions, RNA quality control, and reduction-oxidation (redox) regulation. Knock-down of APE1/Ref-1 affects all of these functions. We therefore examined whether the redox function is responsible for the APE1/Ref-1-mediated regulation of hypoxia signaling pathways using an APE1/Ref-1 specific redox inhibitor that does not affect other APE1/Ref-1 functions (44, 45) and is slated for clinical trial in the summer of 2016. We previously showed that APX3330 decreases CA9 mRNA levels in Panc-1 and MIA-PaCa2 cells exposed to hypoxia (21). Here we expand these results to primary cells and CAF cells, as well as 3D co-cultures. Following treatment with APX3330 and exposure to hypoxia, CA9 mRNA levels in Pa03C cells and in pancreatic CAF cells were attenuated in a dose-dependent manner (Figure 3C–D). Additionally, CA9 protein expression was measured in a 3D co-culture model following inhibition of APE1/Ref-1 with APX3330. While CA9 was not detected under normoxic conditions in the patient-derived Pa03C cells in monolayer, when grown as spheroids, these cells now express CA9. Tumor spheroids grown in the presence of CAFs more strongly upregulated CA9 expression ~3-fold, likely due to larger spheroids containing larger regions of hypoxia, as well as the more complex signaling present with the stromal elements. Inhibition of APE1/Ref-1 redox signaling with APX3330 led to decreased CA9 expression in 3D tumor cultures in a dose-dependent manner, both in the presence and absence of CAF cells (Figure 3E). These data support the use of the 3D co-culture system for preclinical studies validating novel _targets like CA9 and APE1/Ref-1 in PDAC.
Importantly, we evaluated CA9 and APE1/Ref-1 protein expression following exposure to hypoxia (0.2% oxygen) in three PDAC cell lines and found that, while CA9 levels increased over time, APE1/Ref-1 levels did not change significantly (Supplemental Figure 6), indicating that hypoxia-induced CA9 expression is not secondary to APE1/Ref-1 upregulation.
Dual-_targeting of CA9 and APE1/Ref-1 acidifies PDAC cells and inhibits cell viability under hypoxia
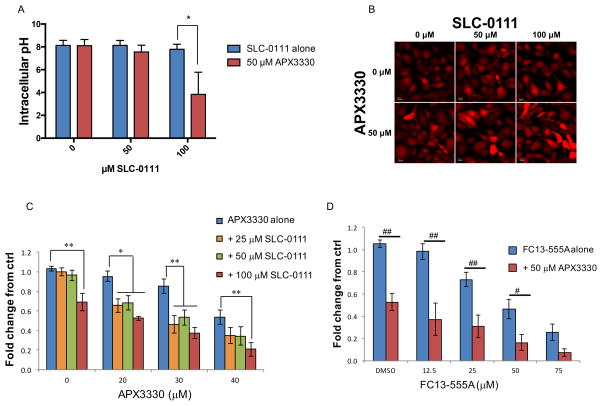
CA9 regulates intracellular pH under hypoxic conditions, and APE1/Ref-1 redox activity contributes to hypoxia-induced CA9 expression. We analyzed intracellular pH in hypoxia-exposed PDAC cells following treatment with CA9 inhibitor, SLC-0111 and the APE1/Ref-1 redox inhibitor, APX3330 using the pHrodo Red AM fluorescent pH indicator as a functional endpoint for carbonic anhydrase activity under hypoxic conditions. The analysis of intracellular pH was performed at an early timepoint (48 hr exposure to inhibitors and hypoxia) to avoid drastic changes in cell survival. Still, some viability changes were observed (Supplemental Figure 7), so these changes were taken into account in the analysis and normalization of the intracellular pH data. Dual-treatment with SLC-0111 and APX3330 results in a greater decrease in intracellular pH than treatment with either inhibitor alone (Figure 4A–B).
Figure 4. Dual-_targeting of CA9 and APE1/Ref-1 acidifies PDAC cells and inhibits cell viability under hypoxia.
A. Panc10.05 cells were treated with APX3330 and SLC-0111 and exposed to hypoxia (0.2% O2) for 48 hrs prior to analysis of intracellular pH (Avg ± SE, n=3). Representative images from pH experiments are shown in B. C–D. Viability assay of Pa02C cells treated with the indicated concentrations of APX3330 and CA9 inhibitors, SLC-0111 or FC13-555A and exposed to hypoxia (0.2%) for six days (Avg ± SE, n=6). Fold change refers to the comparison of each data point to the fluorescence of untreated tumor cells. *p<0.05 & **p<0.01 (Dunnett’s Multiple Comparisons Test); #p<0.05 & ##p<0.01 (Sidak’s Multiple Comparisons Test). Additionally, differences in nonlinear regression curves between treatment groups were confirmed using extra-sum-of-squares F tests followed by Bonferroni Corrections in each experiment (p<0.05 for all dual-treatment curves vs. single-agent curves).
Inhibition of APE1/Ref-1 redox activity results in a dose-dependent decrease in PDAC cell viability following treatment of cells with APX3330 and hypoxia. Remarkably, the effect of APE1/Ref-1 inhibition on cell viability is greatly enhanced by treating with the CA9 inhibitor, SLC-0111 in addition to APX3330 treatment under hypoxia (Figure 4C). In support of these results, new CA9 inhibitors are being developed and the combination of APX3330 with SLC-0111 analog, FC13-555A is also significantly effective at killing PDAC cells in monolayer (Figure 4D), supporting the hypothesis that blockade of hypoxia signaling proteins will be deleterious to PDAC cells.
Dual-_targeting of CA9 and APE1/Ref-1 inhibits PDAC tumor growth in a 3D co-culture model
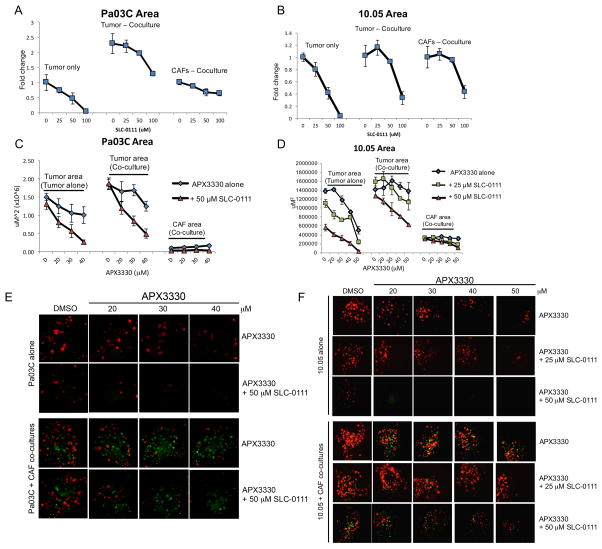
In order to more accurately mimic the tumor microenvironment, we utilized a three-dimensional co-culture model of PDAC that included the low passage patient derived tumor cells as well as cancer-associated fibroblasts. As demonstrated above, the levels of CA9 were greater in these tumor spheroids when grown with CAF cells, and CA9 expression was attenuated by treatment with APX3330 (Figure 3E). Inhibition of CA9 with SLC-0111 was more potent in the 3D model with dramatic effects on tumor cell killing observed at lower doses than in monolayer, as measured by reductions in area of patient-derived cells (Figure 5A–B). Cell killing was more dramatic in the tumor cells than in the CAFs, especially when CAF19 cells were in co-culture with Pa03C cells. Although these CAFs have aberrantly activated signaling pathways, they are non-tumorigenic and suggest that tumor cells are more greatly affected by the dual _targeting approach as compared to normal cells. Similar trends were seen when measuring fluorescence intensity (data not shown). Importantly, inhibition of CA9 can effectively kill tumor cells even when in the protective environment of the CAFs.
Figure 5. Dual-_targeting of CA9 and APE1/Ref-1 inhibits PDAC tumor growth in a 3D co-culture model.
Pa03C (A, C, & E) and Panc10.05 (B, D, & F) tumor cells were grown in 3D cultures in the presence and absence of CAFs. Spheroids were treated with SLC-0111 alone (A & B) and in combination with APX3330 (C & D), and the area of tumor (red) and CAF (green) were quantified following 12 days in culture, n=3–4. Representative images from dual-treatment experiments are shown in E and F. Differences in nonlinear regression curves between treatment groups were confirmed using extra-sum-of-squares F tests followed by Bonferroni Corrections in dual-treatment experiments (p<0.01 for each curve vs. the curve for APX3330 alone in tumor cells alone; p<0.01 for curves with 50 μM SLC-0111 vs. the curve for APX3330 alone in tumor + CAF co-cultures).
To determine if blockade of STAT3 and HIF-mediated transcription alongside the inhibition of CA9 activity would potentiate PDAC cell death, we combined APX3330 and SLC-0111 in the 3D co-culture model. We can assess the effects of dual _targeting on both the tumor alone and the tumor and CAFs in co-culture due to the different fluorescent labels in each cell type. As seen in hypoxia-exposed 2D cultures, addition of CA9 inhibition to APE1/Ref-1 redox inhibition resulted in dramatic potentiation of the cell killing in the tumor spheroids. Spheroids composed of patient-derived PDAC cells (Pa03C or Panc10.05 – labeled red) and CAF cells (labeled green) were treated with APX3330 and SLC-0111 (Figure 5E–F), and the graphical representation i.e. area of red and green fluorescence were evaluated separately as markers for each cell type and is shown in Figure 5C–D. Dramatic enhancement of the APX3330-induced blockade of spheroid growth was observed with the addition of CA9 inhibition. The observed decrease in tumor cell area with APX3330 treatment was significantly different in the presence of SLC-0111, validating the effects seen in hypoxia-exposed 2D cultures. Similar trends were seen when measuring red and green fluorescence intensity (data not shown).
Discussion
Elevated APE1/Ref-1 expression is associated with numerous cancers, including pancreatic, ovarian, gastric, breast, lung, glioblastoma, liver, and colon (18, 19, 46), and analysis of publicly available data from The Cancer Genome Atlas (TCGA, cancergenome.nih.gov) reveals a significant decrease in survival of PDAC patients with elevated APE1/Ref-1 expression (Supplemental Figure 8A, more information in Supplemental Methods) (47, 48). In tumor cells, reduction-oxidation (redox) of thiols of cysteines in various tumor-promoting transcription factors such as STAT3, NFκB, and HIF-1 by APE1/Ref-1 is a crucial step in the activation of these factors. These TFs are all important _targets in cancer therapy and particularly PDAC, but have been shown to be particularly hard to drug (18, 20, 21, 46, 49, 50). While previous studies by us and others (23, 51, 52) have demonstrated the effect of APE1/Ref-1 knockdown on tumor cell growth and survival, additional studies have demonstrated the effectiveness of a small molecule, APX3330, in _targeting and blocking the redox signaling activity of APE1/Ref-1. APX3330 has been shown in multiple in vitro and in vivo models of pancreatic cancer to be effective in reducing tumor volume and metastases as both a single agent and in combination with gemcitabine (20, 21, 53). The mechanism of action has been extensively investigated and the drug has a direct and selective interaction with APE1/Ref-1 as demonstrated by chemical footprinting, mass spectrometry, and other biochemical analyses (33, 44, 54). Importantly, while APX3330 blocks APE1/Ref-1’s redox function, it has no effect on APE1/Ref-1 endonuclease DNA repair activity (45). Although multiple pathways may be modulated, unacceptable toxicity following APE1/Ref-1 inhibition has not been observed in animal or human studies (21, 55). APX3330 is slated for a Phase 1a/1b clinical trial of a dose-escalation study of APX3330 in patients with advanced solid tumors and a dose-expansion cohort of patients with advanced PDAC in mid-2016 in PDAC patients. Therefore, _targeting APE1/Ref-1 with APX3330 has a great deal of potential in cancer therapy.
We previously demonstrated that APE1/Ref-1 contributes to STAT3 activation and the consequent tumor-promoting effects of STAT3 in PDAC cells (20). The cooperative activities of STAT3 and HIF-1 have been demonstrated in a variety of cancers (56, 57); however the finding that APE1/Ref-1 binding to STAT3 is stimulated by exposure to hypoxia in PDAC cells, presented here for the first time, further indicates the importance of both APE1/Ref-1 and STAT3 as potential therapeutic _targets in PDAC. These findings will be further pursued with preclinical STAT3 inhibitors that are being developed for eventual clinical trials (42). Furthermore, our results demonstrating that APX3330 treatment decreases hypoxia-induced HIF-1 transcriptional activity and CA9 mRNA levels (21) is exciting since CA9 inhibitors are either entering or are in clinical trials. This latter finding is of great interest, not only because it is closer to patient applicability, but builds upon our strategy of blocking various signaling pathways at multiple points along pathways influenced by APE1/Ref-1.
CA9 is one of only two tumor-associated carbonic anhydrase isoenzymes known, and it has similarly been established as a potential therapeutic cancer _target (25, 26, 30). CA9 is not detected in most normal tissues, but its expression in renal and other cancers often indicates locally advanced, hypoxic tumors and poor treatment response (58, 59). Variations in CA9 expression by region in tumor samples make whole-tissue analysis difficult (60, 61), but analysis of publicly available data from Oncomine (oncomine.org) using microdissected samples reveals upregulation of CA9 in PDAC tissue samples, as compared to normal pancreas and pancreatic cancer precursor samples (Supplemental Figure 8B–D) (62, 63). Furthermore, the CA9 inhibitor SLC-0111 is in clinical trials to evaluate its safety and efficacy in patients with advanced solid tumors, including pancreatic cancer (clinicaltrials.gov ID: NCT02215850). Therefore, our strategy of inhibiting the HIF-CA9 axis at two points; blocking HIF-1 production of CA9 with APX3330 as well as blocking the activity of any CA9 that is produced using SLC-0111 (Figure 6), is a novel approach to the _targeting of hypoxic PDAC cells. That being said, not only hypoxic PDAC cells will be sensitive to the combination of APX3330 and SLC-0111. APX3330 is _targeting other signaling pathways that are activated in tumor cells that are fully oxygenated, and SLC-0111 can also inhibit CA12, another tumor-associated carbonic anhydrase (31). Our findings establish this strategy resulting in additive cell acidification and inhibition of hypoxic PDAC cell proliferation. This combination approach is similar to the results we previously published using APX3330 and STAT3 inhibition as a dual hit strategy in PDAC cells (20) and APX3330 (HIF-1 inhibition) and Avastin (bevacizumab) for VEGF-signaling inhibition as an anti-angiogenesis combination strategy (45, 64, 65).
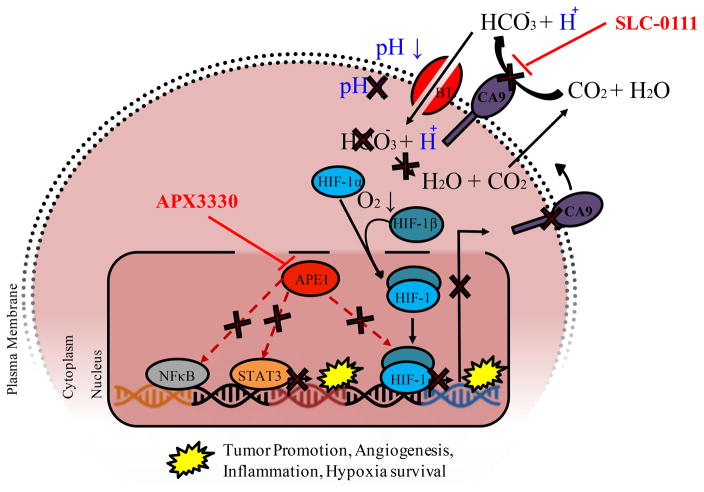
Figure 6. Schematic demonstrating the effects of APE1/Ref-1 - CA9 dual _targeting on cellular acidification and downstream signaling.
Inhibition of APE1/Ref-1 redox signaling with APX3330 results in decreased DNA binding of transcription factors including HIF-1 (as well as STAT3 and NFκB). HIF-1 transactivation is stimulated under low oxygen conditions, resulting in transcription of tumor-promoting factors, including CA9. CA9 catalyzes the conversion of carbon dioxide and water to bicarbonate and hydrogen ions, resulting in stabilization of intracellular pH, which promotes cell survival under hypoxic conditions. Inhibition of CA9 activity with SLC-0111 promotes cell killing via acidification, and this inhibition is enhanced by APX3330 due to a decrease in CA9 expression as well as inhibition of other key signaling pathways via STAT3, NFκB, and AP-1.
As discussed, APX3330 will soon be entering clinical trials for general solid tumors as well as pancreatic cancer. Since SLC-0111 is already in clinical trials with other CA9 inhibitors queued up to enter the clinic, our data demonstrating a novel two pronged approach _targeting highly hypoxic pancreatic cancer could be clinically applicable in the foreseeable future. Additionally, given data demonstrating elevated expression of APE1/Ref-1, STAT3, and CA9 in a variety of other solid tumors (59, 66), these combination approaches should have applicability beyond pancreatic cancer (30, 32, 59).
In conclusion, the data presented here provides continued evidence of the close relationship between APE1/Ref-1, STAT3, and HIF-1 signaling and CA9 production in PDAC as well as the first evidence that the combination of two small molecule inhibitors, each showing minimal toxicity, may be an important next step in the treatment of PDAC, a disease for which effective treatment remains elusive.
Supplementary Material
Acknowledgments
Financial Support: Financial support for this work was provided by the National Cancer Institute CA122298 (M.L. Fishel), CA167291 to M.L. Fishel, M. Korc, M. Ivan and M.R. Kelley, NCI CA075059 to M. Korc, and the National Institutes of Health, R21NS091667 (M.R. Kelley). Additional financial support was provided by Ralph W. and Grace M. Showalter Research Trust Fund (M.L. Fishel), the Earl and Betty Herr Professor in Pediatric Oncology Research, Hyundai Hope on Wheels, Jeff Gordon Children’s Foundation and the Riley Children’s Foundation (M.R. Kelley). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Dr. Bryan Schneider, Department of Hematology/Oncology for use of laboratory equipment and Dr. Malgorzata Kamocka for help with cell imaging. We also thank Christopher Below from Faculty 6 – Natural Sciences at Brandenburg University of Technology for help with figure design.
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- APE1/Ref-1
AP endonuclease-1/redox effector factor 1
- APE1/Ref-1
hypoxia inducible factor 1 alpha
- CA9
carbonic anhydrase IX
- STAT3
signal transducer and activator of transcription 3
- CAFs
cancer-associated fibroblasts
- VHL
von Hippel-Lindau protein
- HRE
hypoxia-response element
- NFκB
nuclear factor kappa-B
- IL-6
interleukin 6
- RPLP0
ribosomal protein large P0
- B2M
beta-2-microglobulin
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- MEF
mouse embryonic fibroblast
- EGFP
enhanced green fluorescent protein RGF, reduced growth factor
- TNFα
tumor necrosis factor alpha
- BER
base escision repair
- TCGA
the cancer genome atlas
- VEGF
vascular endothelial growth factor
Footnotes
Conflict of Interest: Mark R. Kelley has licensed APX3330 though Indiana University Research & Technology Corporation to ApeX Therapeutics.
References
- 1.Sclafani F, Iyer R, Cunningham D, Starling N. Management of metastatic pancreatic cancer: Current treatment options and potential new therapeutic _targets. Critical reviews in oncology/hematology. 2015;95:318–36. doi: 10.1016/j.critrevonc.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 2.McIntyre CA, Winter JM. Diagnostic evaluation and staging of pancreatic ductal adenocarcinoma. Semin Oncol. 2015;42:19–27. doi: 10.1053/j.seminoncol.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Thota R, Pauff JM, Berlin JD. Treatment of metastatic pancreatic adenocarcinoma: a review. Oncology (Williston Park, NY) 2014;28:70–4. [PubMed] [Google Scholar]
- 4.Tamburrino A, Piro G, Carbone C, Tortora G, Melisi D. Mechanisms of resistance to chemotherapeutic and anti-angiogenic drugs as novel _targets for pancreatic cancer therapy. Frontiers in pharmacology. 2013;4:56. doi: 10.3389/fphar.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarroll JA, Naim S, Sharbeen G, Russia N, Lee J, Kavallaris M, et al. Role of pancreatic stellate cells in chemoresistance in pancreatic cancer. Frontiers in physiology. 2014;5:141. doi: 10.3389/fphys.2014.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–76. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo A, Wang LC, Scholler J, Monslow J, Avery D, Newick K, et al. Tumor-Promoting Desmoplasia Is Disrupted by Depleting FAP-Expressing Stromal Cells. Cancer Res. 2015;75:2800–10. doi: 10.1158/0008-5472.CAN-14-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell. 2014;25:735–47. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z, Pothula SP, Wilson JS, Apte MV. Pancreatic cancer and its stroma: a conspiracy theory. World journal of gastroenterology: WJG. 2014;20:11216–29. doi: 10.3748/wjg.v20.i32.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bijlsma MF, van Laarhoven HW. The conflicting roles of tumor stroma in pancreatic cancer and their contribution to the failure of clinical trials: a systematic review and critical appraisal. Cancer metastasis reviews. 2015;34:97–114. doi: 10.1007/s10555-014-9541-1. [DOI] [PubMed] [Google Scholar]
- 11.Arora S, Bhardwaj A, Singh S, Srivastava SK, McClellan S, Nirodi CS, et al. An undesired effect of chemotherapy: gemcitabine promotes pancreatic cancer cell invasiveness through reactive oxygen species-dependent, nuclear factor kappaB- and hypoxia-inducible factor 1alpha-mediated up-regulation of CXCR4. J Biol Chem. 2013;288:21197–207. doi: 10.1074/jbc.M113.484576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blum R, Kloog Y. Metabolism addiction in pancreatic cancer. Cell death & disease. 2014;5:e1065. doi: 10.1038/cddis.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao T, Ren H, Li J, Chen J, Zhang H, Xin W, et al. LASP1 is a HIF1alpha _target gene critical for metastasis of pancreatic cancer. Cancer Res. 2015;75:111–9. doi: 10.1158/0008-5472.CAN-14-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nature cell biology. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 15.Masoud GN, Li W. HIF-1 pathway: role, regulation and intervention for cancer therapy. Acta pharmaceutica Sinica B. 2015;5:378–89. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SY, Yang EG. Recent Advances in Developing Inhibitors for Hypoxia-Inducible Factor Prolyl Hydroxylases and Their Therapeutic Implications. Molecules (Basel, Switzerland) 2015;20:20551–68. doi: 10.3390/molecules201119717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharbeen G, McCarroll J, Goldstein D, Phillips PA. Exploiting Base Excision Repair to Improve Therapeutic Approaches for Pancreatic Cancer. Frontiers in Nutrition. 2015;2:10. doi: 10.3389/fnut.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fishel ML, Kelley MR. The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive _target. Molecular aspects of medicine. 2007;28:375–95. doi: 10.1016/j.mam.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Kelley MR, Logsdon D, Fishel ML. _targeting DNA repair pathways for cancer treatment: what’s new? Future Oncology. 2014;10:1215–37. doi: 10.2217/fon.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cardoso AA, Jiang Y, Luo M, Reed AM, He Y, Kelley MR, et al. APE1/Ref-1 Redox Function Regulates STAT3 Transcriptional Activity and APE1/Ref-1-STAT3 Dual-_targeting Synergize to Effectively Inhibit Pancreatic Cancer Cell Survival. PloS one. 2012;10:e47462. doi: 10.1371/journal.pone.0047462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fishel ML, Jiang Y, Rajeshkumar NV, Scandura G, Sinn AL, He Y, et al. Impact of APE1/Ref-1 Redox Inhibition on Pancreatic Tumor Growth. Molecular Cancer Therapeutics. 2011;10:1698–708. doi: 10.1158/1535-7163.MCT-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fishel ML, Wu X, Devlin CM, Logsdon DP, Jiang Y, Luo M, et al. Apurinic/apyrimidinic endonuclease/redox factor-1 (APE1/Ref-1) redox function negatively regulates NRF2. J Biol Chem. 2015;290:3057–68. doi: 10.1074/jbc.M114.621995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Zhou S, Sandusky GE, Kelley MR, Fishel ML. Reduced expression of DNA repair and redox signaling protein APE1/Ref-1 impairs human pancreatic cancer cell survival, proliferation, and cell cycle progression. Cancer Investigation. 2010;28:885–95. doi: 10.3109/07357907.2010.512816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poletto M, Malfatti MC, Dorjsuren D, Scognamiglio PL, Marasco D, Vascotto C, et al. Inhibitors of the apurinic/apyrimidinic endonuclease 1 (APE1)/nucleophosmin (NPM1) interaction that display anti-tumor properties. Molecular carcinogenesis. 2015;55:688–704. doi: 10.1002/mc.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedlakova O, Svastova E, Takacova M, Kopacek J, Pastorek J, Pastorekova S. Carbonic anhydrase IX, a hypoxia-induced catalytic component of the pH regulating machinery in tumors. Frontiers in physiology. 2014;4:400. doi: 10.3389/fphys.2013.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McIntyre A, Patiar S, Wigfield S, Li JL, Ledaki I, Turley H, et al. Carbonic anhydrase IX promotes tumor growth and necrosis in vivo and inhibition enhances anti-VEGF therapy. Clin Cancer Res. 2012;18:3100–11. doi: 10.1158/1078-0432.CCR-11-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaluz S, Kaluzova M, Liao SY, Lerman M, Stanbridge EJ. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: A one transcription factor (HIF-1) show? Biochimica et biophysica acta. 2009;1795:162–72. doi: 10.1016/j.bbcan.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swietach P, Wigfield S, Cobden P, Supuran CT, Harris AL, Vaughan-Jones RD. Tumor-associated carbonic anhydrase 9 spatially coordinates intracellular pH in three-dimensional multicellular growths. J Biol Chem. 2008;283:20473–83. doi: 10.1074/jbc.M801330200. [DOI] [PubMed] [Google Scholar]
- 29.Schoppmann SF, Jesch B, Friedrich J, Jomrich G, Maroske F, Birner P. Phosphorylation of signal transducer and activator of transcription 3 (STAT3) correlates with Her-2 status, carbonic anhydrase 9 expression and prognosis in esophageal cancer. Clin Exp Metastasis. 2012;29:615–24. doi: 10.1007/s10585-012-9475-3. [DOI] [PubMed] [Google Scholar]
- 30.Supuran CT, Winum JY. Designing carbonic anhydrase inhibitors for the treatment of breast cancer. Expert opinion on drug discovery. 2015;10:591–7. doi: 10.1517/17460441.2015.1038235. [DOI] [PubMed] [Google Scholar]
- 31.Supuran CT. How many carbonic anhydrase inhibition mechanisms exist? Journal of enzyme inhibition and medicinal chemistry. 2016;31:345–60. doi: 10.3109/14756366.2015.1122001. [DOI] [PubMed] [Google Scholar]
- 32.Fishel ML, Colvin ES, Luo M, Kelley MR, Robertson KA. Inhibition of the redox function of APE1/Ref-1 in myeloid leukemia cell lines results in a hypersensitive response to retinoic acid-induced differentiation and apoptosis. Experimental hematology. 2010;38:1178–88. doi: 10.1016/j.exphem.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su DG, Delaplane S, Luo M, Rempel DL, Vu B, Kelley MR, et al. Interactions of APE1 with a redox inhibitor: Evidence for an alternate conformation of the enzyme. Biochemistry. 2011;50:82–92. doi: 10.1021/bi101248s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, NY) 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter K, Omura N, Hong SM, Griffith M, Goggins M. Pancreatic cancer associated fibroblasts display normal allelotypes. Cancer Biol Ther. 2008;7:882–8. doi: 10.4161/cbt.7.6.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HS, Guo C, Thompson EL, Jiang Y, Kelley MR, Vasko MR, et al. APE1, the DNA base excision repair protein, regulates the removal of platinum adducts in sensory neuronal cultures by NER. Mutat Res. 2015;779:96–104. doi: 10.1016/j.mrfmmm.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang Y, Guo C, Fishel ML, Wang ZY, Vasko MR, Kelley MR. Role of APE1 in differentiated neuroblastoma SH-SY5Y cells in response to oxidative stress: use of APE1 small molecule inhibitors to delineate APE1 functions. DNA Repair (Amst) 2009;8:1273–82. doi: 10.1016/j.dnarep.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rankin EB, Wu C, Khatri R, Wilson TL, Andersen R, Araldi E, et al. The HIF signaling pathway in osteoblasts directly modulates erythropoiesis through the production of EPO. Cell. 2012;149:63–74. doi: 10.1016/j.cell.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sempere LF, Gunn JR, Korc M. A novel 3-dimensional culture system uncovers growth stimulatory actions by TGFbeta in pancreatic cancer cells. Cancer biology & therapy. 2011;12:198–207. doi: 10.4161/cbt.12.3.15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arpin CC, Mac S, Jiang Y, Cheng H, Grimard M, Page BD, et al. Applying Small Molecule Signal Transducer and Activator of Trancription-3 (STAT3) Protein Inhibitors as Pancreatic Cancer Therapeutics. Mol Cancer Ther. 2016;15:794–805. doi: 10.1158/1535-7163.MCT-15-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindblom P, Berg AL, Zhang H, Westerberg R, Tugwood J, Lundgren H, et al. Tesaglitazar, a dual PPAR-alpha/gamma agonist, hamster carcinogenicity, investigative animal and clinical studies. Toxicologic pathology. 2012;40:18–32. doi: 10.1177/0192623311429972. [DOI] [PubMed] [Google Scholar]
- 44.Zhang JLM, Marasco D, Logsdon D, LaFavers KA, Chen Q, Reed A, Kelley MR, Gross ML, Georgiadis MM. Inhibition of apurinic/apyrimidinic endonuclease I’s redox activity revisited. Biochemistry. 2013;52:2955–66. doi: 10.1021/bi400179m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo M, Delaplane S, Jiang A, Reed A, He Y, Fishel M, et al. Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule inhibition of the redox function of Ape1. Antioxid Redox Signal. 2008;10:1853–67. doi: 10.1089/ars.2008.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of _targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Current molecular pharmacology. 2012;5:36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dibra D, Mishra L, Li S. Molecular mechanisms of oncogene-induced inflammation and inflammation-sustained oncogene activation in gastrointestinal tumors: An underappreciated symbiotic relationship. Biochimica et biophysica acta. 2014;1846:152–60. doi: 10.1016/j.bbcan.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prabhu L, Mundade R, Korc M, Loehrer PJ, Lu T. Critical role of NF-kappaB in pancreatic cancer. Onco_target. 2014;5:10969–75. doi: 10.18632/onco_target.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3:679–86. [PubMed] [Google Scholar]
- 52.Cun Y, Dai N, Xiong C, Li M, Sui J, Qian C, et al. Silencing of APE1 enhances sensitivity of human hepatocellular carcinoma cells to radiotherapy in vitro and in a xenograft model. PloS one. 2013;8:e55313. doi: 10.1371/journal.pone.0055313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Zou GM, Maitra A. Small-molecule inhibitor of the AP endonuclease 1/REF-1 E3330 inhibits pancreatic cancer cell growth and migration. Mol Cancer Ther. 2008;7:2012–21. doi: 10.1158/1535-7163.MCT-08-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo M, Zhang J, He H, Su D, Chen Q, Gross ML, et al. Characterization of the redox activity and disulfide bond formation in apurinic/apyrimidinic endonuclease. Biochemistry. 2012;51:695–705. doi: 10.1021/bi201034z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vasko MR, Guo C, Thompson EL, Kelley MR. The repair function of the multifunctional DNA repair/redox protein APE1 is neuroprotective after ionizing radiation. DNA Repair (Amst) 2011;10:942–52. doi: 10.1016/j.dnarep.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1alpha cooperatively activate HIF1 _target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33:1670–9. doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gariboldi MB, Ravizza R, Monti E. The IGFR1 inhibitor NVP-AEW541 disrupts a pro-survival and pro-angiogenic IGF-STAT3-HIF1 pathway in human glioblastoma cells. Biochemical pharmacology. 2010;80:455–62. doi: 10.1016/j.bcp.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 58.McDonald PC, Winum JY, Supuran CT, Dedhar S. Recent developments in _targeting carbonic anhydrase IX for cancer therapeutics. Onco_target. 2012;3:84–97. doi: 10.18632/onco_target.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oosterwijk E. Carbonic anhydrase expression in kidney and renal cancer: implications for diagnosis and treatment. Sub-cellular biochemistry. 2014;75:181–98. doi: 10.1007/978-94-007-7359-2_10. [DOI] [PubMed] [Google Scholar]
- 60.Juhasz M, Chen J, Lendeckel U, Kellner U, Kasper HU, Tulassay Z, et al. Expression of carbonic anhydrase IX in human pancreatic cancer. Alimentary pharmacology & therapeutics. 2003;18:837–46. doi: 10.1046/j.1365-2036.2003.01738.x. [DOI] [PubMed] [Google Scholar]
- 61.Kivela AJ, Parkkila S, Saarnio J, Karttunen TJ, Kivela J, Parkkila AK, et al. Expression of transmembrane carbonic anhydrase isoenzymes IX and XII in normal human pancreas and pancreatic tumours. Histochemistry and cell biology. 2000;114:197–204. doi: 10.1007/s004180000181. [DOI] [PubMed] [Google Scholar]
- 62.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–57. [PubMed] [Google Scholar]
- 63.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer cell. 2009;16:259–66. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Liu X, Zhou T, Kelley MR, Edwards P, Gao H, et al. Inhibition of APE1/Ref-1 redox activity rescues human retinal pigment epithelial cells from oxidative stress and reduces choroidal neovascularization. Redox biology. 2014;2:485–94. doi: 10.1016/j.redox.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang A, Gao H, Kelley MR, Qiao X. Inhibition of APE1/Ref-1 Redox Activity with APX3330 Blocks Retinal Angiogenesis in vitro and in vivo. Vision Res. 2011;51:93–100. doi: 10.1016/j.visres.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furtek SL, Backos DS, Matheson CJ, Reigan P. Strategies and approaches of _targeting STAT3 for cancer treatment. ACS chemical biology. 2016;11:308–18. doi: 10.1021/acschembio.5b00945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.