ABSTRACT
Rabbit monoclonal antibodies (RabMAbs) can recognize diverse epitopes, including those poorly immunogenic in mice and humans. However, there have been only a few reports on RabMAb humanization, an important antibody engineering step usually done before clinical applications are investigated. To pursue a general method for humanization of RabMAbs, we analyzed the complex structures of 5 RabMAbs with their antigens currently available in the Protein Data Bank, and identified antigen-contacting residues on the rabbit Fv within the 6 Angstrom distance to its antigen. We also analyzed the supporting residues for antigen-contacting residues on the same heavy or light chain. We identified “HV4” and “LV4” in rabbit Fvs, non-complementarity-determining region (CDR) loops that are structurally close to the antigen and located in framework 3 of the heavy chain and light chain, respectively. Based on our structural and sequence analysis, we designed a humanization strategy by grafting the combined Kabat/IMGT/Paratome CDRs, which cover most antigen-contacting residues, into a human germline framework sequence. Using this strategy, we humanized 4 RabMAbs that recognize poorly immunogenic epitopes in the cancer _target mesothelin. Three of the 4 humanized rabbit Fvs have similar or improved functional binding affinity for mesothelin-expressing cells. Interestingly, 4 immunotoxins composed of the humanized scFvs fused to a clinically used fragment of Pseudomonas exotoxin (PE38) showed stronger cytotoxicity against tumor cells than the immunotoxins derived from their original rabbit scFvs. Our data suggest that grafting the combined Kabat/IMGT/Paratome CDRs to a stable human germline framework can be a general approach to humanize RabMAbs.
KEYWORDS: Humanization, mesothelin, paratope, rabbit monoclonal antibody, recombinant immunotoxin, structure alignment
Abbreviations
- Ag
antigen
- CDR
complementary-determining region
- FR
framework region
- RabMAb
rabbit monoclonal antibody
- scFv
single-chain variable fragments
Introduction
Rabbits have long been used as a source of antibodies used as research tools. They can potentially generate antibodies _targeting uncommon epitopes, including interesting epitopes that are less immunogenic in mice and humans.1 Knight's group described the rabbit hybridoma technology by generating a fusion partner in 1995.2 In recent years, several commercial services have been available for producing rabbit monoclonal antibodies (RabMAb), including rabbit hybridoma technology (Epitomics or Abcam),2 the serum immunoproteomics-based NG-XMT™ technology (Cell Signaling),3 and phage display-based rabbit variable fragment (Fv) library.1,4 Like the mouse counterparts, the RabMAbs generated from hybridoma have high specificity and affinity.5 It has been reported that rabbit hybridoma technology may have a higher success rate, particularly for those difficult antigens or epitopes that are poorly immunogenic in mice.6 While RabMAbs are routinely used as research or diagnostic tools, none are approved as antibody therapeutics for cancer and other diseases. APX005M, a humanized RabMAb _targeting CD40, is currently being evaluated in a Phase 1 clinical study for the treatment of patients with advanced solid tumors (NCT02482168).7
To enable the successful development of RabMAbs as therapeutics, the antibodies should be humanized, which can reduce the chance that patients will produce neutralizing antibodies to the non-human molecules.8 The current methods for mouse antibody humanization include complementary-determining region (CDR) grafting,9-12 specificity-determining residues (SDR) grafting,13 phage display,14 and de-immunization.8,15,16 The CDR grafting is the first-used and clinically validated humanization technique.9,12,17 This method was successfully used to humanize murine antibody 4D5 (anti-human epidermal growth factor receptor 2), which was then developed as trastuzumab (Herceptin®, Genentech, Inc., South San Francisco, California).17 Trastuzumab has tolerable immunogenicity that does not affect its serum concentration and the pathologic complete response of patients.18,19 It would be useful to explore whether RabMAbs can be directly humanized by comparison with human sequences, in a similar way to humanization of mouse antibodies. In one study, humanized anti-VEGF RabMAbs have been made by substituting non-critical residues with human residues within both the frameworks and CDR regions.20 Our laboratory has humanized a RabMAb by grafting combined Kabat/IMGT CDRs.6 Although RabMAbs appear more similar to human than mouse in terms of heavy chain CDR3 lengths,21 there are only a few reports on RabMAb humanization. The sequences of RabMAbs have important features that are different from human and mouse antibodies. Rabbits preferentially use a single family (VH1) in heavy chain VDJ rearrangement,22 while mice and humans use multiple VH gene families. The rabbit Vκ (K1 isotype) often contain an additional cysteine in position 80 (Kabat numbering) to form a disulfide bond with the constant region in the kappa light chain.23 Therefore, a robust and general approach for RabMAb humanization is required in the field to develop them as a new class of therapeutics for the treatment of cancer and other diseases.
In both CDR and specificity-determining residues (SDR) grafting, a human antibody framework replaces large fragments of non-essential non-human framework, restricting the non-human sequence to small and isolated fragments that are important for antigen-binding, therefore reducing immunogenicity. Accurate differentiation of the antigen-binding site (the paratope) and the non-essential framework is critical to success. By definition, the CDR contains the paratope in most antibodies. Several different methods have been used to identify CDRs. In the 1970s, Kabat, et al. identified CDR based on the presumed criteria that statistically they have the most variable sequences in immunoglobulins.24,25 In the 1980s, Chothia et al. re-defined the CDR based on the loop position on antibody structures, under the assumption that the structural “loop” contains the antigen-binding site.26 The international ImMunoGeneTics database (IMGT) defined IMGT CDR, taking into account the definition of the Kabat CDR, structural data by Satow et al. and the characterization of the hypervariable loops by Chothia et al.27 In the 1990s, Padlan, et al. defined SDR as the residues that directly bind the antigen, and they defined 2 residues to bind each other as the distance between their closest atoms ≤ the sum of their van der Waals's radii +0.5 Å.28 In 2012, Ofran and colleagues defined an antigen-binding region (ABR; we will refer to this as “Paratome CDRs” to be consistent with other methods) as a “structural binding consensus,” defining bind (to the antigen) as closest atoms ≤ 6 Å apart, and consensus as the same position in at least 10% of the analyzed antibodies that binds the antigen.29 They confirmed the previous presumptions that virtually all antigen binding residues lie in regions of structural consensus across antibodies and this consensus is identifiable from the sequence of the antibody. They also showed that 15–21% of antigen binding residues are located outside Kabat, IMGT, and Chothia CDRs.29,30 In contrast, only 6% of antigen binding residues are located outside Paratome CDRs. Although Paratome CDRs more accurately define the paratope in mouse and human antibodies compared with other CDRs, Paratome CDRs have not been widely used. In particular, Ofran's structure database (Paratome) does not contain rabbit antibody structures.
Here, we analyzed the complex crystal structures of 5 RabMAbs with their antigens available in the Protein Data Bank (PDB) and identified antigen-contacting residues along with their supporting residues. We then compared our findings with Kabat, IMGT, and Paratome CDRs. Based on our analysis, we decided to humanize RabMAb via grafting combined Kabat/IMGT/Paratome CDRs into human germline framework sequences and successfully humanized 4 RabMAbs specific for mesothelin without back mutations.
Results
RabMAb structure and sequence analysis
To humanize RabMAbs, we need to analyze antibody structures and sequences to identify antigen binding structural consensus. To this end, we analyzed and aligned a set of crystal structures containing 5 available RabMAbs with their antigens (not shown) from the PDB database (Figs. 1, 2 and 3; Table 1). All the VH sequences match the most similar rabbit VH1 germline (IGHV1) in IMGT/DomainGapAlign analysis (Table 1). The haplotypes usually contain rabbit strain information, and they were also obtained from IMGT/DomainGapAlign analysis. All of the Vκ sequences match germline sequences that were isolated from haplotypes “K1 b4,” suggesting that they were from allotype b4 rabbits, but the 2×7L light chain matched to J gene IGKJ1–2*04, which is seen in haplotype K1 b9. The rabbit strain information from the literature is listed together with the reference in Table 1.
Figure 1.
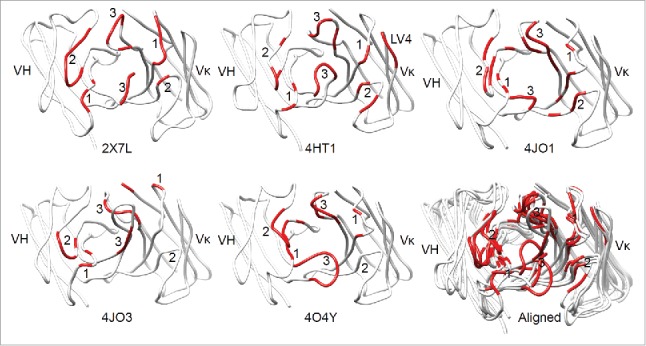
The antigen-contacting residues on the structures of 5 RabMAbs in the PDB. An antigen-contacting residue (highlighted in red) contains at least one atom that is ≤ 6 Å away from an atom on the antigen (not shown). The approximate locations of CDRs are labeled. Aligned: RabMAbs are structurally aligned with 2×7L by Dali pairwise comparison and visualized by Chimera.
Figure 2.
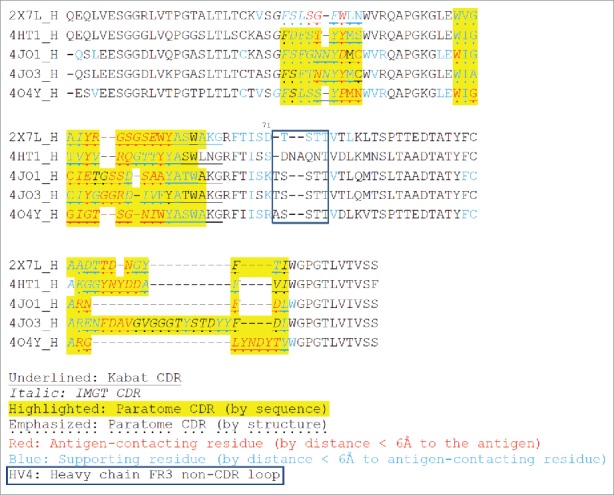
Multiple structure alignment of RabMAb VHs by Strap. Antigen-contacting residues, their supporting residues, and Kabat/IMGT/Paratome CDRs are indicated in protein sequences. HV4, a non-CDR loop in heavy chain FR3.
Figure 3.
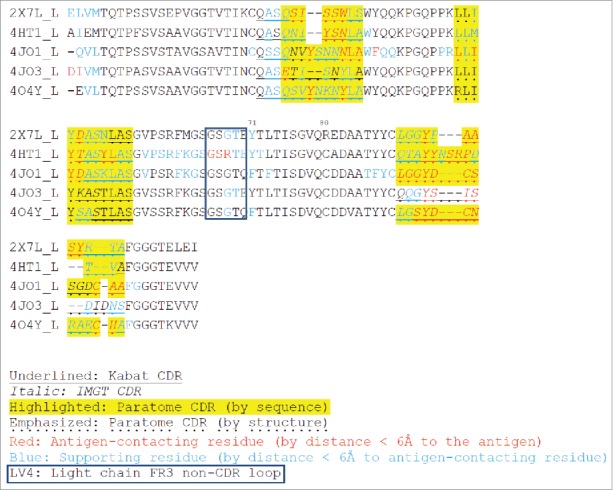
Multiple structure alignment of RabMAb Vκs by Strap. Antigen-contacting residues, their supporting residues, and Kabat/IMGT/Paratome CDRs are indicated in protein sequences. LV4: a non-CDR loop in light chain FR3.
Table 1.
Identification of the most similar rabbit germline sequences for the 5 rabbit Fvs in the PDB database by IMGT/DomainGapAlign.
| PDB ID | Antibody | Antigen | VH | J | VL | J | References |
|---|---|---|---|---|---|---|---|
| 2×7L | NA | HIV-1 Rev | IGHV1S69*01 | IGHJ6*02 | IGKV1S3*02 | IGKJ1–2*04 | 48,49 |
| 4HT1 | TW305chi | TWEAK | IGHV1S7*01 | IGHJ6*02 | IGKV1S3*01 | IGKJ1–2*01 | 50 New Zealand White rabbit |
| 4JO1 | R56 | HIV-1 gp120 V3 | IGHV1S40*01 | IGHJ6*01 | IGKV1S5*01 | IGKJ1–2*01 | 51 New Zealand White rabbit |
| 4JO3 | R20 | HIV-1 gp120 V3 | IGHV1S40*01 | IGHJ4*01 | IGKV1S3*01 | IGKJ1–2*01 | 51 New Zealand White rabbit |
| 4O4Y | 2095–2 | IdeS-cleaved human IgG1 hinge | IGHV1S69*01 | IGHJ2*01 | IGKV1S2*02 | IGKJ1–2*01 | 52 |
TWEAK, tumor necrosis factor-like weak inducer of apoptosis; NA, not available.
We defined the antigen-contacting residues to contain at least one atom that is ≤ 6 Å away from an atom on the antigen, and identified them via contact map analysis using CCP4 with the CONTACT/ACT program. We used multiple structure alignment (Figs. 2 and 3) and calculated the numbers of antigen-contacting residues in CDRs. Our calculation shows that antigen-contacting residues are found in all CDRs except 2/5 light chain CDR2, and that heavy chain CDR2/CDR3 and light chain CDR3 contains the most antigen-contacting residues (Table 2).
Table 2.
The number of antigen-contacting residues in CDRs.
| Number of antigen-contacting residues |
|||||||
|---|---|---|---|---|---|---|---|
| H-CDR1 | H-CDR2 | H-CDR3 | L-CDR1 | L-CDR2 | L-CDR3 | Total | |
| 2×7L | 3 | 9 | 3 | 5 | 1 | 5 | 26 |
| 4HT1 | 2 | 4 | 5 | 3 | 4 | 5 | 23 |
| 4JO1 | 2 | 9 | 4 | 5 | 4 | 10 | 34 |
| 4JO3 | 2 | 4 | 4 | 1 | 0 | 4 | 15 |
| 4O4Y | 3 | 11 | 8 | 2 | 0 | 7 | 31 |
| Utilization rate | 100% | 100% | 100% | 100% | 60% | 100% | |
| Average | 2.4 | 7.4 | 4.8 | 3.2 | 1.8 | 6.2 | 25.8 |
We compared the CDR sequences using 3 online tools (Kabat, IMGT and Paratome) and found that most of the antigen-contacting residues we identified were located in the combined Kabat, IMGT and Paratome CDRs, but often fell out of Kabat CDR or IMGT CDR alone (Figs. 2 and 3). The Paratome CDR can be identified by either structure or sequence alignment using online tools. Although the Paratome tools, especially structure alignment, can be more accurate to identify RabMAb CDRs, RabMAb structures are generally not available. Moreover, the Paratome sequence alignment sometimes are unable to identify certain RabMAb CDRs (e.g., CDR-1 in the heavy chain and CDR3 in light chain); in this case, we found that Kabat/IMGT CDRs can be useful to define those CDRs and that combined Kabat/IMGT heavy chain CDR-1 and light chain CDR3 are almost the same as Paratome CDRs (Fig. 2 and Fig. 3).
Beyond CDRs, we found that the first 2 residues in 4JO3 light chain and a loop between CDR2 and CDR3 (Kabat numbering 66–70) in 4HT1 light chain also contain antigen-contacting residues. We named the loop between CDR2 and CDR3 in the heavy chain “HV4” (Kabat numbering 72–75 or 76), and the corresponding region in the light chain “LV4.”
We hypothesized that the non-antigen-contacting residues may support antigen-contacting residues (paratope). Thus, we identified such paratope-supporting residues (Figs. 2 and 3) based on 2 criteria: 1) they are within ≤ 6 Å from the antigen-contacting residues, and 2) they reside on the same chain as the antigen-binding residues. Most of such paratope-supporting residues are located in the CDRs and their flanking sequences, N-terminus of VH/VL, and around HV4/LV4. Kabat position 71 was suggested to be important in previous studies because it affected the orientation of CDR-H2 and CDR-H1.31,32 In our analysis, they are supporting residues in all the heavy chain and 4/5 of the light chain of the 5 RabMAbs.
Humanization of 4 RabMAbs by grafting combined Kabat/IMGT/Paratome CDRs
Since the combined KABAT/IMGT/Paratome CDRs cover most antigen-contacting residues, we hypothesized that grafting the combined CDRs to a stable human Ig germline framework will likely preserve the antigen-binding affinity and provide a robust humanization strategy for RabMAbs. To test this hypothesis, we grafted the combined Kabat/IMGT/Paratome CDRs of 4 anti-human mesothelin RabMAb to the framework of human germline IGHV3–66*01, IGHJ4*01, IGKV1–27*01 and IGKJ4*01 (Fig. 4, 5). The frameworks used in this study were proven to humanize YP218 (hYP218 first version) in our previous study with no affinity loss.6 We decided to use the same framework to observe the robustness of this grafting method, noting that they are not the most similar human germline frameworks for YP3, YP158 and YP223. The YP218 were humanized again to test the current grafting strategy, in which we grafted the Paratome CDR in addition to the previously grafted Kabat/IMGT CDRs. 6 To compare current human germline templates with a humanization template previously reported by ESBATech,33 we also included the FW1.4 and FW1.4 gen sequences in the alignment (Fig. 5).
Figure 4.
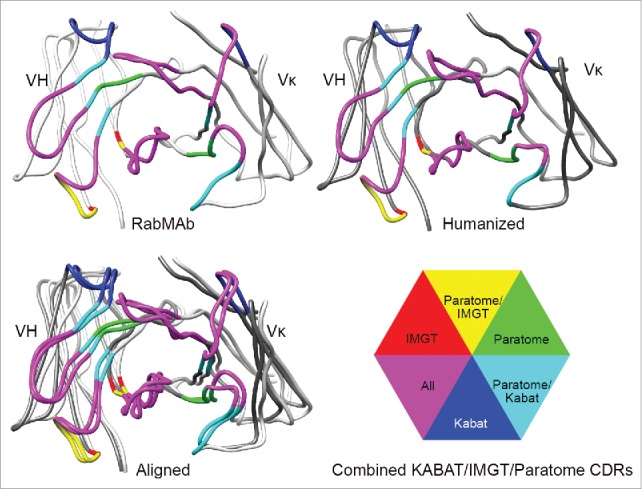
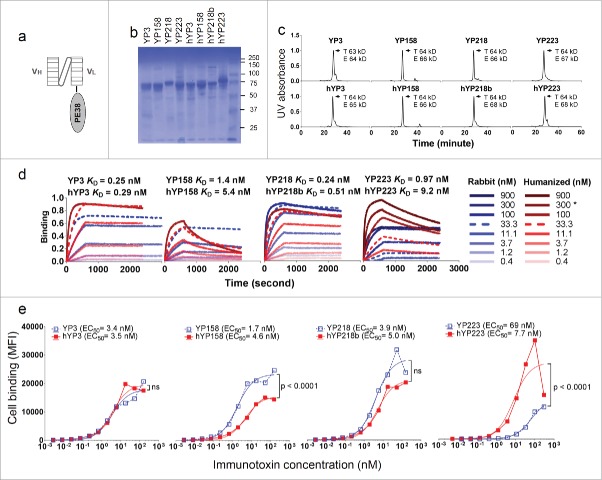
Humanization of RabMAbs via grafting combined Kabat/IMGT/Paratome CDRs. The YP218 RabMAb and its humanized antibody (hYP218) structures were modeled by I-TASSER, aligned by Dali pairwise comparison, and visualized by Chimera.
Figure 5.

The sequence alignment of rabbit Fv and human germline sequences used as templates. Human germline IGHV3–66*01, IGHJ4*01, IGKV1–27*01 and IGKJ4*01 were used as the templates for humanization. FW1.4 and FW1.4 gen were used as the templates in a previous RabMAb humanization report.33. The additional cysteine in position 80 (Kabat numbering) on the rabbit Vκ is replaced with proline in human germline template (IGKV1–27*01).
We made the humanized Fv in the format of scFv-Pseudomonas exotoxin (PE38) fusion protein as described before (Fig. 6a, b). 6,34 The purified proteins were analyzed by an analytical size-exclusion chromatogram (SEC) coupled with a UV detector and multi-angle light scattering (MALS) detector (Fig. 6c). We found that the experimental molecular weights of the dominant protein peaks were consistent with the theoretical molecular weights of the immunotoxins (Fig. 6c), indicating that these immunotoxin molecules are monovalent.
Figure 6.
The characterization of immunotoxins with humanized scFvs and original RabMAb scFvs. (a) Schematic cartoon of a monovalent scFv-based immunotoxin. (b) SDS-PAGE of purified immunotoxins. (c) Size-exclusion chromatogram of purified proteins. T: theoretical molecular weight calculated from the protein sequence. E: experimental molecular weight calculated by the MALS. (d) Association and dissociation curve on purified proteins, measured on the Octet platform. *: for hYP223IT, 282.6 nM instead of 300 nM immunotoxin protein was used. (e) Immunotoxin binding curve on H9 cells by flow cytometry. BL22 is an irrelevant immunotoxin (anti-CD22) used as a negative control.
We measured the binding kinetics of monovalent scFv-based immunotoxins on the Octet platform (Fig. 6d). With the exception of hYP223, the overall changes in kon, koff and KD after humanization are moderate (Table 4). We also compared the cell surface binding (Fig. 6e) and cytotoxicity (Fig. 7) with a mesothelin-expressing cell line (H9). As shown in Fig. 6 and Table 4, the EC50 measured on cell-surface mesothelin and the KD measured with Octet using mesothelin protein are comparable, although again with the exception hYP223. Compared to their original RabMAb scFv, hYP3 preserved the original binding on cells, hYP218b showed an insignificant decrease in the binding on cells, and hYP158 showed a modest decrease (3-fold) in the binding on cells. Interestingly, although hYP223 showed a decrease in the binding on mesothelin protein, the humanized Fv showed significantly improved binding for cell-surface mesothelin (9-fold),
Table 4.
The binding kinetics of purified immunotoxins containing rabbit or humanized scFv. The binding kinetics were measured by Octet and calculated by Graphpad Prism. The cell surface binding EC50 on H9 cells are listed for comparison.
| Binding kinetics |
Flow cytometry on H9 cells | ||||||
|---|---|---|---|---|---|---|---|
| Immunotoxin | koff | koff error | kon | kon error | KD (M) | Bmax | EC50 (M) |
| YP3 | 5.38E-05 | 3.58E-08 | 2.12E+05 | 2.97E+02 | 2.54E-10 | 0.72 | 3.44E-09 |
| hYP3 | 4.38E-05 | 4.80E-08 | 1.51E+05 | 1.85E+02 | 2.90E-10 | 0.9111 | 3.45E-09 |
| YP158 | 1.61E-04 | 1.25E-07 | 1.17E+05 | 1.53E+02 | 1.37E-09 | 0.59 | 1.7E-09 |
| hYP158 | 8.35E-04 | 7.51E-07 | 1.55E+05 | 3.06E+02 | 5.38E-09 | 0.6207 | 4.56E-09 |
| YP218 | 7.85E-05 | 6.01E-08 | 3.32E+05 | 5.18E+02 | 2.36E-10 | 0.8536 | 3.88E-09 |
| hYP218b | 9.52E-05 | 4.35E-08 | 1.88E+05 | 2.39E+02 | 5.07E-10 | 0.7814 | 4.95E-09 |
| YP223 | 1.26E-05 | 5.70E-08 | 1.30E+04 | 4.11E+01 | 9.67E-10 | 0.5179 | 6.85E-08 |
| hYP223 | 2.63E-04 | 1.11E-07 | 2.86E+04 | 5.96E+01 | 9.18E-09 | 0.8815 | 7.73E-09 |
Figure 7.
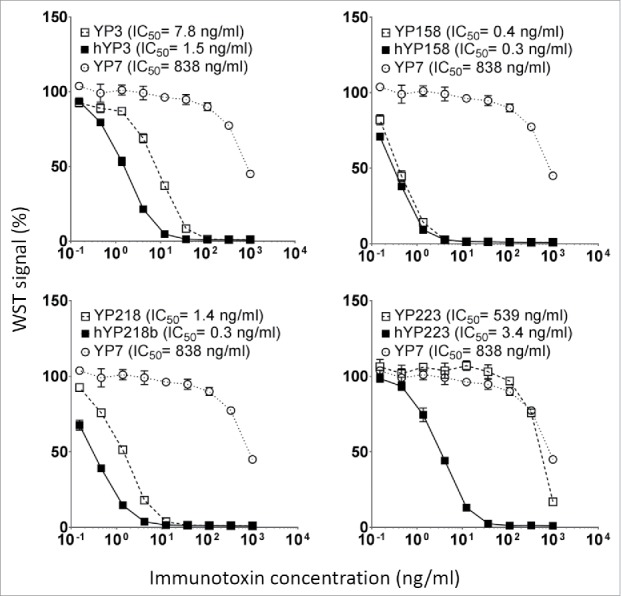
The cytotoxicity induced by immunotoxins with humanized scFvs and original RabMAb scFvs. They were tested on H9 cells in a WST assay. Error bars indicate standard errors. YP7, an irrelevant immunotoxin to GPC3.
All the immunotoxins with humanized scFvs showed stronger or similar cytotoxicity to the H9 cell than the ones with original RabMAb scFvs (Fig. 7). The IC50 value of YP3 immunotoxin decreased from the original 7.8 ng/ml to 1.5 ng/ml, YP158 immunotoxin from 0.4 ng/ml to 0.3 ng/ml, YP218 immunotoxin from 1.4 ng/ml to 0.3 ng/ml, and YP223 immunotoxin from 539 ng/ml to 3.4 ng/ml. After humanization, the percentage of human residues in VH and Vκ was determined by IMGT/DomainGapAlign: the percentage in Vκ is 90.5∼92.3%, and the percentage in VH is 77.8∼83.8%. Taken together, our results indicate that we successfully humanized all the anti- mesothelin RabMAbs without back mutations, and validated their comparable binding affinity and improved anti-tumor activity in the format of monovalent immunotoxins.
Discussion
We designed and tested a robust humanization method for RabMAbs by grafting the combined Kabat/IMGT/Paratome CDRs. The design was based on our findings using the RabMAbs from the PDB crystal structure database in which the combined CDRs cover most antigen-contacting residues. Human and mouse antibody CDRs (Paratome CDRs) were recently defined by Ofran et al.29 Our findings are consistent to Ofran's observations that antigen-contacting residues are located in regions of structural consensus across antibodies, and that structural consensus are identifiable from the sequence of the antibody. The Paratome CDRs identified by sequence alignment more accurately cover antigen-contacting residues than Kabat/IMGT. However, certain rabbit CDRs (VH-CDR1 and VL-CDR3) cannot be identified by Paratome sequence analysis. In this case, if RabMAb structures or structural models are available, the CDRs can be identified by Paratome structure analysis; if RabMAb structures are not available, our data indicates that combined Kabat and IMGT CDRs are as accurate as combined Kabat, IMGT, and Paratome CDRs for rabbit VH-CDR1 and VL-CDR3. Based on our analysis, we decided to assess the humanization strategy of grafting combined Kabat/IMGT/Paratome CDRs. CDRs from different RabMAbs were grafted to the same human germline framework, and the resulting humanized scFvs had similar affinities compared with the original rabbit scFv, suggesting that the method may be applicable generally to other RabMAbs that do not yet have structural information present.
We also identified potential back-mutation sites in cases when the first humanization is unsatisfactory. Beyond the 6 major CDRs, the first 2 residues in the light chain also contact antigens in one of the 5 rabbit antibodies (4JO3), as they do in some human and mouse antibodies,30 which may explain why they have a high back-mutation rate in the mouse antibody humanization literature.35 In addition to this site, the loop LV4 (Kabat position 66–70) in 1/5 RabMAbs (structure id: 4HT1) may interact with the antigen. The counterpart in the heavy chain is HV4 (Kabat numbering 72–75 or 76). Similarly, LV4 and HV4 in human antibodies occasionally interact with the protein antigen (not small epitopes such as peptides or haptens),36 and have a high back-mutation rate in humanization of mouse antibodies (H75, L69–70).35 Therefore, the first few residues at the N terminus or HV4/LV4 sequences could be the sites for back mutation if humanized RabMAbs need to be further optimized.
Besides grafting antigen-binding residues, successful humanization also relies on a suitable human framework. The human germline IGHV3–66*01, IGHJ4*01, IGKV1–27*01 and IGKJ4*01 provided a successful framework to humanize 4 RabMAbs in this study.
To understand more about the role of framework residues, we identified the RabMAb residues that directly contact the paratope within the same chain as their “supporting residues.” The murine CDR-contacting residues (closest atoms ≤ 6 Å apart) have higher back-mutation rates as analyzed by Haidar et al.35,37 Our method is different from Haidar's method35 because we identified supporting residues for antigen-contacting residues, whereas Haidar identified the supporting residues for MacCallum's CDRs. The first and end residues in MacCallum's CDR satisfy the following criteria: 1) it is buried > 1.0 A˚2 from solvent by antigen binding in at least one member of their structure database; and 2) the average buried surface by antigen exceed 1% surface area by antigen binding.37 Some residues within MacCallum's CDR may detach from antigen. In RabMAbs, we found that these “supporting” residues cluster together, mostly around the antigen-contacting residues. Some parts of the supporting regions are within the combined Kabat/IMGT/Paratome CDRs and may contribute to the conformation reservation after CDR grafting. Additionally, the N terminus of the light chains and the regions around position 71 (Kabat numbering) in both chains of RabMAb are also CDR-supporting regions. This is similar to Haidar's findings in murine antibodies,35 suggesting a similarity between RabMAb and murine antibodies.
The importance of keeping position 71 in murine antibody humanization was summarized by Haidar et al., who showed that position 71 were back mutated in 33/89 VH and 17/89 VL of humanized murine antibodies, both being the most frequently back-mutated sites in their respective chains.35 Murine VH position 71 (Kabat numbering) has been shown to affect the relative positions and orientations of CDR-H2 and H1 in a mouse Fv (PDB id: 1BBJ),32 and was suggested to be a major determinant of the position and conformation of the CDR-H2.31 Interestingly, given the choice between human and rabbit sequences at position 71 in phage panning, rabbit sequence was favored at H71, but not L71 (Table 3).4 RabMAb humanization literature and our RabMAb humanization studies do not clearly support an essential role of position 71 in antigen binding affinity (Table 3). Overall, we have not seen obvious correlation between the mutation in position 71 and the affinity so far.
Table 3.
The effect of position 71 on RabMAb humanizations.
| H71 |
L71 |
|||||
|---|---|---|---|---|---|---|
| RabMAb | rabbit | humanized | rabbit | humanized | Affinity after humanization | Reference |
| YP3 | K | R | F | F | unchanged in protein and cell binding | |
| YP158 | K | R | Y | F | similar in protein and cell binding | |
| YP218 | K | R | Y | F | similar in protein and cell binding | 6 and the present study |
| YP223 | R | R | F | F | increased in cell binding and decreased in protein binding | |
| EBV321 | K | R | F | F | increased | 20 |
| anti-A33 | L | given the choice between L and R, resulting sequences have L/F = 4/2 | Y | given the choice between Y and F, resulting sequence have Y/F = 3/3 | clones selected based on high affinity | 4 |
The rabbit VK (K1 isotype) often have an extra cysteine in position 80 (Kabat numbering) that forms disulfide bonds with the constant region in the kappa light chain.23 This Cys 80 is removed in previous RabMAb humanization literature.1,6,20 Here, we also showed that replacing VK Cys 80 in scFv of immunotoxins does not affect affinities and may even stabilize scFv, given that the cytotoxicity of humanized scFv-containing immunotoxins increased.
A previous report showed that conserving rabbit Thr-H23, Gly-H49, Thr-H73, Val-H78, and Arg-H94 in human framework FW1.4 provide a generic humanization template (FW1.4 gen) to accept rabbit Kabat CDR and combine Kabat/IMGT CDR at CDR-H1.33 The template sequences are aligned with our template sequences in Fig. 5. Among them, Gly-H49 and Arg-H94 are within the combined Kabat/IMGT/Paratome CDRs, Thr-H23 is not included in combined CDRs, and is not an antigen-contacting residue or a supporting residue in known RabMAb structure complexes, Thr-H73 is within and Val-H78 is next to the HV4 loop, and they are supporting residues for antigen-contacting residues in some of the RabMAb structure complexes we analyzed.
One advantage of the combined Kabat/IMGT/Paratome CDR-grafting humanization is its high success rate. The resulting percentage of human residues in Vκ, identified by IMGT/DomainGapAlign, is 90.5∼92.3%, which is comparable to approved fully human antibodies and qualifies the name “-umab” according to the current World Health Organization (WHO) International Nonproprietary Name (INN) definitions; whereas the percentage of human residues in VH is 77.8∼83.8%, which is similar to the currently approved humanized antibodies (-zumab), but is less than 85% and could also fall into the “chimeric” category (-ximab).38 To further reduce the immunogenicity, one can consider removing T-cell epitopes8,39,40 from the grafted combined Kabat/IMGT/Paratome CDRs, where it is the major immunogenic site that we have seen in humanized murine antibodies.8
Anti-mesothelin immunotoxins have been developed for clinical trials.41 The RG7787/LMB-100 immunotoxin currently being evaluated in Phase 1 clinical studies 42 contains a humanized mouse SS1 Fab fragment that binds to the N terminus (Region I) of cell-surface mesothelin.43 Region I of mesothelin also interacts with other proteins that may interfere with the binding and function of anti-mesothelin region I antibodies. For example, MUC16/CA125, a protein that is often present in the serum of patients with mesothelin-related cancers, interacts with mesothelin via its Region I and can compete with antibody therapeutics _targeting Region I. The humanized rabbit scFv-based immunotoxins reported here can be important alternative immunotoxins for clinical development because they bind rare and novel epitopes in Region II (hYP223), Region III (hYP218), and a conformational epitope (hYP3) of mesothelin on tumor cells.6
Materials and methods
Cell culture
The H9 cell line is an A431 cell line overexpressing human mesothelin,44 and was cultured in DMEM (ThermoFisher Scientific Catalog number 11995) 10% fetal bovine serum supplemented with 1x GlutaMAX (Gibco, ThermoFisher Scientific, Catalog number 35050061) and 1x penicillin/streptomycin (Gibco, ThermoFisher Scientific, Catalog number 10378016).
Annotation with Kabat, IMGT CDRs and Paratome CDRs
Kabat CDR were annotated with Igblast tool from NCBI (http://www.ncbi.nlm.nih.gov/igblast/), IMGT CDRs were annotated with IMGT/DomainGapAlign tool from IMGT (www.imgt.org/3Dstructure-DB/cgi/DomainGapAlign.cgi), the Paratome CDRs were annotated with the Paratome tool (ofranservices.biu.ac.il/site/services/paratome/index.html).
Generation of multiple structure alignment and contact map
To identify the structural consensus of antigen-contacting residues, we analyzed rabbit antibody-antigen complex structures using a strategy similar to Ofran's29: we obtained the PDB files of the non-redundant rabbit Ab-Ag complex in PDB, made multiple structure alignments with the Aligner3D algorithm in Strap, defined contact residues as the shortest atom distance between 2 residues is no more than 6 Å, generated interchain contact maps by CCP445 with the CONTACT/ACT program, identified antigen-contacting residues and their supporting residues using Excel, and highlighted the antigen-contacting residues on antibody structure alignments (Fig. 3 and 4). The antigen-contacting residues of the antibody contact the antigen; the supporting residues contact antigen-contacting residues on the same rabbit heavy or light chain.
The structure alignment shown in Fig. 1 were made with Dali pairwise comparison by aligning heavy and light chains of each RabMAb with 2×7L and the figures were made with Chimera.46 In Fig. 4, the structure model of RabMAb YP223 and its humanized version were made using I-TASSER47 and aligned with Dali pairwise comparison.
Immunotoxin production and analysis
The scFvs were expressed as fusion proteins with scFv on the N terminus, and PE38, a truncated form of pseudomonas exotoxin, on the C terminus as described previously.6,34 This protocol includes a SEC step using column TSK-GEL® G3000SW 7.5 mm ID × 60 cm 10 µm (TOSCH Bioscience part No. 05103). The resulting proteins were scFv-based monovalent immunotoxins. To make immunotoxin constructs, the nucleotide sequence of humanized scFvs were synthesized by Genscript (Piscataway, NJ) and cloned into pRB98 vector to make pMH231(hYP3), pMH232 (hYP158), pMH233 (hYP218b), pMH234 (hYP223) plasmids. The immunotoxins were produced and tested for cell binding by flow cytometry and cytotoxicity by WST cell proliferation assay as described in detail previously6,34 on H9 cells. In flow cytometry analysis, we consecutively used rabbit whole serum containing anti-Pseudomonas exotoxin A antibody (Sigma Catalog number P2318) and R-phycoerythrin-conjugated goat anti rabbit antibody (Invitrogen, Catalog number P-2771MP), both diluted 200 folds. The original RabMAb scFvs were expressed in the same format and have been described previously.6 The purity of purified immunotoxins (4 µg per lane) were checked by SDS-PAGE (Fig. 6b). To calculate the EC50s, the binding of BL22 were subtracted in Excel. In GraphPad Prism 6 “XY analysis,” nonlinear regression with “specific binding” equation was used to measure cell binding EC50s, and the Fit spline/LOWESS with point-to-point curve were used to measure cytotoxicity IC50s. BL22 is an irrelevant anti-CD22 immunotoxin; YP7 immunotoxin is an irrelevant immunotoxin that _targets glypican-3 (GPC3).
Biophysical analysis
To characterize purified proteins, we used size-exclusion column “Superdex 200 Increase 10/300 GL” (GE Healthcare Life Sciences) coupled with Dawn HELEOS-II MALS with QELS DLS from Wyatt Technology in the SEC-MALS setup. The system uses Agilent (HP) 1100 series pumps. The SEC setup is equipped with the Agilent autosampler and diode-array UV/Vis. The proteins were prepared and analyzed in phosphate-buffered saline (PBS) buffer. We measured the binding kinetics in Octet RED96 at 30°C. The assay buffer was PBS 0.05% Tween20 with 0.1% (w/v) BSA. The assay program was 10 minutes' presoak, 180 s baseline establishment, 300 s antigen loading, 60 s baseline establishment, 600 s immunotoxin association, 30 mins or more immunotoxin dissociation. For the YP3, hYP3, YP158, hYP158, YP218 and hYP218 immunotoxins, 10 µg/ml rabbit Fc-Mesothelin-His protein was used to load Ni-NTA biosensor (Fortebio, Catalog No.18–5101); for YP223IT and hYP223IT, 1.25 µg/ml biotinylated rabbit Fc-Mesothelin-His protein was used to load the SA biosensor (Fortebio, Catalog No. 18–5019). Both loading conditions achieved similar readings with the machine. The binding kinetics were calculated with Graphpad Prism. The koff was calculated first, using “Dissociation - One phase exponential decay” equation and the NS (the binding at infinite times) was set to 0. Then the association kinetics were calculated using “Association kinetics - Two or more conc. of hot” equation and the koff was set to calculate kon.
Disclosure of potential conflict of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Grzegorz Piszczek and Stephanie Zalesak in the Biophysics Core at the National Heart, Lung, and Blood Institute for technical advice on SEC-MALS and Octet measurement, NIH Fellows Editorial Board for editorial assistance and Jessica Hong (NCI) for her proof-reading of our manuscript. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Funding
This research was supported by the Intramural Research Program of NIH, NCI, Center for Cancer Research (Z01 BC 010891 and ZIA BC 010891 to MH). This work was also possible in part by the Mesothelioma Applied Research Foundation Craig Kozicki Memorial Grant and by an Ovarian Cancer Research Fund Individual Investigator Award to MH.
References
- 1.Popkov M, Mage RG, Alexander CB, Thundivalappil S, Barbas Iii CF, Rader C. Rabbit immune repertoires as sources for therapeutic monoclonal antibodies: the impact of kappa allotype-correlated variation in cysteine content on antibody libraries selected by phage display. J Mol Biol 2003; 325:325-35; PMID:12488098; http://dx.doi.org/ 10.1016/S0022-2836(02)01232-9 [DOI] [PubMed] [Google Scholar]
- 2.Spieker-Polet H, Sethupathi P, Yam PC, Knight KL. Rabbit monoclonal antibodies: generating a fusion partner to produce rabbit-rabbit hybridomas. Proc Natl Acad Sci USA 1995; 92:9348-52; PMID:7568130; http://dx.doi.org/ 10.1073/pnas.92.20.9348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung WC, Beausoleil SA, Zhang X, Sato S, Schieferl SM, Wieler JS, Beaudet JG, Ramenani RK, Popova L, Comb MJ, et al.. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat Biotechnol 2012; 30:447-52; PMID:22446692; http://dx.doi.org/ 10.1038/nbt.2167 [DOI] [PubMed] [Google Scholar]
- 4.Rader C, Ritter G, Nathan S, Elia M, Gout I, Jungbluth AA, Cohen LS, Welt S, Old LJ, Barbas CF. The rabbit antibody repertoire as a novel source for the generation of therapeutic human antibodies. J Biol Chem 2000; 275:13668-76; PMID:10788485; http://dx.doi.org/ 10.1074/jbc.275.18.13668 [DOI] [PubMed] [Google Scholar]
- 5.Rossi S, Laurino L, Furlanetto A, Chinellato S, Orvieto E, Canal F, Facchetti F, Dei Tos AP. Rabbit monoclonal antibodies: a comparative study between a novel category of immunoreagents and the corresponding mouse monoclonal antibodies. Am J Clin Pathol 2005; 124:295-302; PMID:16040303; http://dx.doi.org/ 10.1309/NR8HN08GDPVEMU08 [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y-F, Phung Y, Gao W, Kawa S, Hassan R, Pastan I, Ho M. New high affinity monoclonal antibodies recognize non-overlapping epitopes on mesothelin for monitoring and treating mesothelioma. Sci Rep 2015; 5:9928; PMID:25996440; http://dx.doi.org/ 10.1038/srep09928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendell JC, Fakih M, Infante JR, Bajor DL, Cristea MC, Tremblay T, Trifan OC, Vonderheide RH. Phase 1 study to evaluate the safety and tolerability of the CD40 agonistic monoclonal antibody APX005M in subjects with solid tumors. ASCO annual meeting; J Clin Oncol 2016; 34 (suppl; abstr TPS3110); http://meetinglibrary.asco.org/content/164672-176 [Google Scholar]
- 8.Harding FA, Stickler MM, Razo J, DuBridge R. The immunogenicity of humanized and fully human antibodies. mAbs 2010; 2:256-65; PMID:20400861; http://dx.doi.org/ 10.4161/mabs.2.3.11641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 1986; 321:522-5; PMID:3713831; http://dx.doi.org/ 10.1038/321522a0 [DOI] [PubMed] [Google Scholar]
- 10.Verhoeyen M, Milstein C, Winter G. Reshaping human antibodies: grafting an antilysozyme activity. Science 1988; 239:1534-6; PMID:2451287; http://dx.doi.org/ 10.1126/science.2451287 [DOI] [PubMed] [Google Scholar]
- 11.Williams D, Matthews D, Jones T. Humanising antibodies by CDR grafting In: Kontermann R, Dübel S, eds. Springer Berlin Heidelberg: Antibody Engineering, 2010:319-39; http://dx.doi.org/ 10.1007/978-3-642-01144-3_21 [DOI] [Google Scholar]
- 12.Queen C, Schneider WP, Selick HE, Payne PW, Landolfi NF, Duncan JF, Avdalovic NM, Levitt M, Junghans RP, Waldmann TA. A humanized antibody that binds to the interleukin 2 receptor. Proc Natl Acad Sci U S A 1989; 86:10029-33; PMID:2513570; http://dx.doi.org/ 10.1073/pnas.86.24.10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashmiri SVS, De Pascalis R, Gonzales NR, Schlom J. SDR grafting—a new approach to antibody humanization. Methods 2005; 36:25-34; PMID:15848072; http://dx.doi.org/ 10.1016/j.ymeth.2005.01.003 [DOI] [PubMed] [Google Scholar]
- 14.Rader C, Cheresh DA, Barbas CF. A phage display approach for rapid antibody humanization: designed combinatorial V gene libraries. Proc Natl Acad Sci USA 1998; 95:8910-5; PMID:9671778; http://dx.doi.org/ 10.1073/pnas.95.15.8910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones T, Crompton L, Carr F, Baker M. Deimmunization of monoclonal antibodies In: Dimitrov AS, ed. Therapeutic Antibodies: Humana Press, 2009:405-23; PMID:19252848; http://dx.doi.org/1350088 10.1007/978-1-59745-554-1_21 [DOI] [PubMed] [Google Scholar]
- 16.Alard P, Desmet J, Lasters I. In silico de-immunization In: Kontermann R, Dübel S, eds. Springer Berlin Heidelberg: Antibody Engineering, 2010:369-76; http://dx.doi.org/ 10.1007/978-3-642-01144-3_24 [DOI] [Google Scholar]
- 17.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA 1992; 89:4285-9; PMID:1350088; http://dx.doi.org/ 10.1073/pnas.89.10.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ismael G, Hegg R, Muehlbauer S, Heinzmann D, Lum B, Kim S-B, Pienkowski T, Lichinitser M, Semiglazov V, Melichar B, et al.. Subcutaneous versus intravenous administration of (neo)adjuvant trastuzumab in patients with HER2-positive, clinical stage I–III breast cancer (HannaH study): a phase 3, open-label, multicentre, randomised trial. Lancet Oncol 2012; 13:869-78; PMID:22884505; http://dx.doi.org/ 10.1016/S1470-2045(12)70329-7 [DOI] [PubMed] [Google Scholar]
- 19.Jackisch C, Kim S-B, Semiglazov V, Melichar B, Pivot X, Hillenbach C, Stroyakovskiy D, Lum BL, Elliott R, Weber HA, et al.. Subcutaneous versus intravenous formulation of trastuzumab for HER2-positive early breast cancer: updated results from the phase III HannaH study. Ann Oncol 2015; 26:320-5; PMID:25403587; http://dx.doi.org/ 10.1093/annonc/mdu524 [DOI] [PubMed] [Google Scholar]
- 20.Yu Y, Lee P, Ke Y, Zhang Y, Yu Q, Lee J, Li M, Song J, Chen J, Dai J, et al.. A humanized anti-VEGF rabbit monoclonal antibody inhibits angiogenesis and blocks tumor growth in xenograft models. PLoS One 2010; 5:e9072; PMID:20140208; http://dx.doi.org/ 10.1371/journal.pone.0009072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavinder JJ, Hoi KH, Reddy ST, Wine Y, Georgiou G. Systematic characterization and comparative analysis of the rabbit immunoglobulin repertoire. PLoS One 2014; 9:e101322; PMID:24978027; http://dx.doi.org/ 10.1371/journal.pone.0101322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight KL, Becker RS. Molecular basis of the allelic inheritance of rabbit immunoglobulin VH allotypes: Implications for the generation of antibody diversity. Cell 1990; 60:963-70; PMID:2317867; http://dx.doi.org/ 10.1016/0092-8674(90)90344-E [DOI] [PubMed] [Google Scholar]
- 23.Mage R. Rabbit immune system In: Vohr H-W, ed. Encyclopedic Reference of Immunot: Springer Berlin Heidelberg, 2005:545-9; http://dx.doi.org/ 10.1007/3-540-27806-0_1248 [DOI] [Google Scholar]
- 24.Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med 1970; 132:211-50; PMID:5508247; http://dx.doi.org/ 10.1084/jem.132.2.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabat EA, Te Wu T, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. Diane Publ Company, 1992; ISBN: 9780941375658. [Google Scholar]
- 26.Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol 1987; 196:901-17; PMID:3681981; http://dx.doi.org/ 10.1016/0022-2836(87)90412-8 [DOI] [PubMed] [Google Scholar]
- 27.Lefranc M-P, Pommié C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 2003; 27:55-77; PMID:12477501; http://dx.doi.org/ 10.1016/S0145-305X(02)00039-3 [DOI] [PubMed] [Google Scholar]
- 28.Padlan EA, Abergel C, Tipper JP. Identification of specificity-determining residues in antibodies. FASEB J 1995; 9:133-9. PMID:7821752. [DOI] [PubMed] [Google Scholar]
- 29.Kunik V, Peters B, Ofran Y. Structural consensus among antibodies defines the antigen binding site. PLoS Comput Biol 2012; 8:e1002388; PMID:22383868; http://dx.doi.org/ 10.1371/journal.pcbi.1002388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sela-Culang I, Kunik V, Ofran Y. The structural basis of antibody-antigen recognition. Front Immunol 2013; 4:302; PMID:24115948; http://dx.doi.org/ 10.3389/fimmu.2013.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tramontano A, Chothia C, Lesk AM. Framework residue 71 is a major determinant of the position and conformation of the second hypervariable region in the VH domains of immunoglobulins. J Mol Biol 1990; 215:175-82; PMID:2118959; http://dx.doi.org/ 10.1016/S0022-2836(05)80102-0 [DOI] [PubMed] [Google Scholar]
- 32.Xiang J, Sha Y, Jia Z, Prasad L, Delbaere LTJ. Framework residues 71 and 93 of the chimeric B72.3 antibody are major determinants of the conformation of heavy-chain hypervariable loops. J Mol Biol 1995; 253:385-90; PMID:7473721; http://dx.doi.org/ 10.1006/jmbi.1995.0560 [DOI] [PubMed] [Google Scholar]
- 33.Borras L, Gunde T, Tietz J, Bauer U, Hulmann-Cottier V, Grimshaw JPA, Urech DM. Generic approach for the generation of stable humanized single-chain fv fragments from rabbit monoclonal antibodies. J Biol Chem 2010; 285:9054-66; PMID:20056614; http://dx.doi.org/ 10.1074/jbc.M109.072876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastan I, Ho M. Recombinant immunotoxins for treating cancer In: Kontermann R, Dubel S, eds. Antibody Engineering. Berlin-Heidelberg: Springer-Verlag, 2010:127-46; http://dx.doi.org/ 10.1007/978-3-642-01147-4 [DOI] [Google Scholar]
- 35.Haidar JN, Yuan Q-A, Zeng L, Snavely M, Luna X, Zhang H, Zhu W, Ludwig DL, Zhu Z. A universal combinatorial design of antibody framework to graft distinct CDR sequences: A bioinformatics approach. Proteins: Struct Funct Bioinform 2012; 80:896-912; PMID:22180101; http://dx.doi.org/22407974 10.1002/prot.23246 [DOI] [PubMed] [Google Scholar]
- 36.Raghunathan G, Smart J, Williams J, Almagro JC. Antigen-binding site anatomy and somatic mutations in antibodies that recognize different types of antigens. J Mol Recognit 2012; 25:103-13; PMID:22407974; http://dx.doi.org/ 10.1002/jmr.2158 [DOI] [PubMed] [Google Scholar]
- 37.MacCallum RM, Martin ACR, Thornton JM. Antibody-antigen interactions: contact analysis and binding site topography. J Mol Biol 1996; 262:732-45; PMID:8876650; http://dx.doi.org/ 10.1006/jmbi.1996.0548 [DOI] [PubMed] [Google Scholar]
- 38.Jones TD, Carter PJ, Plückthun A, Vásquez M, Holgate RGE, Hötzel I, Popplewell AG, Parren PWHI, Enzelberger M, Rademaker HJ, et al.. The INNs and outs of antibody nonproprietary names. mAbs 2016; 8:1-9; PMID:26716992; http://dx.doi.org/ 10.1080/19420862.2015.1114320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roque-Navarro L, Mateo C, Lombardero J, Mustelier G, Fernández A, Sosa K, Morrison SL, Pérez R. Humanization of predicted T-Cell epitopes reduces the immunogenicity of chimeric antibodies: new evidence supporting a simple method. Hybrid Hybridomics 2003; 22:245-57; PMID:14511570; http://dx.doi.org/ 10.1089/153685903322328974 [DOI] [PubMed] [Google Scholar]
- 40.Stickler M, Chin R, Faravashi N, Gebel W, Razo OJ, Rochanayon N, Power S, Valdes AM, Holmes S, Harding FA. Human population-based identification of CD4+ T-cell peptide epitope determinants. J Immunol Methods 2003; 281:95-108; PMID:14580884; http://dx.doi.org/ 10.1016/S0022-1759(03)00279-5 [DOI] [PubMed] [Google Scholar]
- 41.Hassan R, Ho M. Mesothelin _targeted cancer immunotherapy. Eur J Cancer 2008; 44:46-53; PMID:17945478; http://dx.doi.org/ 10.1016/j.ejca.2007.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassan R, Thomas A, Alewine C, Le D, Jaffee E, Pastan I. Mesothelin immunotherapy for cancer: ready for prime time? J Clin Oncol 2016; 34:4171-9. PMID:27863199; http://dx.doi.org/ 10.1200/JCO.2016.68.3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaneko O, Gong L, Zhang J, Hansen JK, Hassan R, Lee B, Ho M. A binding domain on mesothelin for CA125/MUC16. J Biol Chem 2009; 284:3739-49; PMID:19075018; http://dx.doi.org/ 10.1074/jbc.M806776200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho M, Hassan R, Zhang J, Q-c Wang, Onda M, Bera T, Pastan I. Humoral immune response to mesothelin in mesothelioma and ovarian cancer patients. Clin Cancer Res 2005; 11:3814-20; PMID:15897581; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-2304 [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al.. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 2011; 67:235-42; PMID:21460441; http://dx.doi.org/ 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605-12; PMID:15264254; http://dx.doi.org/ 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008; 9:1-8; PMID:18173834; http://dx.doi.org/ 10.1093/bib/bbn041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 Å resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci USA 2010; 107:5810-4; PMID:20231488; http://dx.doi.org/ 10.1073/pnas.0914946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stahl SJ, Watts NR, Rader C, Dimattia MA, Mage RG, Palmer I, Kaufman JD, Grimes JM, Stuart DI, Steven AC, et al.. Generation and characterization of a chimeric rabbit/human Fab for co-crystallization of HIV-1 Rev. J Mol Biol 2010; 397:697-708; PMID:20138059; http://dx.doi.org/ 10.1016/j.jmb.2010.01.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lammens A, Baehner M, Kohnert U, Niewoehner J, von Proff L, Schraeml M, Lammens K, Hopfner K-P. Crystal structure of human TWEAK in complex with the fab fragment of a neutralizing antibody reveals insights into receptor binding. PLoS One 2013; 8:e62697; PMID:23667509; http://dx.doi.org/ 10.1371/journal.pone.0062697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan R, Sampson JM, Chen Y, Vaine M, Wang S, Lu S, Kong X-P. Rabbit anti-HIV-1 monoclonal antibodies raised by immunization can mimic the antigen-binding modes of antibodies derived from HIV-1-infected humans. J Virol 2013; 87:10221-31; PMID:23864637; http://dx.doi.org/ 10.1128/JVI.00843-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malia TJ, Teplyakov A, Brezski RJ, Luo J, Kinder M, Sweet RW, Almagro JC, Jordan RE, Gilliland GL. Structure and specificity of an antibody _targeting a proteolytically cleaved IgG hinge. Proteins: Struct Funct Bioinform 2014; 82:1656-67; PMID:24638881; http://dx.doi.org/ 10.1002/prot.24545 [DOI] [PubMed] [Google Scholar]