Abstract
Covalent modifications of the histone tails and the cross talk between these modifications are hallmark features of gene regulation. The SAGA histone acetyltransferase complex is one of the most well-characterized complexes involved in these covalent modifications. The recent finding that the removal of the ubiquitin group from H2B is performed by a component of SAGA, Ubp8, is intriguing as it assigns two posttranslation modification processes to one complex. In this work, we characterize the association of Ubp8 with SAGA and the effect that acetylation and deubiquitylation have on one another in vitro and in vivo. We found not only that Ubp8 is a part of the SAGA complex, but also that its deubiquitylation activity requires Ubp8's association with SAGA. Furthermore, we found that the Ubp8 association with SAGA requires Sgf11 and that this requirement is reciprocal. We also found that the acetylation and deubiquitylation activities of SAGA are independent of one another. However, we found that preacetylating histone H2B inhibited subsequent deubiquitylation. Additionally, we found that increasing the ubiquitylation state of H2B inhibited the expression of the ARG1 gene, whose repression was previously shown to require the RAD6 ubiquitin ligase. Taken together, these data indicate that the expression of some genes, including ARG1, is regulated by a balance of histone H2B ubiquitylation in the cell.
Chromatin is an important regulator of cellular processes, including transcription, replication, recombination, repair, segregation, chromosomal stability, cell cycle progression, and epigenetic silencing (23). One step that is crucial for regulating these processes is the conversion from the active to inactive chromatin states. The ability to understand the change from euchromatin to heterochromatin is paramount in defining the mechanisms by which genes are active or inactive. Key players in regulating this process are the histone proteins, which cooperate to create the higher order structure of DNA. Histones undergo a variety of posttranslational modifications, including acetylation, phosphorylation, methylation, sumoylation, and ubiquitylation (3). The combination of these modifications is what constitutes the histone code hypothesis. Much work has gone into understanding and further defining the roles of these modifications in transcriptional control (reviewed in reference 4).
A variety of multiprotein complexes function in the cell to perform posttranslational modifications of the histone proteins, many of which have been well characterized in Saccharomyces cerevisiae. The SAGA, NuA4, Nua3, and SAS complexes acetylate a variety of lysine residues on all four histones, while the SNF1 complex phosphorylates serine 10 on histone H3, the RAD6 complex ubiquitylates histone H2B, and SET1, along with SET2 and DOT1, methylates histone H3 (reviewed in references 4, 16, and 26). Much of this work has lent support to the idea of the histone code, in particular the discovery of cross talk between histone ubiquitylation and methylation in yeast and promoter-specific histone modifications at the Igf2R imprinted gene (24, 28, 32).
Recent work from several labs has revealed an additional twist to this already complicated story. The Berger and Grant laboratories have recently shown that in addition to their ability to acetylate histones, the SAGA and SLIK histone acetyltransferase (HAT) complexes function to deubiquitylate histone H2B both in vivo and in vitro (5, 10). They found that this activity is dependent on the Ubp8 (ubiquitin-specific processing protease 8) gene product, since the deletion of UBP8 results in an increase in the overall ubiquitylation state of H2B (5, 10). Additionally, a partial disruption of SAGA via a deletion of SPT20 also leads to an increase in overall H2B ubiquitylation levels, although not to the same level as a Ubp8 deletion, suggesting that Ubp8 may belong to other complexes in addition to SAGA (10) These labs also showed the role of ubiquitylation in transcription and observed that the loss of Ubp8 leads to an overall increase of H3 lysine 36 methylation and a decrease in H3 lysine 4 methylation. They further illustrated that unlike other reversible histone modifications, for which the addition or removal of the modification leads to differential effects on transcription, ubiquitylation and deubiquitylation can both be involved in transcriptional activation (5, 10).
In the S. cerevisiae genome, there are 16 potential UBPs, 14 of which were shown to have ubiquitin-cleaving activity (1, 11). These proteins all have similar Cys and His domains which form the catalytic region. The roles of UBPs are thought to be diverse but remain poorly understood for both mammals and yeast. Surprisingly, none of the yeast UBPs is essential for viability (1). Specific functions have been assigned to very few UBPs, and their specific substrates have been even more rarely identified (30). In addition to the recently identified function of Ubp8, two UBPs, Ubp10p (Dot4p) and Ubp3p, regulate silencing (12, 15). Since the function of most of these deubiquitylating enzymes remains unknown, characterizing the ones that have been identified will lead to a better understanding and definition of the role that these enzymes play in the cell.
We set out to understand how Ubp8 deubiquitylates H2B and how it does this in the context of the SAGA complex. Using proteomic approaches, we identified a novel subunit of the SAGA and SLIK HAT complexes, named Sgf11, whose loss results in the additional loss of Ubp8 from SAGA and SLIK (13, 17). The Link group showed that Sgf11 functions at a subset of SAGA-regulated genes and, at least in the case of Matα1, this function also requires Ubp8 (17). Similar to the loss of Ubp8, the loss of Sgf11 does not affect the HAT activity of these complexes but simply results in the loss of Ubp8 from the complex (13, 17). However, the impact on histone ubiquitin levels as well as the effect on histone methylation has not been addressed.
We set out to further understand the relationship between Ubp8 and Sgf11 and how it relates to the overall function and composition of the SAGA and SLIK complexes. Since SAGA is made up of modules which include a subset of SPT proteins and a subset of TAF proteins (8), we tested whether Ubp8 and Sgf11 also function as a module in the context of the large 2-MDa SAGA complex and whether this association with SAGA is important for deubiquitylation activity in vivo and in vitro.
In addition to the role that H2B ubiquitylation plays in GAL1 and ADH2 transcription, it was previously shown that a K123R mutation of H2B and a RAD6 deletion lead to derepression of the ARG1 gene, presumably due to the loss of ubiquitylation on H2B (27). Since the loss of H2B ubiquitylation leads to an increase in the transcription of ARG1 (27), we tested the effect of increasing H2B ubiquitin levels in the cell on ARG1 transcription under repressing and activating conditions.
MATERIALS AND METHODS
S. cerevisiae strains.
The genotypes of strains used for this study are listed in Table 1. Individual TAP-tagged strains and deletion strains were obtained from Open Biosystems. TAP-tagged strains with deletions were obtained by crossing and dissecting individual TAP-tagged strains and deletion strains. Gene deletions that were not obtained from Open Biosystems were obtained by transformation with a PCR product obtained from the pRS415 vector, which harbors the LEU2 gene. Deletions and epitope-tagged strains were tested by PCR amplification of genomic DNAs or by Western blot analysis.
TABLE 1.
S. cerevisiae strains used for this study
| Strain | Genotype | Reference or source |
|---|---|---|
| By4741 | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| YKL101 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:Ada2-TAP::HIS3 MX6 | Open Biosystems |
| YKL134 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:Ada2TAP::HIS3 MX6 ubp8Δ::KANMX6 | This study |
| YKL128 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:Ada2-TAP::HIS3 MX6 sgf11Δ::KANMX6 | This study |
| YKL117 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:Ubp8-TAP::HIS3 MX6 | Open Biosystems |
| YKL132 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:Ubp8-TAP::HIS3 MX6 sgf11Δ::KANMX6 | This study |
| YKL138 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:Sgf11-TAP::HIS3 MX6 ubp8Δ::KANMX6 | This study |
| YKL60 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:gen5Δ::KANMX6 | Open Biosystems |
| YKL120 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:ubp8Δ::KANMX6 | Open Biosystems |
| YKL97 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:sgf11Δ::KANMX6 | Open Biosystems |
| YKL136 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:gen5Δ::KANMX6 ubp8Δ::KANMX6 | This study |
| YKL116 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:gen5Δ::KANMX6 sgf11Δ::KANMX6 | This study |
| YKL137 | matahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0:ubp8Δ::KANMX6 sgf11Δ::KANMX6 | This study |
| YKH045 | MATaura3-1 leu2,3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (Flag-HTB1-CEN-TRP1) pRG145 (GAPDHprom-3HA-UB14-URA3 integrative) | 10 |
| YKH046 | MATaura3-1 leu2,3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (Flag-htb1K123R-CEN-TRP1) pRG145 (GAPDHprom-3HA-UB14-URA3 integrative) | 10 |
| YKH047 | MATaura3-1 leu2,3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (Flag-HTB1-CEN-TRP1) ubp8Δ::KanMx pRG145 (GAPDHprom-3HA-UB14-URA3 integrative) | 10 |
| YKL142 | MATaura3-1 leu2,3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (Flag-HTB1-CEN-TRP1) sgf11Δ::LEU2 pRG145 (GAPDHprom-3HA-UB14-URA3 integrative) | This study |
| YKL143 | MATaura3-1 leu2,3,-112 his3-11,-15 trp1-1 ade2-1 htb1-1 htb2-1 pRS314 (Flag-HTB1-CEN-TRP1) gen5Δ::LEU2 pRG145 (GAPDHprom-3HA-UB14-URA3 integrative) | This study |
| Fy2034 | MATa HA-SPT7-TAP::TRP1 ura3Δ0 leu2Δ1 his3Δ200 gen5::HIS3 trp1Δ63 lys2-173R2 | 33 |
| YJW589 | MATa HA-SPT-TAP::TRP1 ura3Δ0 leu2Δ1 his3Δ200 gen5DBr::KANMX6 trp1Δ63 lys2-173R2 | Mark Chandy |
TAP purification.
The purification of Ada2-TAP-containing complexes was performed as previously described in order to separate the GCN5-containing HAT complexes ADA, SLIK, and SAGA (13). Purifications of Ubp8-TAP, Sgf11-TAP, and Spt7-TAP were performed as previously described, but with several modifications (19). All steps were performed at 4°C. Briefly, 6 liters of cells were grown to approximately 2 × 107 cells/ml in yeast extract-peptone-dextrose (YPD) and then washed in 10 ml of extract buffer (40 mM HEPES [pH 7.4], 350 mM NaCl, 10% glycerol, 0.1% Tween 20, 2 μg of pepstatin A/ml, 2 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Whole-cell extracts were prepared by bead beating in a bead beater (BioSpec). Cellular debris was removed by spinning the extracts at 12,000 × g for 30 min. The extracts were cleared by ultracentrifugation at 100,000 × g for 90 min. For TAP purification, 500 μl of immunoglobulin G-Sepharose (Amersham, Uppsala, Sweden) beads was incubated with clarified lysate overnight at 4°C. The beads were washed with 20 ml of extract buffer followed by 10 ml of TEV cleavage buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 10% glycerol, 1 mM dithiothreitol [DTT]) and then were resuspended in 1 ml of TEV cleavage buffer. TEV cleavage was performed by the use of 10 μl (100 U) of TEV (Gibco BRL) overnight at 4°C. TEV-cleaved products were added to 3 ml of calmodulin binding buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, 10% glycerol, 0.5 mM DTT) along with 3 μl of 1 M CaCl2 for each milliliter of TEV elution. Calmodulin-Sepharose (Amersham) purification was performed by binding of the eluate to 500 μl of beads overnight at 4°C. Proteins bound to calmodulin beads were eluted in 0.25-ml fractions by the use of calmodulin elution buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM EGTA, 0.1% NP-40, 10% glycerol, 0.5 mM DTT). TAP-purified complexes were resolved in a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and visualized by silver staining.
MudPIT.
TAP-purified protein complexes were digested with endoproteinase Lys-C and trypsin (Roche) as previously described (29, 31). Peptide mixtures were loaded on three-phase MudPIT microcapillary 100-μm-diameter columns as described previously (14). Tandem mass spectra (MS/MS) were acquired for the eluting peptides by use of a Deca-XP ion-trap mass spectrometer equipped with a nano-liquid chromatography (nano-LC) electrospray ionization source (ThermoFinnigan). SEQUEST (7) was used to match MS/MS spectra to peptides in a database containing S. cerevisiae protein sequences downloaded from the National Center for Biotechnology Information website. Spectrum-peptide matches were only retained if they had a normalized difference in cross-correlation scores (ΔCn) of at least 0.08 and minimum cross-correlation scores (XCorr) of 1.8 for singly, 2.5 for doubly, and 3.5 for triply charged spectra. In addition, the peptides had to be at least seven amino acids long and at least half-tryptic. DTASelect (25) was used to select and sort peptide-spectrum matches passing this set of criteria. Peptide hits from multiple runs were compared with CONTRAST software (25).
Isolation of ubiquitylated histones.
The relative levels of ubiquitylated histone H2B (ubH2B) in different strain backgrounds (YKH045, YKH046, YKH047, YKL142, and YKL143) were tested by use of an N-terminally FLAG-tagged histone H2B to aid in purification and detection with an anti-FLAG-horseradish peroxidase (HRP) antibody (Sigma) as described previously (20). Purifications of the ubH2B substrate from strains YKH047 and YKL142 for the deubiquitylation assays described below were also performed as described above.
Deubiquitylation assay.
The FLAG-tagged H2B substrate (containing ubH2B and unmodified H2B) was obtained as described previously (20). Between 250 and 500 ng of this substrate was incubated at 30°C for 60 min in DUB buffer (10 mM Tris-HCl [pH 8.0], 1 mM DTT, 1 μM PMSF, 1 μg of aprotinin and pepstatin A/ml) with 1% TAP-purified fractions. As a control, the substrate was also incubated in DUB buffer to which only calmodulin elution buffer was added. The reaction was stopped by freezing in liquid nitrogen, followed by boiling in 1 volume of 2× SDS sample buffer for 5 min and electrophoresis in an SDS-18% polyacrylamide gel. Gels were transferred to polyvinylidene membranes (Immobilon), and Western blot analysis was performed with anti-FLAG-HRP (to detect ubH2B and H2B) and anti-hemagglutinin (HA)-HRP (to detect ubiquitin) antibodies.
Experiments involving reconstitution of the in vitro deubiquitylation activity were performed as described above, except that equal amounts of complexes were mixed 1:1 or 1:1:1 for 1 h at 30°C prior to the deubiquitylation reaction.
HAT assays.
HAT assays were performed essentially as described previously (6), with the following changes: purified ubH2B and unmodified H2B were used as substrates for HAT reactions, which were performed with either wild-type (WT) SAGA or SAGA lacking Ubp8 and Sgf11 (for assays involving the effect of preacetylating the histone template on subsequent deubiquitylation). The HAT reactions were carried out in a buffer containing 10 mM Tris-HCl (pH 8.0), 1 mM DTT, and 1 mM PMSF.
RNA analyses.
Saturated cultures were grown in YPD and diluted 1/100 in YPD or minimal medium. The cells were grown at 30°C to an A600 of ∼1.0 (∼107 cells/ml in a 10-ml total volume), and RNAs were extracted by the use of Trizol reagent (Invitrogen). For reverse transcription-PCR (RT-PCR) analysis, 2.5 μg of total RNA was used. The reverse transcriptase reaction was carried out in a 50°C water bath for 1 h with a random hexamer primer and Superscript III (Invitrogen), followed by incubation at 95°C for 5 min to inactivate the enzyme. Control RT reactions were carried out in the absence of reverse transcriptase. Five microliters of the RT reaction mixture was subsequently used in a 25-μl PCR. The ARG1 primers (5′-GTTGGGTACCTCTTTGGCAA-3′ and 5′-GCCCAGAATGATGACGTTACCC-3′) (18) generated a 700-bp product, and the ACT1 primers (5′-GTGAACGATAGATGGACCAC-3′ and 5′-TTGGGTTTGGAATCTGCCGG-3′) generated a 300-bp product.
RESULTS
Ubp8 is only associated with SAGA and SLIK, and this association requires Sgf11.
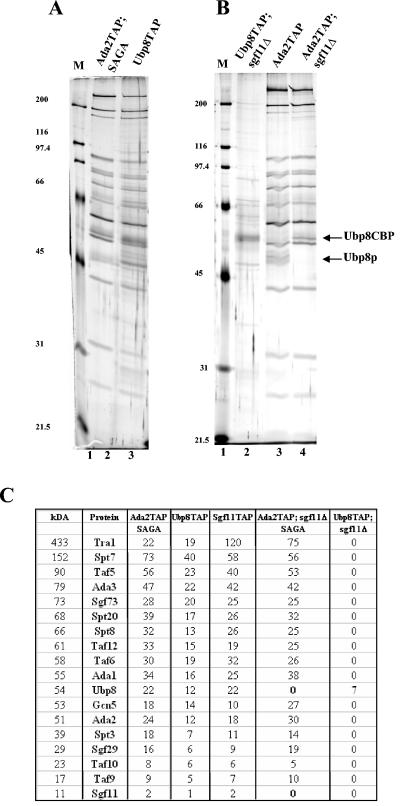
In order to understand how Ubp8 functions as a part of SAGA, we first needed to determine whether Ubp8 was associated with any other complexes in the cell. To this end, we tagged Ubp8 with TAP and subsequently isolated protein complexes associated with Ubp8 and showed that Ubp8 copurifies with HAT activity (Fig. 1A; also data not shown). The purification of Ubp8 looked similar to purifications of SAGA (Fig. 1A, compare lanes 2 and 3). In order to confirm this, we performed mass spectrometry analysis (MudPIT) of the purification product and confirmed that Ubp8 is only associated with proteins that were previously identified in the SAGA, SLIK, and ADA HAT complexes (Fig. 1C) (22). We also used a modified TAP purification of Ubp8 that allowed us to separate the various Gcn5-containing HAT complexes and confirmed that Ubp8 is part of both the SAGA and SLIK complexes, but not the ADA complex (5; also data not shown).
FIG. 1.
TAP purification of Ubp8 reveals that it is only associated with SAGA. (A) Silver-stained gel showing protein profiles, comparing Ubp8 purification with conventional SAGA purification. Lane 1, molecular weight marker; lane 2, Ada2-TAP, SAGA purification; lane 3, Ubp8-TAP purification. (B) Silver-stained gel comparing Ada2-TAP sgf11Δ strain with Ubp8-TAP sgf11Δ strain, showing that in the absence of Sgf11, Ubp8 is unable to associate with any other proteins in the cell. Lane 1, molecular weight marker; lane 2, Ubp8-TAP sgf11Δ; lane 3, Ada2-TAP; lane 4, Ada2-TAP sgf11Δ. (C) Proteins purified from various purifications were identified by MudPIT mass spectrometry. The numbers indicate nonredundant spectra identifying each protein.
We previously showed that the deletion of Sgf11 results in the preferential loss of Ubp8 from the SAGA complex, so we tested the effects of deleting Sgf11 from the Ubp8-TAP strain. Since Ubp8 copurified with SAGA components and its association with SAGA depends on Sgf11, we were not surprised to find that upon deletion of Sgf11, we were only able to isolate the Ubp8 protein (Fig. 1B, lane 2, and C). These purified complexes were further used in the deubiquitylation experiments described below.
Sgf11 association with SAGA/SLIK requires Ubp8.
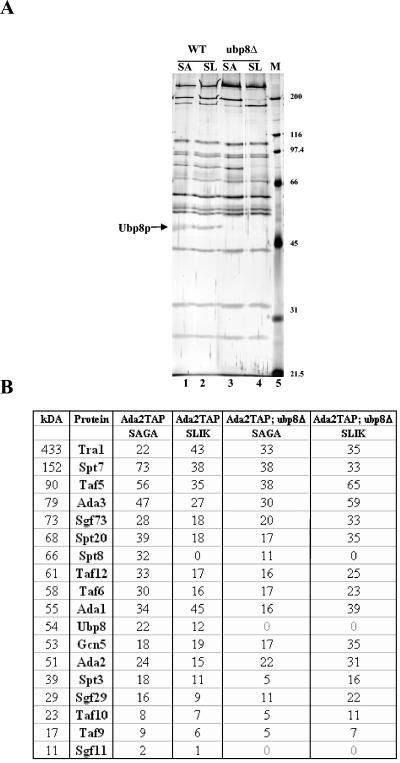
Since Ubp8 requires Sgf11 for its association with SAGA, we tested whether there was a reciprocal requirement. Using a TAP-tagged Ada2 strain, we purified both SAGA and SLIK from a strain lacking Ubp8 (Fig. 2A, lanes 5 and 6). It was previously shown that the loss of Ubp8 does not affect HAT activity and therefore that Ubp8 is not required for the integrity of the SAGA or SLIK HAT complex (10; also data not shown). Through protein gel analysis, we could not determine if any components, including Sgf11, were missing from the SAGA or SLIK complexes. Therefore, we went on to analyze these purified SAGA and SLIK complexes by MudPIT and found that in the absence of Ubp8, the only other subunit lost was Sgf11 (Fig. 2B). Therefore, Ubp8 and Sgf11 are reciprocally required for their stable associations with SAGA and SLIK.
FIG. 2.
Deletion of Ubp8 from SAGA/SLIK results in the loss of Sgf11, indicating that the two proteins require each other for their association with SAGA/SLIK. (A) Silver-stained gel comparing wild-type SAGA/SLIK to complexes lacking Ubp8 (note that these complexes were purified as previously described [13]). Lane 1, wild-type SAGA purification; lane 2, wild-type SLIK purification; lane 3, SAGA lacking Ubp8; lane 4, SLIK lacking Ubp8; lane 5, molecular weight marker. The approximate migration of SAGA components is indicated on the right. (B) Proteins purified from various purifications were identified by MudPIT mass spectrometry. The numbers indicate nonredundant spectra identifying each protein.
In previous work, Powell et al. purified complexes that associate with Sgf11 through TAP purification (17). They found that most of the subunits that purify with Sgf11 are part of the SAGA complex, but their mass spectrometry analysis also showed proteins that are independent of SAGA and that copurified with Sgf11, in particular, TATA-binding protein (17). In our purification of Sgf11-TAP, only proteins associated with SAGA and SLIK were identified (13). In order to resolve this difference, we set out to identify proteins that were associated with Sgf11 independently of Ubp8 by deleting the UBP8 gene from the Sgf11-TAP strain. In contrast to the stability of Ubp8-TAP in the absence of Sgf11, Sgf11-TAP was unstable in the absence of Ubp8 (data not shown). Possible explanations for this apparent instability are discussed below.
Deletion of Sgf11 results in an increase in the overall level of H2B ubiquitylation.
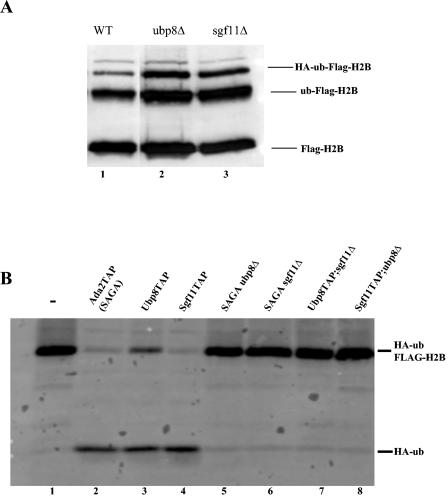
Our data indicate that Ubp8 and Sgf11 function together in SAGA as a subcomplex that carries out the deubiquitylation of histone H2B. However, the exact role that Sgf11 plays in this process is not clear. Previous data demonstrated that the overall cellular histone H2B ubiquitylation levels are increased in the absence of Ubp8. Moreover, this increase was apparent in a yeast strain carrying a mutation in the catalytic domain of Ubp8, indicating that Ubp8 alone is responsible for the deubiquitylation of H2B in vivo. To determine if this is indeed the case, we deleted Sgf11 from a yeast strain harboring FLAG-tagged H2B and HA-tagged ubiquitin that was previously used to assay H2B ubiquitylation levels. We found that the deletion of Sgf11 increased the overall H2B ubiquitylation levels to the same extent observed for a Ubp8 deletion (Fig. 3A, compare lanes 1 and 3). Therefore, both Ubp8 and Sgf11 are important for maintaining wild-type H2B ubiquitin levels in the cell.
FIG. 3.
Deubiquitylation requires Ubp8 and Sgf11 in the context of SAGA. (A) The overall ubiquitylation level of H2B is increased in sgf11Δ and ubp8Δ strains. FLAG purification of H2B from wild-type, ubp8Δ, and sgf11Δ strains was followed by an anti-FLAG antibody to monitor both unmodified and modified FLAG-tagged H2B in the cell. Lane 1, wild type (YKH045); lane 2, ubp8Δ (YKH047); lane 3, sgf11Δ (YKH142). (B) In vitro deubiquitylation assay. Lane 1, mock; lane 2, SAGA; lane 3, Ubp8-TAP; lane 4, Sgf11-TAP; lane 5, Ada2-TAP ubp8Δ; lane 6, Ada2-TAP sgf11Δ; lane 7, Ubp8-TAP sgf11Δ; lane 8, Sgf11-TAP ubp8Δ.
Ubp8 is necessary, but not sufficient, for deubiquitylation of H2B in vitro.
Since we were able to purify Ubp8 away from SAGA through the deletion of Sgf11, we wanted to determine if Ubp8 alone was able to deubiquitylate H2B in vitro. We found that although Ubp8 is necessary for deubiquitylation (10), it is not able to carry out this reaction on its own (Fig. 3B, lane 7). This finding is intriguing, as it suggests that it is not only the protein but also the context in which the protein is located that allows it to perform its function. We also found that SAGA purified in the absence of Sgf11 was unable to deubiquitylate H2B in vitro since it also lacked Ubp8p (Fig. 3B, lane 6). Additionally, both Ubp8-TAP and Sgf11-Tap purified complexes were able to deubiquitylate H2B (Fig. 3B, lanes 3 and 4). However, we did find that Ubp8-TAP purified complexes were less efficient than Sgf11-TAP or Ada2-TAP purified complexes in deubiquitylation. This was probably due to the presence of the TAP tag on Ubp8 causing some partial inhibition of activity.
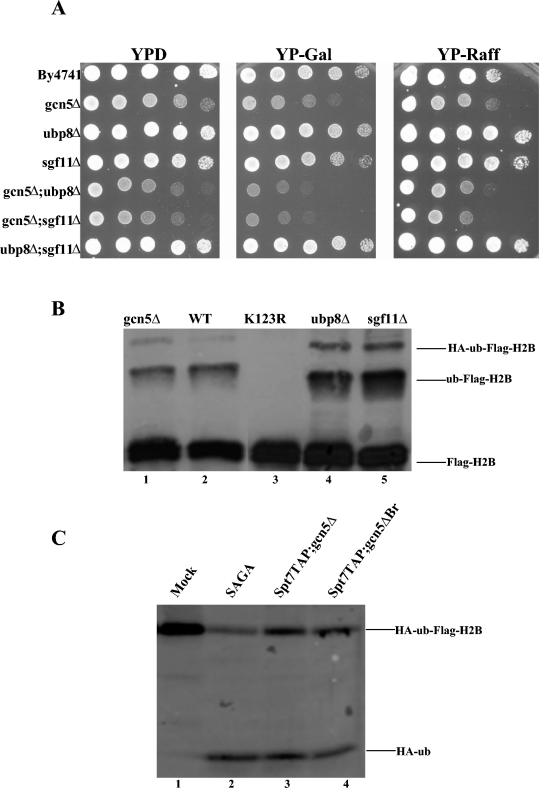
GCN5 is not required for H2B deubiquitylation in vitro or in vivo.
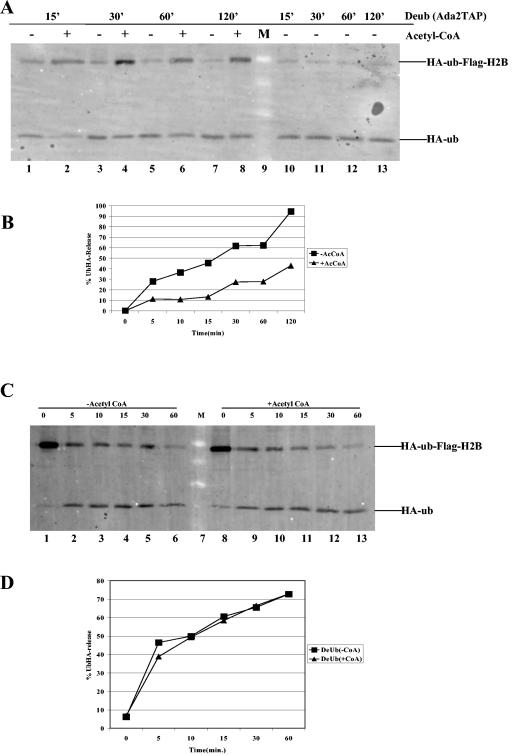
Previous work indicated that the deletion of Ubp8 together with GCN5 resulted in severe defects in the transcription of both GAL1 and ADH2 (10). Instead of examining the role that Ubp8 or Sgf11 plays in SAGA, we set out to see if GCN5 and, more specifically, acetylation by SAGA, plays a role in deubiquitylation. We first tested to see if the deletion of Sgf11 alone or in combination with GCN5 had similar growth defects as those seen for the UBP8 GCN5 double deletion. We found this to be the case. The synthetic phenotype upon the deletion of Sgf11 and Gcn5 may result from the loss of the two enzymatic activities from the SAGA complex. However, it may also arise if the double deletion is more disruptive to the complex than either single deletion alone. These possibilities are currently being investigated. The deletion of Sgf11 in combination with Ubp8 had no growth defects on galactose, indicating that these two proteins function together and partially redundantly with GCN5 to regulate GAL1 (Fig. 4A). We next examined if a deletion of GCN5 resulted in a change in the overall H2B ubiquitylation level in vivo. We found that unlike the deletion of Sgf11 and Ubp8, the deletion of GCN5 allowed wild-type levels of H2B ubiquitylation, the first indication that these two SAGA activities are independent (Fig. 4B, compare lanes 1 and 2). We then checked to see if SAGA purified from a strain lacking GCN5 or lacking the GCN5 bromodomain was able to deubiquitylate H2B in vitro. We found that both GCN5 mutant complexes were able to deubiquitylate H2B, similar to wild-type SAGA (Fig. 4C). We next tested whether the deubiquitylation of H2B was affected by acetylation. First, we looked at the effect of acetylating purified FLAG-H2B or FLAG-ubH2B with SAGA lacking Sgf11 (to separate acetylation from deubiquitylation) and, surprisingly, found that preacetylating purified FLAG-H2B or FLAG-ubH2B slightly inhibited subsequent deubiquitylation by wild-type SAGA (Fig. 5A and B). This was a specific effect of acetylation and not a steric effect of the SAGA added to the initial HAT reaction, as the presence of this complex in the absence of acetyl-coenzyme A (CoA) did not inhibit the reaction (Fig. 5A, compare lanes 1, 3, 5, 7, 9, and 12). We also examined the effect of the addition of acetyl-CoA on deubiquitylation and found that the two enzymatic activities of SAGA were able to function normally when occurring at the same time (Fig. 5C and D).
FIG. 4.
Similar to Ubp8 deletion, the deletion of Sgf11 with GCN5 results in a GAL phenotype, but deubiquitylation does not require GCN5. (A) Cell growth and expression in sgf11Δ strain. Fourfold serial dilutions were spotted onto plates containing rich medium with dextrose (YPD), with galactose (YP-Gal), and with raffinose (YP-Raff). The wild type and mutants were grown at 30°C for ∼2 days. (B) Level of H2B ubiquitylation is unaffected by a GCN5 deletion, as determined by Western blotting of FLAG. Lane 1, gcn5Δ; lane 2, WT; lane 3, K123R; lane 4, ubp8Δ; lane 5, sgf11Δ. (C) SAGA complexes purified from strains mutated in GCN5 are able to deubiquitylate in vitro. Lane 1, mock; lane 2, wild-type SAGA; lane 3, gcn5Δ; lane 4, gcn5ΔBr.
FIG. 5.
Effect of acetylation on deubiquitylation in vitro. (A) Purified FLAG-H2B (modified and unmodified) was acetylated with Ada2-TAP sgf11Δ for 60 min prior to being subjected to deubiquitylation with wild-type SAGA for the indicated times. HAT assays were done either in the presence or in the absence of acetyl-CoA. Lanes 10 to 13 did not have acetyl-CoA and Ada2-TAP sgf11Δ as a control for deubiquitylation. (B) Semiquantitative analysis of the effects of preacetylating the FLAG-H2B templates prior to deubiquitylation. (C) Simultaneous acetylation and deubiquitylation of purified FLAG-H2B with wild-type SAGA. Lanes 1 to 6, deubiquitylation in the absence of acetyl-CoA; lane 7, molecular weight marker; lanes 8 to 13, deubiquitylation in the presence of acetyl-CoA. (D) Semiquantitative analysis of the effect of simultaneous acetylation and deubiquitylation by SAGA. Acetylation was monitored by the incorporation of [3H]acetyl-CoA, and deubiquitylation was monitored by the release of HA-ubiquitin from FLAG-ubH2B-HA.
Sgf11 and Ubp8 are required for the expression and/or repression of ARG1.
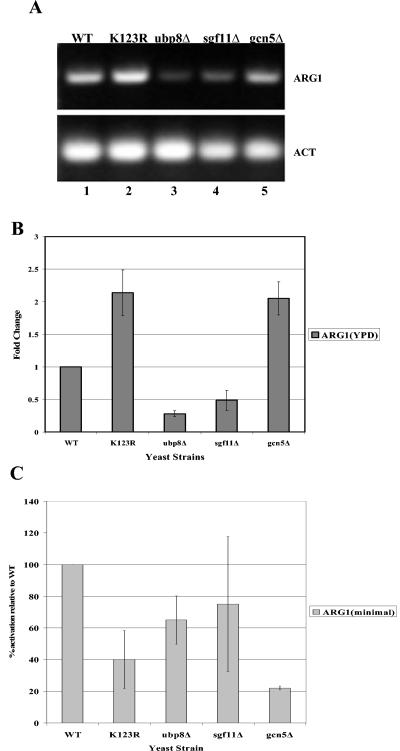
Since SAGA and ubiquitylation function in the activation and repression of ARG1, we examined the effect of increased cellular H2B ubiquitylation levels on ARG1 transcription (18, 27). Turner et al. demonstrated that ARG1 is derepressed by a RAD6 deletion and by a K123R mutation in H2B, indicating that the ubiquitylation of H2B is required for the repression of ARG1 (27). They also showed that GCN5 is required for the activation and repression of ARG1. Since deubiquitylation has also been implicated in gene activation, we performed an RT-PCR analysis of ARG1 transcription in both Ubp8 and Sgf11 deletion strains under repressing conditions (Fig. 6A and B). We found that under repressing conditions (growth in YPD), the deletion of either Ubp8 or Sgf11 resulted in a decrease in ARG1 expression to levels below wild-type expression; however, the deletion of Sgf11 had less of an effect on ARG1 expression than the deletion of Ubp8 (fivefold versus twofold) (Fig. 6B). We observed only a modest twofold increase in ARG expression in the K123R H2B mutation due to the fact that our strain had a high level of ARG1 expression even under repressive conditions (Fig. 6A, lane 1). However, the high level of ARG1 allowed us to see the repression caused by both Ubp8 and Sgf11 deletion. These two findings indicate that Ubp8 and Sgf11 antagonize RAD6 and BRE1 to create a balance of histone ubiquitylation that allows the proper transcription of ARG1. Additionally, we looked at the effects of these same mutations on ARG1 activation (Fig. 6C). We found that the K123R mutation as well as the Ubp8 and Sgf11 deletions resulted in an approximately twofold decrease in ARG1 expression (Fig. 6C). However, as previously shown, the deletion of GCN5 resulted in a larger decrease in the amount of ARG1 transcripts. Therefore, H2B ubiquitylation seems to have less of an effect on ARG1 activation than it does on ARG1 repression.
FIG. 6.
Effects of Ubp8Δ and Sgf11Δ on ARG1 transcription. (A) RNAs isolated from WT strain and from K123R, ubp8Δ, sgf11Δ, and gcn5Δ mutant strains grown under repressive conditions (YPD) were used as templates for RT-PCRs. The products from cycle 22 were separated in a 1% agarose gel and stained with ethidium bromide. (B) Data from three independent RNA isolations were quantitated with Image Quant software. The fold change indicates the increase or decrease in ARG1 compared to the wild type, which is set as 1. (C) Data for three independent RNA isolations under transcriptional activation conditions were quantitated with Image Quant software. WT ARG1 expression was arbitrarily set to 100%, and the effects of mutations on ARG1 activity are expressed as percentages of the WT.
DISCUSSION
In this study, we have presented evidence that supports the idea of context-dependent activities being associated with the SAGA HAT complex. Although Ubp8 is required genetically for deubiquitylation, we demonstrated that it is not Ubp8 alone that is able to perform this function. We found that Ubp8 and Sgf11 reciprocally require each other for an association with SAGA and that the loss of Sgf11 causes the same increase in H2B ubiquitylation as that seen in a Ubp8 deletion strain. We found that Ubp8 in the absence of SAGA and Sgf11 does not possess deubiquitylation activity in vitro or in vivo. Although Ubp8 is still present in the Sgf11 deletion strain, it is unable to deubiquitylate H2B effectively, indicating that Ubp8 must be part of SAGA in order to function. However, we were unable to address whether a complex containing just Ubp8 and Sgf11 would be able to deubiquitylate H2B in vitro, since we do not yet know what holds those two proteins as a module within SAGA. One possibility is that SPT20 may help to tether Ubp8 and Sgf11 to SAGA, since the loss of SPT20 increases H2B ubiquitylation levels, but not quite to the extent of a Ubp8 deletion (10). This indicates that there is still some activity present and that this activity may be due to a Ubp8/Sgf11 heterodimer. At present, we cannot differentiate whether Sgf11 is required for Ubp8 deubiquitylation activity directly or indirectly by simply holding Ubp8 to SAGA.
What role does SAGA play in regulating the deubiquitylation of H2B? One possibility is that Ubp8/Sgf11 uses SAGA to gain access to specific _target genes or promoters. This makes sense since Sgf11 is known to regulate a subset of SAGA-regulated genes (17). It is also possible that the acetylation activity of SAGA plays a role in the deubiquitylation activity. Our results indicate that the acetylation activity of SAGA is independent of its deubiquitylation activity, since the deletion of GCN5 did not affect the overall H2B ubiquitin levels in vivo. Additionally, SAGA complexes lacking GCN5 or the GCN5 bromodomain deubiquitylate H2B to wild-type levels in vitro. The observation that prior acetylation by SAGA reduces the deubiquitylation activity of the complex suggests that deubiquitylation may precede acetylation and that the two processes may not be totally independent. Lastly, it is also possible that Ubp8 is more stable as a part of SAGA. For our purification of Ubp8 in the absence of Sgf11, MudPIT analysis revealed several heat shock proteins and other chaperones associated with Ubp8, indicating that Ubp8 is unstable and/or unfolded when it is not a part of SAGA. Therefore, we cannot rule out that the effect that we saw with an Sgf11 deletion on H2B ubiquitylation was simply a consequence of Ubp8 degradation.
The concept of modularity and contextual functions within SAGA applies to its acetylation activity as well. Although GCN5 alone possesses acetyltransferase activity in vitro, it is only active on core histones and requires Ada2 and Ada3 in order to acetylate nucleosomes (2). Additionally, we know that the loss of the TAFs from SAGA also reduces its ability to acetylate nucleosomes (9). Therefore, our findings that deubiquitylation requires the presence of an intact SAGA complex are in agreement with previous findings about the importance of not just the individual components of SAGA, but of the complex as a whole. A three-dimensional reconstruction of SAGA lended more support to the modularity of SAGA, but the location of Ubp8 was not studied, and when that work was done, Sgf11 had not yet been identified (33). It will be interesting to learn where Ubp8 and Sgf11 are located within the complex in order to further understand how deubiquitylation is carried out in the context of SAGA.
The results presented here also indicate that, like Ubp8, Sgf11 is important for transcription. First, we found that as in the case of Ubp8, the deletion of Sgf11 in combination with GCN5 results in a severe phenotype when grown in the presence of galactose, implying that GAL1 transcription is impaired. This is consistent with the possibility that there is a link between acetylation and deubiquitylation and that this leads to more defects in transcription than either mutant on its own. Alternatively, while SAGA is intact in the absence of Gcn5 or Ubp8, the double deletion removing both proteins may reduce the stability of the complex and thereby affect other functions.
Is deubiquitylation also involved in transcriptional repression? In order to address this point, we looked at the effects of deleting Sgf11 and Ubp8 on ARG1 transcription. It was previously shown that mutations which result in a decrease in H2B ubiquitylation result in the derepression of ARG1 under conditions in which ARG1 transcription is not active (18). Similarly, we found that the K123R mutation in H2B results in an increase in ARG1 transcription. We also found that the deletion of Ubp8 or Sgf11 results in a decrease in ARG1 transcription. These results indicate that the level of histone H2B ubiquitylation is important for regulating ARG1 transcription. Although our wild-type yeast strain contained high levels of ARG1, we were still able to observe increases in ARG1 transcription, but more importantly it allowed us to see decreases in ARG1 transcription that were associated with the loss of Ubp8 and Sgf11 and the resulting increase in H2B ubiquitylation. Since there is cross talk between ubiquitylation and methylation, it would be interesting to test whether the repression of ARG1 transcription is also carried out by SET1/2 or if this effect is a methylation-independent effect of ubiquitylation.
As we continue to increase our understanding of multiprotein complexes such as SAGA, we are presented with new questions. We are now left with merging the concept of the histone code hypothesis with protein complexes that are able to perform multiple types of histone modifications. Understanding how the complexes that perform these modifications communicate will facilitate a better understanding of epigenetic gene regulation.
Acknowledgments
We thank Mary Ann Osley and Fred Winston for yeast strains used in this study. We thank Michael Carrozza, Philippe Prochasson, Bing Li, and Laurie Krom for helpful discussions and critical comments on the manuscript. We thank Samantha Pattenden for reagents and advice on RT-PCR experiments. We thank Mark Chandy for providing the Gcn5ΔBr strain and Erin Smith for constructing the Sgf11-TAP ubp8Δ strain.
This work was supported by a postdoctoral fellowship to K.K.L. from the Damon Runyon Cancer Research Foundation (1751-03) and by NIH grant GM46787 to J.L.W.
REFERENCES
- 1.Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 5.Daniel, J. A., M. S. Torok, Z. W. Sun, D. Schieltz, C. D. Allis, J. R. Yates III, and P. A. Grant. 2004. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 279:1867-1871. [DOI] [PubMed] [Google Scholar]
- 6.Eberharter, A., T. Lechner, M. Goralik-Schramel, and P. Loidl. 1996. Purification and characterization of the cytoplasmic histone acetyltransferase B of maize embryos. FEBS Lett. 386:75-81. [DOI] [PubMed] [Google Scholar]
- 7.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 8.Grant, P. A., L. Duggan, J. Côté, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 9.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 10.Henry, K. W., A. Wyce, W. S. Lo, L. J. Duggan, N. C. Emre, C. F. Kao, L. Pillus, A. Shilatifard, M. A. Osley, and S. L. Berger. 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17:2648-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 12.Kahana, A., and D. E. Gottschling. 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6608-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, K. K., P. Prochasson, L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2004. Proteomic analysis of chromatin-modifying complexes in Saccharomyces cerevisiae identifies novel subunits. Biochem. Soc. Trans. 32:899-903. [DOI] [PubMed] [Google Scholar]
- 14.McDonald, W. H., R. Ohi, D. T. Miyamoto, T. J. Mitchison, and J. R. Yates III. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 219:245-251. [Google Scholar]
- 15.Moazed, D., and D. Johnson. 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86:667-677. [DOI] [PubMed] [Google Scholar]
- 16.Osley, M. A. 2004. H2B ubiquitylation: the end is in sight. Biochim. Biophys. Acta 1677:74-78. [DOI] [PubMed] [Google Scholar]
- 17.Powell, D. W., C. M. Weaver, J. L. Jennings, K. J. McAfee, Y. He, P. A. Weil, and A. J. Link. 2004. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol. Cell. Biol. 24:7249-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ricci, A. R., J. Genereaux, and C. J. Brandl. 2002. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell. Biol. 22:4033-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 20.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 21.Sadygov, R. G., J. Eng, E. Durr, A. Saraf, H. McDonald, M. J. MacCoss, and J. R. Yates III. 2002. Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1:211-215. [DOI] [PubMed] [Google Scholar]
- 22.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 24.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 25.Tabb, D. L., W. H. McDonald, and J. R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner, B. M. 2003. Memorable transcription. Nat. Cell Biol. 5:390-393. [DOI] [PubMed] [Google Scholar]
- 27.Turner, S. D., A. R. Ricci, H. Petropoulos, J. Genereaux, I. S. Skerjanc, and C. J. Brandl. 2002. The E2 ubiquitin conjugase Rad6 is required for the ArgR/Mcm1 repression of ARG1 transcription. Mol. Cell. Biol. 22:4011-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vu, T. H., T. Li, and A. R. Hoffman. 2004. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum. Mol. Genet. 13:2233-2245. [DOI] [PubMed] [Google Scholar]
- 29.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson, K. D. 1997. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11:1245-1256. [DOI] [PubMed] [Google Scholar]
- 31.Wolters, D. A., M. P. Washburn, and J. R. Yates III. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73:5683-5690. [DOI] [PubMed] [Google Scholar]
- 32.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 33.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15:199-208. [DOI] [PubMed] [Google Scholar]