Summary
Prior studies suggested that SAGA and TFIID are alternative factors that promote RNA polymerase II transcription with about 10% of genes in S. cerevisiae dependent on SAGA. We reassessed the role of SAGA by mapping its genome-wide location and role in global transcription in budding yeast. We find that SAGA maps to the UAS elements of most genes, overlapping with Mediator binding and irrespective of previous designations of SAGA or TFIID-dominated genes. Disruption of SAGA through mutation or rapid subunit depletion reduces transcription from nearly all genes, measured by newly-synthesized RNA. We also find that the acetyltransferase Gcn5 synergizes with Spt3 to promote global transcription and that Spt3 functions to stimulate TBP recruitment at all tested genes. Our data demonstrate that SAGA acts as a general cofactor required for essentially all RNA polymerase II transcription and is not consistent with the previous classification of SAGA and TFIID-dominated genes.
Introduction
Formation of the transcription preinitiation complex (PIC), containing RNA polymerase II (Pol II) and general transcription factors (GTFs) is a major regulatory step in eukaryotic gene expression. TFIID is a GTF that binds to promoters and is known to nucleate PIC formation. One important class of factors that regulate PIC formation and function are coactivator complexes. Coactivators directly interact with the basal transcription machinery and/or open the chromatin structure at promoter regions through either remodeling activities or post-translational histone modifications (Hahn and Young, 2011; Thomas and Chiang, 2006). Several coactivators were shown to act at specific subsets of genes, whereas others appear to have a more global role in Pol II transcription. For example, the coactivator Mediator has been recognized as a general factor required for essentially all Pol II transcription, interacts directly with Pol II and other basal factors, and plays a key role in PIC stabilization and Pol II activation (Ansari et al., 2009; Holstege et al., 1998; Thompson and Young, 1995).
To determine the specific regulatory network of distinct coactivators, several studies used genome-wide transcriptome analyses to quantify the levels of steady-state mRNA in different mutant strains. Using this approach, a seminal study analyzed the contribution of the SAGA coactivator complex to gene expression in S. cerevisiae (Huisinga and Pugh, 2004). The yeast SAGA complex is thought to activate Pol II transcription through both the recruitment of the TATA binding protein (TBP) and via chromatin modifications mediated by its histone acetyltransferase (HAT) and deubiquitinase (DUB) activities [reviewed in (Koutelou et al., 2010; Rodriguez-Navarro, 2009; Weake and Workman, 2012)]. Organization of these activities in distinct structural modules of SAGA allows the inactivation of one specific function without altering others (Lee et al., 2011). Upon deletion of SPT3, a TBP-interacting subunit of SAGA, levels of total mRNA were reduced for about 10% of yeast genes (Huisinga and Pugh, 2004). This set of Spt3-regulated genes was compared to those regulated by TBP associated factor 1 (Taf1), a subunit of the TFIID complex also suggested to deposit TBP at promoters. This led to the distinction between two different genes classes: (i) the SAGA-dominated genes, which are positively regulated by Spt3, but are essentially independent of Taf1 and (ii) the larger class (90%) of TFIID-dominated genes which are more dependent on Taf1 than on Spt3 (Huisinga and Pugh, 2004). However, the inactivation of both Taf1 and Spt3 induced a severe decrease in the steady-state mRNA levels of almost all yeast genes, which was used as an indication that Spt3 function is partially redundant with that of TFIID. Different gene features were compared in these two gene classes leading to the conclusion that SAGA-dominated promoters tend to have consensus TATA box, are more stress regulated/inducible genes and tend to be more tightly regulated (Basehoar et al., 2004). As a general model, it was proposed that TBP recruitment is primarily dependent on SAGA at TATA-containing promoters, but dominated by TFIID at the TATA-like (or TATA-less) promoters (reviewed in (Tora and Timmers, 2010)).
Further studies reported different chromatin organization at the two classes of genes accounting for the higher plasticity in expression of SAGA-dominated genes (Rhee and Pugh, 2012; Tirosh and Barkai, 2008). The TFIID-dominated genes, which have less expression variability, have a large nucleosome depleted region just upstream of the transcription start site (TSS) and well-positioned flanking nucleosomes, which may play a role in PIC assembly. The presence of a consensus TATA-box is more important for PIC assembly at SAGA-dominated promoters, where less well-positioned nucleosomes appear to compete with transcription factors. A more recent study systematically analyzed transcriptional changes upon deletion of all non-essential SAGA subunits (Lenstra et al., 2011). This analysis suggested that even fewer genes were SAGA-dependent as steady state mRNA levels for only ~150 genes were either up- or down-regulated in a spt3Δ strain. Surprisingly, when compared with genome-wide localization of the same SAGA subunits (Venters et al., 2011), very little correlation was found between SAGA location and SAGA-dominated genes (Lenstra and Holstege, 2012).
Recent observations provide putative explanations to the seemingly conflicting findings on SAGA location and transcriptional effects. Several studies revealed that mRNA buffering through widespread mRNA stabilization is a common response to a global reduction of Pol II transcription (Haimovich et al., 2013; Plaschka et al., 2015; Rodriguez-Molina et al., 2016; Sun et al., 2012). As the steady-state mRNA levels are the net result of both mRNA synthesis and decay, the quantification of total RNA may not appropriately reflect mRNA synthesis rates upon global changes of Pol II transcription. The recruitment of yeast SAGA at a subset of upstream activating sequences (UASs), as determined by ChIP (van Werven et al., 2008; Venters et al., 2011), contrasted with a broad distribution of the HAT or DUB activities of SAGA either acetylating the promoter or deubiquitinating the transcribed region of almost all Pol II transcribed genes (Bonnet et al., 2014). This discrepancy could be explained by an inherent difficulty to ChIP the SAGA complex (HTM. Timmers, unpublished observations), which can be due to its very dynamic interaction with chromatin (Vosnakis et al., 2017).
To understand these seemingly contradictory observations, we assessed the genome-wide localization of SAGA and its role in transcription using different and independent methodologies, which are not affected by the potential biases indicated above. We show here that SAGA is recruited to the UASs at a majority of yeast genes, similarly to Mediator. In good agreement, SAGA was found to be required for RNA synthesis of essentially all genes transcribed by Pol II. We observed a compensatory increase of the half-life of a majority of mRNAs upon SAGA depletion explaining the limited changes in steady-state mRNA levels in the different SAGA mutant strains. We further analyzed the relative contributions of the different SAGA activities to Pol II transcription, thereby revealing a synergistic role for Spt3 and the acetyltransferase Gcn5. Our data lead us to propose that SAGA is a general cofactor for PIC recruitment that is required for transcription of the vast majority of Pol II genes.
Results
SAGA is recruited to the regulatory regions of both SAGA- and TFIID-dominated genes in yeast
While a gene-specific function for SAGA has been previously reported (Basehoar et al., 2004; Huisinga and Pugh, 2004), the genome-wide localization of SAGA remains unclear (Robert et al., 2004; van Werven et al., 2008; Venters et al., 2011). Here we used chromatin endogenous cleavage coupled with high-throughput sequencing (ChEC-seq), a method orthogonal to ChIP-seq, to generate a high-resolution genome-wide binding pattern for SAGA in S. cerevisiae (Grunberg et al., 2016; Grunberg and Zentner, 2017; Zentner et al., 2015). Micrococcal nuclease (MNase) was C-terminally fused to four SAGA specific subunits: Spt7 is required for the structural integrity and thus functions of SAGA; Spt3 and Spt8 interact with TBP and have been suggested to deposit TBP at promoters of SAGA-dominated genes (Bhaumik and Green, 2001, 2002; Dudley et al., 1999; Laprade et al., 2007; Larschan and Winston, 2001; Mohibullah and Hahn, 2008); and Ubp8, the catalytic subunit of the SAGA deubiquitination module. Fusion of the MNase to the respective SAGA subunits did not result in a growth phenotype under the conditions tested in this study, suggesting that the tagging did not affect SAGA function (data not shown).
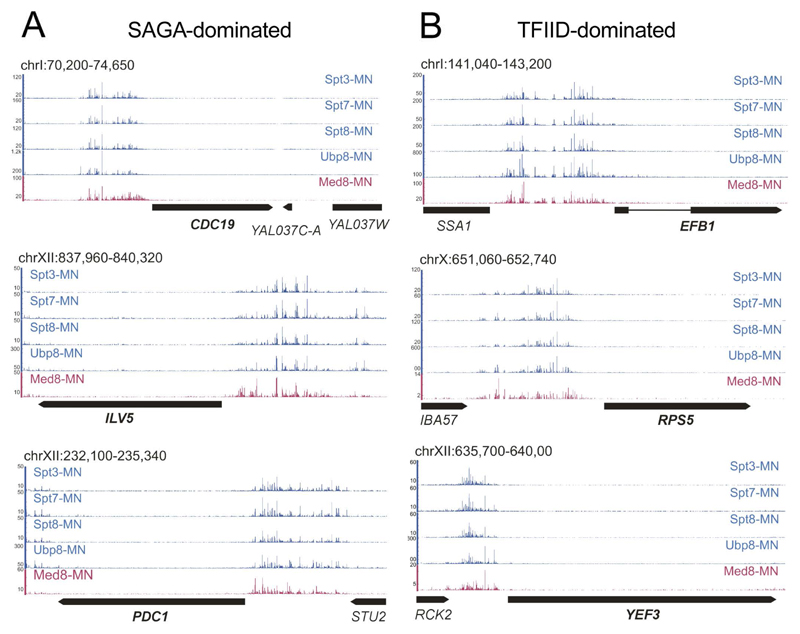
Permeabilized cells were treated with calcium to stimulate MNase-activity and promote DNA cleavage in proximity of the tagged SAGA-subunits. The cleaved fragment ends were sequenced and mapped to the budding yeast genome (Figures 1 and S1). Recent ChEC-seq mapping of Mediator, as exemplified by the Med8 subunit, was used as a reference for a co-activator complex associated with UASs genome-wide (Grunberg et al., 2016). As the cleavage patterns were almost identical after 5 and 15 min of MNase activation (ρ = 0.9938; Figure S1A), only the 5 min samples were used in the following analyses. We found strong SAGA-MNase dependent cleavages upstream of an average of ~ 2700 genes, showing little preference for genes previously characterized as SAGA- or TFIID-dominated (61% of SAGA- and 49% of TFIID-dominated genes were _targeted by SAGA (Xu et al., 2009)). At three exemplary highly expressed SAGA-dominated (CDC19, ILV5 and PDC1) (Figure 1, left panels) and three highly expressed TFIID-dominated genes (EFB1, RPS5 and YEF3) (Figure 1, right panels), we observed strong DNA cleavage upstream of the TSSs, with negligible background signal in the coding region using all four MNase fusion proteins. Enrichment of specific cleavages was dependent on fusion of MNase to SAGA subunits, as it was not observed in a strain expressing untethered MNase under control of the Spt3 promoter after 5 and 15 min incubation (Figure S1A).
Figure 1. ChEC-seq profiling of SAGA specific subunits.
Signal tracks showing cleavages induced by Spt3-MNase, Spt7-MNase, Spt8-MNase and Ubp8-MNase at (A) three representative SAGA-dominated genes (CDC19, ILV5 and PDC1) and (B) three TFID-dominated genes (EFB1, RPS5 and YEF3). Med8-MNase cleavage sites are shown as a reference for the coactivator Mediator binding at UASs (Grunberg et al., 2016). See also Figure S1.
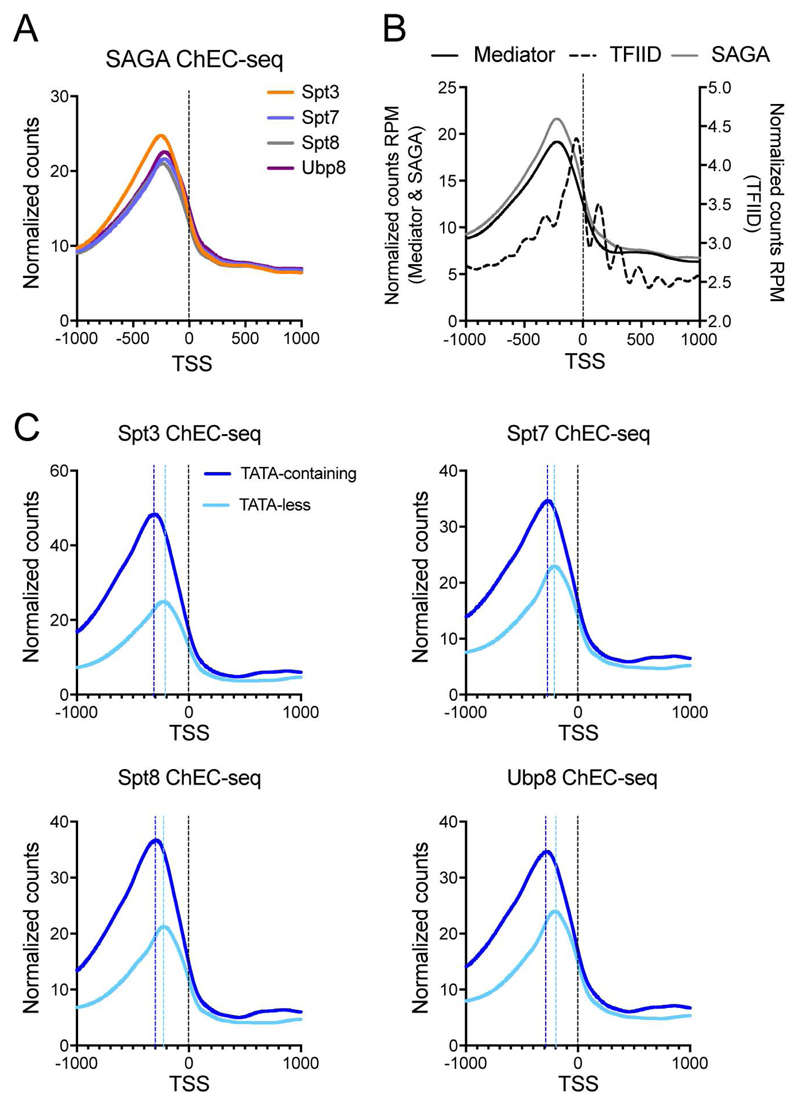
We next analyzed the localization of SAGA relative to the TSSs. The average profiles for the four SAGA subunits were almost identical, suggesting that these analyses reveal the location of the whole SAGA complex (Figure 2A). This was confirmed by pairwise correlations of genes _targeted by SAGA-MNase subunits tested, resulting in Spearman’s rank correlation coefficients of ρ > 0.96 (Figure S1B). When we compared the plots for SAGA (Spt3-MNase) and our previous ChEC-seq data obtained for the Mediator (Med8-MNase) and TFIID (Taf1-MNase) complexes, we found a striking coincidence between SAGA and Mediator position at UASs (Figure 2B) (Grunberg et al., 2016). In agreement with previous ChIP and ChEC-seq data, the peak summit of average SAGA cleavages relative to the TSS at TATA-containing genes was generally more distal (average 286 bp) than at TATA-less genes (average 217 bp) (Grunberg et al., 2016; van Werven et al., 2008; Venters et al., 2011)(Figure 2C). In addition, we found that > 99% of genes bound by SAGA were also _targeted by TFIID (Figure S1C) (Grünberg et al., 2016).
Figure 2. SAGA associates with UASs of SAGA- and TFIID-dominated genes.
(A) Plots of average SAGA subunit cleavages relative to the TSSs of all annotated Pol II genes. (B) Plots of average SAGA (Spt3-MNase, grey line), Mediator (Med8-MNase, black line), and TFIID (Taf1-MNase, dotted line) cleavage at the TSSs of all genes transcribed by Pol II. (C) Plots of average cleavage of the single subunits at TATA-containing (dark blue line) or TATA-less (light blue line) gene promoters. The dotted lines represent the peak summit to TSS distance at TATA-containing (dark blue) or TATA-less (light blue) genes. See also Figure S1.
Previous mapping of Mediator and expression analysis suggest that occupancy of factors is broadly uncoupled from active gene expression (Grunberg et al., 2016; Lenstra et al., 2011). Like Mediator, levels of SAGA-directed DNA cleavage were higher at UASs of SAGA-dominated genes compared to TFIID-dominated genes, which are generally expressed at significantly higher levels (Churchman and Weissman, 2011). Our data indicate that SAGA, like Mediator, binds to the UASs of many genes, independent of gene classification, and broadly uncoupled from gene activation.
Global down-regulation of Pol II transcription upon deletion of SAGA structural subunits
The global recruitment of SAGA at the UASs of many Pol II transcribed genes is in good agreement with the previously reported decrease of Pol II recruitment at most gene promoters in spt20Δ yeast strains (Bonnet et al., 2014). To determine whether SAGA acts as a general cofactor for Pol II, we compared the steady-state and newly-synthesized mRNA levels in SAGA mutant strains at all genes transcribed by Pol II. Wild-type, spt7Δ and spt20Δ strains cultured in rich medium, were pulse labeled with 4-thiouracil (4tU) and mixed in a fixed ratio with labeled S. pombe cells for normalization. Purified total RNA (steady-state) and labeled RNA (newly-synthesized) were hybridized to Affymetrix microarrays containing probes for S. cerevisiae and S. pombe transcripts.
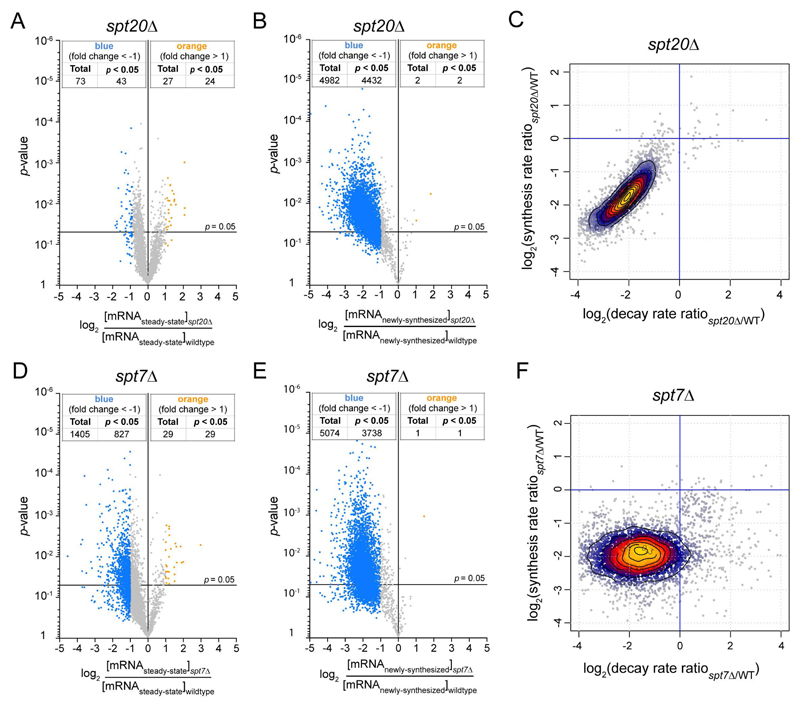
When applying a 2-fold change threshold and a p-value of 0.05 after normalization to the S. pombe spiked-in signal, significant changes for only a very limited number of genes were observed in steady-state mRNA levels in the spt20Δ strain (43 genes down-regulated and 24 genes up-regulated) (Figure 3A). These data are reminiscent of earlier studies suggesting that SAGA regulates a limited number of genes (Lenstra et al., 2011). Importantly, genes with altered steady-state mRNA levels in the SAGA mutant do not constitute a clearly discrete class of genes based on their expression profile, but appear more as the outliers in a Gaussian distribution of gene expression fold changes (Figure 3A). In contrast, the comparison of newly-synthesized mRNA levels between spt20Δ and wild-type strains revealed a decrease in mRNA synthesis for a vast majority of genes (4432 genes down-regulated and 2 genes upregulated) (Figure 3B). Strikingly different profiles were also observed when comparing levels of total or labeled RNA purified from the spt7Δ strain (Figures 3D and 3E). Consistently, the number of down-regulated genes and the extent of down-regulation dramatically increased when analyzing newly-synthesized mRNA as compared to steady-state mRNA indicating a global decrease of Pol II transcription in the spt20Δ and spt7Δ strains.
Figure 3. Compensation of an overall decrease of Pol II transcription by a global change in mRNA decay in SPT20 and SPT7 deletion strains.
Volcano plots showing fold changes in steady-state mRNA levels (A,D) or newly-synthesized mRNA levels (B,E) relative to their significance (p value). Fold changes (FC) were calculated as the Log2 of the ratio of the expression value of each gene after normalization to S. pombe signal in the spt20Δ strain (A,B) or the spt7Δ (D,E) strain versus the expression value of the same gene in wild-type S. cerevisiae. 5385 genes were analyzed and thresholds of 2-fold change (blue dots: more than 2-fold decrease; yellow dots: more than 2-fold increase) and 0.05 p values were considered. cDTA profiles for spt20Δ (C) and spt7Δ (F) strains. For all analyzed genes, changes in synthesis rates were plotted against the changes in mRNA decay rates. Changes were calculated as the Log2 of the ratio between mutant and wild-type. 90% of genes are contained within the outer contour. Yellow and red dots correspond to 60% of genes. For each strain, results were obtained from at least two independent biological replicates. See also Figure S2.
mRNA levels are buffered by increased mRNA half-life in SAGA mutant strains
As the analysis of newly-synthesized mRNA quantifies mRNA synthesis uncoupled from RNA degradation, the very limited changes in steady-state mRNA levels might be explained by compensatory changes in global mRNA decay. To address this hypothesis, we used comparative Dynamic Transcriptome Analysis (cDTA) (Sun et al., 2012), which allows the calculation of mRNA synthesis and decay rates for all S. cerevisiae coding genes. In spt20Δ or spt7Δ strains, we observed a concomitant decrease in both mRNA synthesis and decay rates, when compared to the wild-type strain (Figures 3C and 3F). This compensation was almost complete in the spt20Δ strain with a mean decrease in synthesis by about 3.8-fold and in decay by about 4.1-fold, which accounts for the small number of changes in steady-state mRNA (Figures 3A and 3C). In the spt7Δ strain, the decrease in mRNA synthesis was very similar, but the changes in decay rates were more dispersed (Figure 3F). As a result of an imperfect compensation, the steady-state mRNA levels of a higher number of genes were modified in this mutant strain (Figure 3D).
To verify this global mRNA stabilization using an independent method, we compared mRNA decay following transcription inhibition with thiolutin in wild-type and SAGA mutant strains. In the wild-type strain, the calculated mRNA half-lives were very similar to those previously reported (Geisberg et al., 2014). In good agreement with our cDTA measurements, the mRNA half-lives of five randomly selected genes increased (1.5- to 4-fold) in the spt7Δ and spt20Δ strains (Figure S2). As expected for a gene transcribed by Pol III and not affected by SAGA inactivation, there was no significant change of the long-lived scR1 RNA (Figure S2). Our results show that yeast cells buffer a global decrease in Pol II transcription following SAGA depletion by increasing mRNA half-lives, as previously reported for mutations in Pol II and Mediator, or inhibition of the kinase activity in TFIIH (Plaschka et al., 2015; Rodriguez-Molina et al., 2016; Sun et al., 2012).
SAGA equally contributes to the expression of SAGA- and TFIID-dominated genes
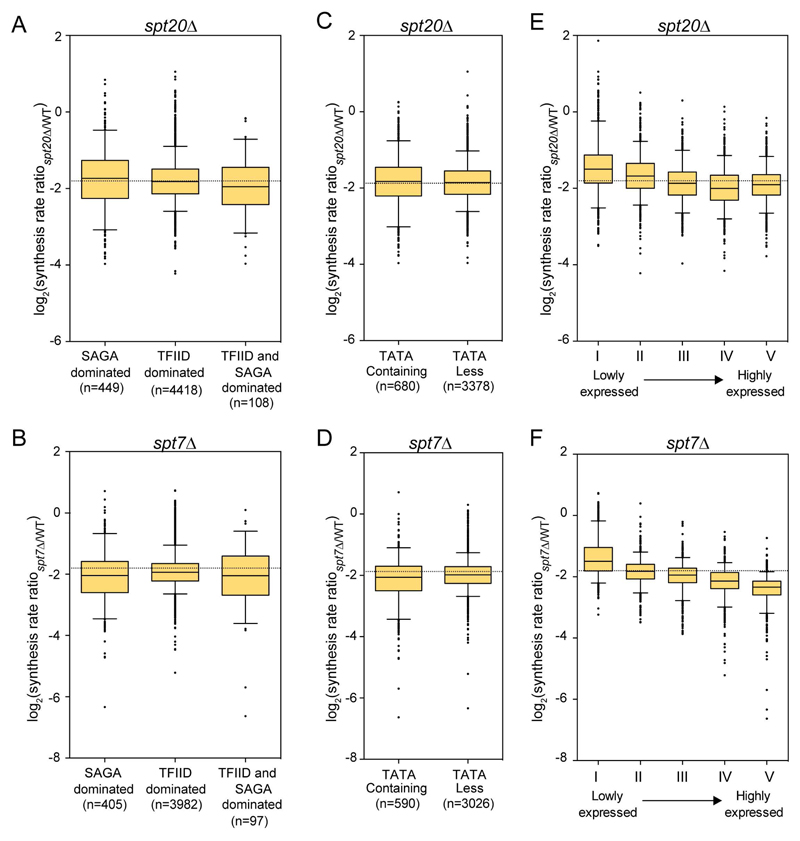
Our ChEC-seq analysis revealed that the cleavage levels at TATA-containing (or SAGA-dominated) genes were significantly higher than observed at TATA-less (or TFIID-dominated) genes (Figure 2C). We therefore asked whether the expression of these two classes of genes would be differentially controlled by SAGA. For each gene category, changes in synthesis rates between mutant and wild-type strains were plotted and their distributions in the different gene classes were compared. Inactivation of the SAGA complex through deletion of either SPT20 or SPT7 induced a similar decrease in Pol II transcription for either the SAGA-or the TFIID-dominated genes (Figures 4A and 4B). Similarly, no difference was observed depending on whether or not these genes contain a TATA box in their promoters (Figures 4C and 4D). We did, however, find that the most highly expressed genes in wild-type cells showed the strongest decrease in mRNA synthesis in both SPT20 and SPT7 deleted strains (Figures 4E and 4F).
Figure 4. Changes in mRNA synthesis rates for different classes of genes in spt20Δ and spt7Δ.
Box plots showing the distribution of changes in mRNA synthesis rates between spt20Δ (A,C,E) or spt7Δ (B,D,F) and wild-type strains. Very similar changes were seen whether genes were described as SAGA- or TFIID-dominated (A,B) or whether their promoter were TATA-containing or TATA-less (C,D). Genes were divided in quintiles according to their expression levels in wild-type cells and the changes in synthesis rates were plotted for each quintile (E,F). Boxes contain genes between the 25th and the 75th centiles, the line indicates to the median and the whiskers correspond to 5th and 95th centiles.
The global decrease of Pol II transcription in SAGA mutants is a primary event
Growth of S. cerevisiae strains deleted for SPT20 or SPT7 is impaired in rich medium, which might cause indirect effects on transcription. Indeed, slow growth of S. cerevisiae deletion strains has been linked to altered cell cycle distribution leading to a common gene expression signature (O'Duibhir et al., 2014). Although our observations of a global decrease in Pol II transcription did not reproduce this signature, a conditional depletion strategy was used to rule out any effect due to the slow growth of the SAGA mutant strains. We used the anchor-away strategy to deplete C-terminally FRB-tagged Spt7 from the nucleus by addition of rapamycin in a strain expressing RPL13A-FKBP12 (Haruki et al., 2008). Nuclear-cytoplasmic fractionation of cells treated or untreated with rapamycin for 30 min demonstrated an efficient nuclear depletion of the fused Spt7 protein, in agreement with the delayed growth of the SPT7-FRB strain (Figures S3A and S3B). Viability of the SPT7-FRB strain was not affected after 30 or 60 min of rapamycin treatment (Figure S3C). Total and newly-synthesized mRNA levels of all tested genes were unchanged between the parental and the SPT7-FRB strain, indicating that the FRB fusion by itself did not induce any detectable phenotype (Figures S3D and S3E).
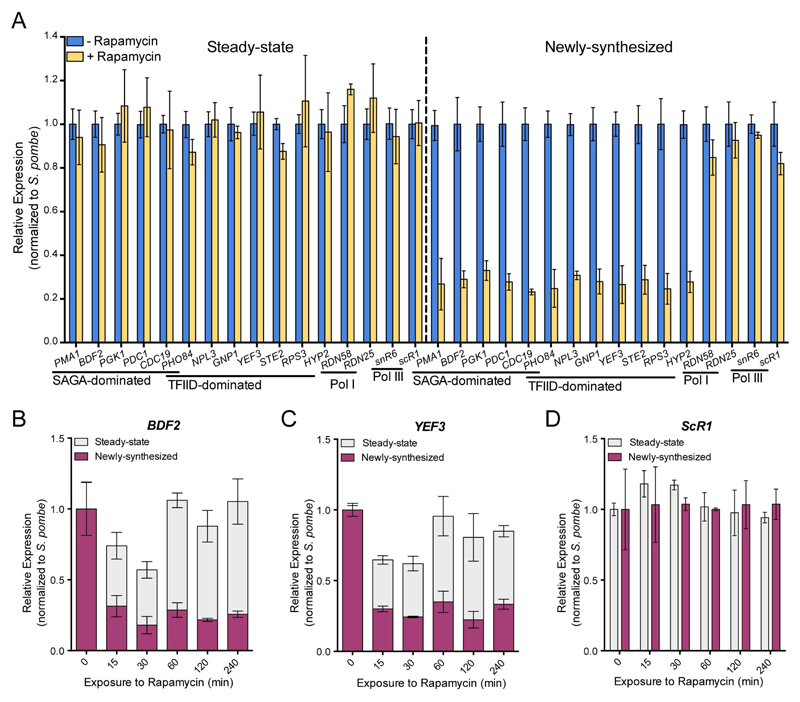
To determine the effects of Spt7 depletion on transcription, we quantified steady-state and newly-synthesized mRNA for a number of selected genes after 60 min of rapamycin treatment and a pulse of RNA labeling using 4tU. Total and labeled RNA were analyzed by RT-qPCR and mRNA levels for both fractions were normalized to the spiked-in S. pombe signal. While the steady-state mRNA levels did not change significantly, the newly-synthesized mRNA levels of all tested genes were reduced by 3- to 4-fold in the rapamycin treated cells when compared to non-treated cells (Figure 5A). The range of decrease in mRNA synthesis was very similar amongst the five SAGA- and the six TFIID-dominated genes tested. The RNA levels of genes transcribed by Pol I or Pol III were not significantly modified upon nuclear depletion of Spt7 (Figure 5A). Accordingly with the almost complete nuclear depletion of Spt7, the extent of gene expression changes highly resembles that observed in the spt7Δ strain.
Figure 5. Conditional nuclear depletion of Spt7 decreased transcription of both SAGA- and TFIID-dominated genes.
(A) Spt7 anchor away strain, untreated or treated with rapamycin for 60 min, were labeled with 4tU. mRNA levels from 6 SAGA- , 6 TFIID-dominated genes and RNA levels from four control genes transcribed by Pol I and Pol III were quantified by RT-qPCR. Expression values (mean ± SD of three independent experiments) were normalized to spiked-in S. pombe signal and set to 1 in the untreated sample. (B-D) Time course analysis of changes in steady-state and newly-synthesized RNA for a SAGA-dominated (B), a TFIID-dominated (C) and a control gene transcribed by Pol III (D) upon Spt7 nuclear depletion. See also Figures S3 and S4.
To better appreciate how Spt7 nuclear depletion affects transcription, we compared steady-state and newly-synthesized mRNA levels for a number of selected genes in a time course experiment. Cells were exposed to rapamycin for different periods (0 to 240 min) and labeled with 4tU for 6 min. Quantification of labeled RNA revealed that mRNA synthesis was strongly reduced as early as 15 min following induction of Spt7 depletion and remained stable over time (Figures 5B, 5C and S4). Interestingly, steady-state mRNA levels of the same genes were slightly reduced at early time points (15 and 30 min) and they returned to normal levels at later time points. The observed profiles are best explained by an early decrease in mRNA synthesis, whereas efficient buffering of mRNA levels would occur at later time points (from 60 min on). The RNA levels of scR1, a gene transcribed by Pol III were unchanged in the total and labeled RNA fractions during the same time-course experiment (Figure 5D). Thus, our observations indicate that the global down-regulation of Pol II transcription upon SAGA inactivation is a primary event, which cannot be explained by indirect effects of SPT7 or SPT20 deletions, and which seems to be a direct consequence of SAGA acting at the promoters of genes.
Activities of the SAGA complex required for Pol II transcription
The budding yeast SAGA complex is composed of 19 different subunits, which are organized in five functional and structural modules (Han et al., 2014; Lee et al., 2011; Setiaputra et al., 2015). To better understand the role of SAGA in global mRNA synthesis as revealed by the analysis of strains deleted for SAGA structural subunits, we wanted to determine which individual activities of SAGA may play a role in Pol II transcription and what are their relative contributions. The most obvious candidates are the enzymatic activities of SAGA, which are known to regulate histone marks associated with active transcription, as well as subunits proposed to recruit TBP at promoters (Koutelou et al., 2010). We thus focused our analysis on Gcn5, the catalytic subunit of the HAT module, Ubp8, the catalytic subunit of the DUB module, and the TBP-interactors Spt3 and Spt8 (Dudley et al., 1999; Laprade et al., 2007; Larschan and Winston, 2001; Mohibullah and Hahn, 2008). Global levels of newly-synthesized mRNA were significantly decreased (by an average 1.5-fold) in the gcn5Δ strain suggesting that histone acetylation favors transcription initiation (Figures 6A and 6E). UBP8 deletion did not affect mRNA synthesis suggesting that increased H2Bub levels do not affect Pol II transcription by themselves (Figures 6E and S5). From the two subunits interacting with TBP, only Spt3 appeared to play a significant role in Pol II transcription as the newly-synthesized mRNAs were reduced by about 2-fold in the spt3Δ, and they were unaltered in the spt8Δ strain (Figures 6B and S5).
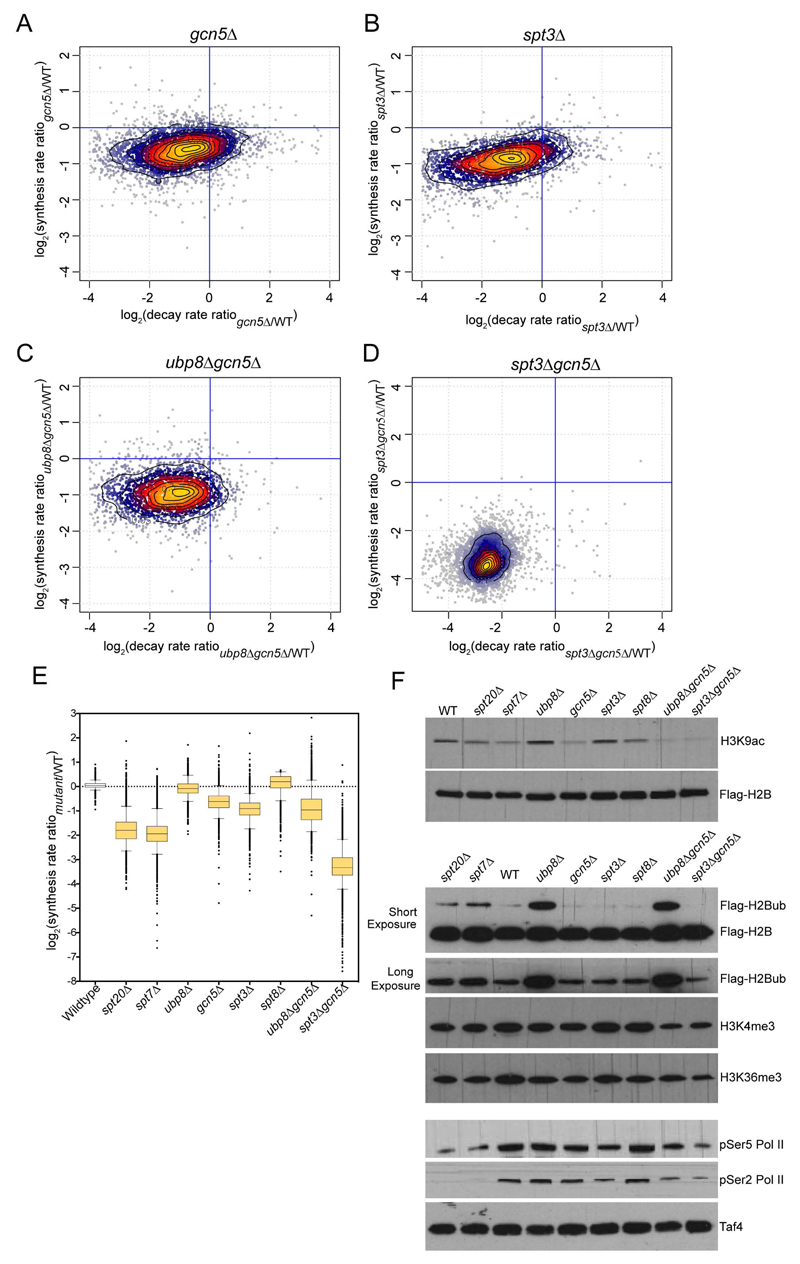
Figure 6. cDTA analyses of different SAGA subunit deletion strains.
Synthesis rates and decay rates were determined for each S. cerevisiae gene in gcn5Δ (A), spt3Δ (B), ubp8Δgcn5Δ (C) and spt3Δgcn5Δ (D). Changes (calculated as the Log2 of the ratio between mutant and wild-type) in synthesis rates were plotted against changes in decay rates. (E) Box plots summarizing the extent of changes in mRNA synthesis for all the analyzed deletion strains. For each strain, cDTA data were obtained from at least two independent biological replicates. (F) Whole cell extracts from wild-type and the indicated deletion strains were revealed with the antibodies corresponding to histones marks regulated by SAGA (H3K9ac, H2Bub) or associated with active transcription (H3K4me3, H3K36me3) or with antibodies specific to the C-terminal domain of Rpb1 phosphorylated on serine 5 (pSer5 RNA Pol II) or on serine 3 (pSer5 RNA Pol II), as indicated. See also Figure S5, S6 and S7.
To recapitulate the transcriptional effects seen upon depletion of SAGA structural subunits (Spt7 and Spt20), budding yeast strains deleted for two SAGA-specific subunits, which are located in distinct modules, were generated. Surprisingly, mRNA synthesis was more affected in the double ubp8Δgcn5Δ mutant (about 2-fold average decrease) than in the single GCN5-deleted strain (Figures 6C and 6E). Most importantly, the cumulative inactivation of TBP-interacting and HAT activities in the spt3Δgcn5Δ strain induced a dramatic decrease in mRNA synthesis by on average 10-fold (Figures 6D, 6E and S5) without affecting the overall SAGA assembly (Figure S6). This global loss in Pol II transcription was larger than the cumulative effect of SPT3 and GCN5 deletion and even greater than deletion of the structural subunits. Altogether, these results point to an important role for TBP-interacting through Spt3 and also for Gcn5 either through acetylation of histones, binding to acetylated histones through the Gcn5 bromodomain, or an alternative mechanism. To further support that these global transcriptional effects are directly related to SAGA function, we compared all our datasets with the previously reported “slow growth gene expression signature” (O'Duibhir et al., 2014) and did not observe any relevant correlation (Figure S7).
For all deletion strains used in this study, we analyzed the global levels of histone marks directly regulated by SAGA (H3K9ac and H2Bub) or associated with active transcription (H3K4me3 and H3K36me3). As expected, H3K9 acetylation strongly decreased upon GCN5 deletion and to a lesser extent upon depletion of SAGA structural subunits (Figure 6F). As previously reported, H2Bub levels slightly increased upon SPT7 or SPT20 deletion and they were dramatically increased in either the ubp8Δ or the ubp8Δgcn5Δ strains (Figure 6F) (Henry et al., 2003). Less pronounced changes were observed for marks enriched at active promoters (H3K4me3) or at transcribed gene regions (H3K36me3), which correlated with global gene expression changes in the corresponding strains (Figure 6F). In addition, the levels of serine 5 and serine 2 phosphorylation on the C-terminal domain of the largest subunit of Pol II (Rpb1) were slightly reduced in gcn5Δ, spt3Δ and ubp8Δgcn5Δ and strongly decreased in spt3Δgcn5Δ, spt7Δ and spt20Δ strains (Figure 6F). Importantly, these analyses revealed a strong correlation of alterations in Pol II transcription with Rpb1 phosphorylation levels.
Role of SAGA in the genome-wide recruitment of TBP
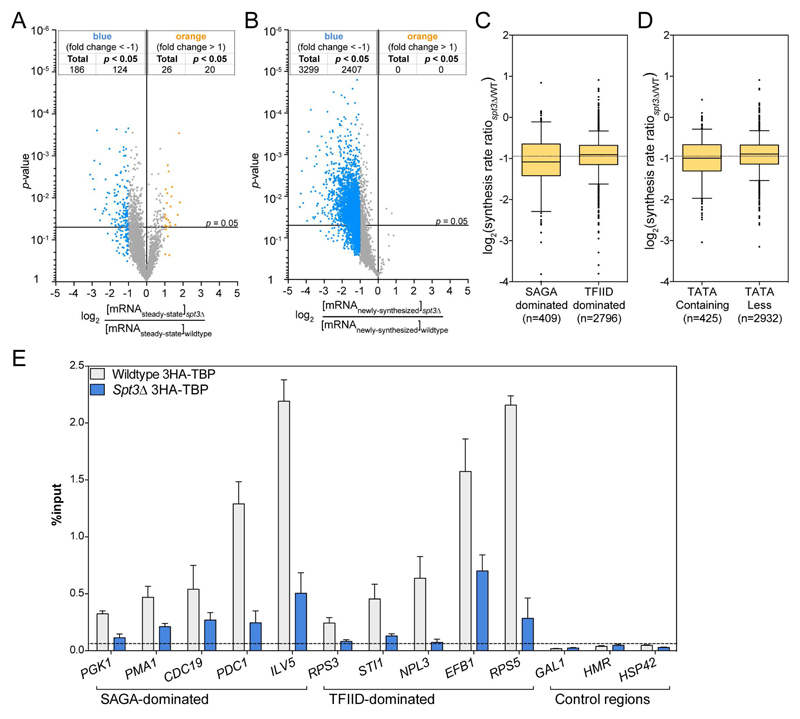
The distinction of the two proposed classes of SAGA-dominated and TFIID-dominated genes was substantiated by the idea that each complex can independently recruit TBP to the corresponding promoters. In line with this hypothesis, it is predicted that in a SPT3-deleted strain, TBP recruitment should be more affected at the SAGA-dominated genes compared to recruitment at the TFIID-dominated genes. Analysis of newly-synthesized RNA in a spt3Δ strain revealed that mRNA synthesis was significantly reduced for a majority of genes with an average 2-fold decrease in mRNA levels, whereas very few changes were detected in steady-state mRNA levels (Figures 7A and 7B). The changes in synthesis rates between spt3Δ and wild-type strains did not discriminate between the different classes of genes (SAGA- versus TFIID-dominated; TATA-containing versus TATA-less promoters) (Figures 7C and 7D). We next investigated whether the SAGA requirement at these different genes was due to different mechanisms. TBP recruitment at selected genes was measured by ChIP in 3HA-TBP and spt3Δ 3HA-TBP strains. At the promoters from four SAGA- and five TFIID-dominated genes, TBP recruitment was similarly reduced upon SPT3 deletion, whereas only background levels could be detected at control regions (Figure 7E). These results suggest that SAGA plays an important role in TBP recruitment or stabilization and subsequent Pol II recruitment at almost all active promoters and that this function is independent of the promoter architecture.
Figure 7. Reduced TBP occupancy at promoters of both SAGA- and TFIID-dominated genes in SPT3 deletion strain.
(A,B) Volcano plots showing changes in steady-state (A) and newly-synthesized mRNA levels between spt3Δ and wild type S. cerevisiae. (C,D) Box plots representing the distribution of changes in synthesis rates upon SPT3 deletion for SAGA- versus TFIID-dominated genes (C) and for TATA-containing versus TATA-less genes (D). (E) TBP enrichment at promoters from 5 SAGA- and 6 TFIID-dominated genes as well as at three control regions was determined by HA ChIP in a spt3Δ 3HA-TBP strain and the parental 3HA-TBP strain and quantified by real time PCR. The values (mean ± SD of three independent ChIP experiments) are expressed as percentage of input DNA signal.
Discussion
In this study, we provide a combination of complementary results indicating that SAGA is recruited at most Pol II transcribed genes in S. cerevisiae, where it is plays a critical role in mRNA synthesis. In striking contrast to the current model for SAGA function, in which SAGA predominantly regulates the expression of only a limited subset of genes, we propose that SAGA, like Mediator, acts as a general co-factor for Pol II transcription. Previous observations suggesting a global role for SAGA in Pol II transcription were: (i) SAGA-dependent changes in H3K9 acetylation revealed that SAGA, through its Gcn5 subunit, acetylates most active promoters; (ii) SAGA is required for Pol II recruitment at a vast majority of active promoters (Bonnet et al., 2014). This conclusion is coherently supported by our new findings including: (i) direct localization studies by ChEC-seq indicate recruitment of SAGA to UASs of genes previously categorized as SAGA- or TFIID-dominated; (ii) all genes bound by SAGA are also _targeted by TFIID; (iii) SAGA is required for TBP recruitment at active promoters of both SAGA and TFIID-dominated genes; and (iv) disruption of the SAGA complex led to a global decrease in mRNA synthesis.
Steady-state mRNA levels do not accurately reflect SAGA activity in Pol II transcription
Earlier studies reporting a gene-specific activity for the SAGA complex were based on the evaluation of steady-state mRNA levels upon deletion of SAGA subunits (Huisinga and Pugh, 2004; Lenstra et al., 2011). By analyzing newly-transcribed RNA, we provide evidence that these studies underestimated the role of SAGA on Pol II transcription. In addition, comparison of steady-state with newly-synthetized mRNA revealed that major and general perturbations of Pol II transcription can lead to a compensatory mechanism, as reported for other coactivator complexes such as Mediator (Plaschka et al., 2015). This compensation emerges as a way to cope with an abrupt decrease in RNA synthesis that would ultimately lead to a decrease of cellular mRNA. This mechanism is also supported by the increased half-life of mRNAs in Spt7- and Spt20-depleted cells resulting in longer-lived and more stable transcripts. Hence, through a mechanism that remains elusive, a simultaneous decrease in mRNA decay ultimately leads to virtually unchanged RNA levels (Haimovich et al., 2013; Sun et al., 2013). A prime candidate for this would be the CCR4-NOT complex, which is a global regulator of mRNA decay and has been linked to the TFIID, SAGA and Mediator complexes by genetic means (Collart and Timmers, 2004; Villanyi and Collart, 2015). Importantly, our experiments indicate that the existence of this compensatory mechanism has occluded the full spectrum of SAGA action and that this co-activator has a critical role in optimal Pol II transcription.
SAGA activities act in a synergistic manner
SAGA is a multifunctional complex with at least five activities: HAT, DUB, TBP-interacting, activator binding and nucleosome binding [reviewed in (Koutelou et al., 2010; Rodriguez-Navarro, 2009; Weake and Workman, 2012)]. As the depletion of different SAGA subunits affected a distinct and relatively small subset of genes, it was suggested that each SAGA subunit makes specific and unique contributions to the function of the complete complex (Lee et al., 2000). Further gene expression analyses of steady-state RNA showed that subunits of a same module share similar transcriptional effects, but each SAGA activity regulated a different set of genes (Lenstra et al., 2011). From the twelve SAGA-specific subunits, we measured the transcriptional effects of four subunits affecting the HAT, the DUB or TBP-interacting activities as well as two structural subunits. Except for SPT8 and UBP8, we observed a global effect on Pol II transcription in all other mutants, suggesting that most SAGA activities have a general regulatory function at all genes. The decrease in global mRNA synthesis observed in a GCN5 and SPT3 double deletion strain was much higher than the sum of any single mutant. Therefore, these subunits having distinct functions (histone acetylation and TBP-interacting) and found in different locations within SAGA (Setiaputra et al., 2015) act in a synergistic manner on Pol II transcription. This underscores that different activities, which might appear as relatively independent (HAT, DUB and TBP-interacting) are joined within a single complex to coordinate their action on Pol II transcription.
Surprisingly, the transcriptional effects seen in the spt3Δgcn5Δ strain were even more severe than that observed upon deletion of SAGA structural subunits, which were expected to compromise multiple, if not all SAGA functions (Grant et al., 1997; Roberts and Winston, 1997; Sterner et al., 1999). However, recent studies demonstrated that upon depletion of these structural subunits, smaller assemblies or sub-complexes still form (Lee et al., 2011). This is supported by the milder increase of H2Bub levels in spt7Δ or spt20Δ when compared with ubp8Δ, which suggest the formation of a partially active DUB module in the absence of Spt7 or Spt20. Therefore, in all mutant strains analyzed, residual activities of such sub-complexes might fulfill some functions of the whole SAGA complex, although we cannot exclude that they may have negative effects on transcription through sequestration of PIC components. Interestingly, systematic proteomic analyses of deletion strains and our co-immunoprecipitation experiments predict that a SAGA complex lacking only Spt3 and Gcn5 would be assembled in the spt3Δgcn5Δ strain (Lee et al., 2011) (Figure S6). Although this incomplete SAGA complex might retain some activities of the wild type SAGA, mRNA synthesis was dramatically reduced in this strain (by an average 10-fold). This is in agreement with the slower growth of this double-mutant strain when compared to spt20Δ cells (Sterner et al., 1999). Hence, further suppression of all SAGA activities may lead to an almost complete suppression of Pol II transcription and cell lethality.
A minor reduction of mRNA synthesis was observed upon deletion of GCN5, which is known to be a subunit of both the SAGA and the ADA complexes. The ADA complex is composed of the four same subunits as the SAGA HAT module with two additional ADA-specific subunits, Ahc1 and Ahc2 (Eberharter et al., 1999; Lee et al., 2011). Depletion of Spt3 which is specific to SAGA on the gcn5Δ background had a much more dramatic effect on transcription with a more than 10-fold average reduction in mRNA synthesis. This suggests that the role of the ADA complex in Pol II transcription is less dominant than that of SAGA, in agreement with the idea that the different SAGA activities have synergistic roles in Pol II transcription. However, this does not discard the hypothesis that both complexes might act cooperatively and that loss of one of them (namely SAGA) potentiates a decreased activity of the other (ADA complex).
An alternative model for SAGA function in PIC assembly
Our study demonstrates that SAGA is recruited to the majority of active promoters where it has a comparable role in transcription independently of the type of gene (TFIID- versus SAGA-dominated; TATA-containing or TATA-less). In this respect, SAGA can be compared to Mediator, a co-activator complex that is required for all Pol II transcription and stimulates PIC formation. Importantly, ChEC-seq analyses revealed that Mediator binds to a majority of UASs clarifying ambiguous results obtained by ChIP-based methods (Grunberg et al., 2016). Widespread binding of Mediator is consistent with the global and dramatic decrease of mRNA synthesis observed upon the loss of Mediator subunits as demonstrated through the analysis of newly-synthesized RNA (Plaschka et al., 2015). The similarities in binding profiles and transcriptional effects of both SAGA and Mediator suggest that the two complexes participate in the assembly and/or stabilization of the PIC at all promoters and should be considered as general cofactors in budding yeast. In addition to the global function of SAGA and Mediator, TFIID was also found at essentially all promoters and was shown to be required for the transcription of both SAGA- or TFIID-dominated genes (Grunberg et al., 2016) (Warfield et al., accompanying manuscript). Therefore, the distinct functions of these three coactivators are needed for Pol II transcription initiation at almost all yeast promoters.
However, as SAGA can contact many different activators and uses several domains to interact with modified histones, SAGA recruitment and dynamics may vary at different gene promoters in yeast. In addition, the different activities of SAGA might have diverse outputs at each gene promoter depending of many additional factors such as promoter architecture or chromatin environment. Indeed, our analyses of various SAGA mutant strains revealed that, although mRNA synthesis of a vast majority of genes was affected, the extent of changes in Pol II activity varied across the genome. However, the distribution of mRNA synthesis changes appears to indicate a continuum of slightly different sensitivities to SAGA action, from which a distinct group of gene could not be identified. Other gene features such as the presence of a TATA-consensus or a TATA-like element allow a better distinction of gene classes characterized by different mechanisms of transcription regulation (Muller and Tora, 2014; Rando and Winston, 2012). Although SAGA had a comparable overall influence on transcription of these two gene classes, our ChEC-seq profiling revealed that SAGA is differently recruited at TATA-containing versus TATA-less genes. SAGA had a higher occupancy and was recruited more upstream relative to TSSs at TATA-containing genes when compared to TATA-less genes, a profile strikingly similar to that of Mediator (Grunberg et al., 2016). This is in agreement with previous studies indicating distinct chromatin structure and different transcriptional plasticity for these two promoter classes (de Jonge et al., 2017; Kubik et al., 2015; Tirosh et al., 2007). It remains to be determined if SAGA and Mediator participate have distinct functions in PIC formation at TATA-containing versus TATA-less promoters. Nevertheless, our data do not support the earlier distinction of two different pathways for PIC assembly with TBP delivery at promoters depending on either SAGA or TFIID. In contrary, our study together with that of the accompanying paper by the Hahn lab demonstrate that in yeast both TFIID and SAGA are required for all mRNA transcription by Pol II.
Star Methods
Key Resources Table
(see separate table).
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Flag M2 antibody | Sigma | Cat# F1804 |
| Rabbit polyclonal anti-H3K4me3 | Abcam | ab8580 |
| Rabbit polyclonal anti-H3K9ac | Abcam | ab4441 |
| Rabbit polyclonal anti-H3K36me3 | Abcam | ab9050 |
| Rat monoclonal anti-RNA Pol II CTD phosphoSer2 | Active Motif | Cat# 61083 |
| Rat monoclonal anti-RNA Pol II CTD phosphoSer5 | Active Motif | Cat# 61085 |
| Rabbit polyclonal anti-Taf4 | P.Anthony Weil | N/A |
| Rabbit polyclonal anti-Taf5 | P.Anthony Weil | N/A |
| Rabbit polyclonal anti-Taf6 | P.Anthony Weil | N/A |
| Rabbit polyclonal anti-Taf10 | P.Anthony Weil | N/A |
| Mouse monoclonal anti-Taf10 | Laszlo Tora | 2D5 |
| Rabbit polyclonal anti-Ada1 | Steven Hahn | 5255 |
| Rabbit polyclonal anti-Spt3 | Steven Hahn | 5113 |
| Rabbit polyclonal anti-Gcn5 | P.Anthony Weil | N/A |
| Rabbit polyclonal anti-Spt7 | Fred Winston | N/A |
| Rabbit polyclonal anti-H3 | Abcam | ab1791 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| cOmplete mini, EDTA-free Protease inhibitor cocktail | Roche | Cat# 11836170001 |
| 4-Thiouracil | Sigma-Aldrich | Cat# 440736 |
| Rapamycin | Euromedex | Cat# SYN-1185 |
| EZ-Link HPDP Biotin | ThermoFisher | Cat# 21341 |
| Thiolutin | Abcam | ab143556 |
| EZview Red anti-HA Affinity Gel | Sigma-Aldrich | Cat# E6779 |
| Protein A-Sepharose 4B, Fast flow | Sigma-Aldrich | Cat# P9424 |
| Critical Commercial Assays | ||
| RiboPure RNA Purification kit, yeast | ThermoFisher | Cat# AM1926 |
| µMACS Streptavidin kit | Miltenyi Biotec | Cat# 130-074-101 |
| Deposited Data | ||
| Raw and analyzed data (Microarrays) | This paper | GEO: GSE96849 |
| Raw and analyzed data (ChEC-seq) | This paper | GEO: GSE97379 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae. Strain background BY4705 (See Table S1) | Brachmann et al., 1998) | N/A |
| SPT3-3FLAGMNase, BY4705 (See Table S1) | This study | N/A |
| SPT7-3FLAGMNase, BY4705 (See Table S1) | This study | N/A |
| SPT8-3FLAGMNase, BY4705 (See Table S1) | This study | N/A |
| UBP8-3FLAGMNase, BY4705 (See Table S1) | This study | N/A |
| S. cerevisiae. Strain background FY406. (See Table S1) | Hirschhorn et al., 1995 | |
| WTyH2B, FY406 (See Table S1) | Bonnet et al., 2014 | N/A |
| ubp8Δ, FY406 (See Table S1) | Bonnet et al., 2014 | N/A |
| gcn5Δ, FY406 (See Table S1) | Bonnet et al., 2014 | N/A |
| spt7Δ, FY406 (See Table S1) | Bonnet et al., 2014 | N/A |
| spt20Δ, FY406 (See Table S1) | Bonnet et al., 2014 | N/A |
| ubp8Δgcn5Δ, FY406 (See Table S1) | This study | N/A |
| spt3Δ, FY406 (See Table S1) | This study | N/A |
| spt8Δ, FY406 (See Table S1) | This study | N/A |
| spt3Δgcn5Δ, FY406 (See Table S1) | This study | N/A |
| SPT7-FRB, BY4742 (See Table S1) | This study | N/A |
| S. cerevisiae. Strain background BY4742 (See Table S1) | Euroscarf | N/A |
| 3HA-TBP, BY4742 (See Table S1) | This study | N/A |
| spt3Δ, 3HA-TBP, BY4742 (See Table S1) | This study | N/A |
| Oligonucleotides | ||
| Primers for RT-qPCR, see Table S3 | This paper | This paper |
| Primers for ChIP-qPCR, see Table S3 | This paper | This paper |
| Recombinant DNA | ||
| pGZ108 (see Table S3) | Zentner et al., 2015 | N/A |
| pFA6a–hphNT1(see Table S3) | Janke et al., 2004 | N/A |
| pFA6a-kanMX6 (see Table S3) | Bähler et al., 1998 | N/A |
| YIplac211-3HA-TBP (see Table S3) | Eyboulet et al., 2015 | N/A |
| Software and Algorithms | ||
| GraphPad Prism, version 6.0 | GraphPad Software | https://www.graphpad.com/ |
| R | R Core Team (2013) | https://www.r-project.org/ |
| RStudio: Integrated Development for R | RStudio Team (2015) | https://www.rstudio.com/ |
| Other | ||
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead author, Didier Devys (devys@igbmc.fr).
Method Details
Yeast strains
Yeast strains for ChEC-seq were generated in the BY4705 background (Brachmann et al., 1998). SAGA subunits were endogenously tagged with 3xFLAG-MNase by transformation with gene-specific PCR products derived from pGZ108 (Zentner et al., 2015). Different yeast strains used in this study were also derived from the previously described FY406 (Hirschhorn et al., 1995). Deletion mutants were generated as described previously (Bahler et al., 1998; Janke et al., 2004) or by transformation with PCR products amplified from genomic DNA of the corresponding strain obtained from the Saccharomyces Genome Deletion (SGD) strain database. The HA-TBP strain expressing N-terminal HA-tagged version of TBP was obtained by inserting 3HA epitopes preceded by URA3 marker using an integrative plasmid (YIplac211-3HA-TBP) (Eyboulet et al., 2015). A complete and descriptive list of the strains and plasmids used in this study can be found in Supplemental Tables S1 and S2, respectively.
ChEC-seq
For ChEC analysis, yeast strains were grown in synthetic media to an OD600 of 0.5-0.7. ChEC reactions were performed as described and quenched after 5 min (Grunberg et al., 2016). Sequencing library preparation, alignment, track visualization, and cleavage pattern analysis were performed as described (Grunberg et al., 2016; Zentner et al., 2015). The scripts used for data processing are available at https://github.com/zentnerlab/chec-seq. HOMER (http://homer.salk.edu) (Heinz et al., 2010) was used to generate reads per million (RPM)-normalized average plots. TFIID- and SAGA-dependent (Huisinga and Pugh, 2004) or TATA-containing or TATA-less genes (Rhee and Pugh, 2012) were assigned to previous TSS annotations (Xu et al., 2009).
RNA labeling and newly-transcribed RNA purification
For each yeast strain and biological replicate, 100mL of wild-type and mutant S. cerevisiae cells were grown in YPD medium at 30°C to an OD600 ≈ 0.8. Newly-synthesized RNAs were labeled for 6 min by adding freshly-prepared 4-thiouracil (Sigma-Aldrich) until a final concentration of 5mM. In parallel, wild-type S. pombe cells were similarly grown in YES medium, at 31°C, and labeled for 6 min to be used as a spike-in across all samples. Upon labeling, cells were immediately pelleted and flash-frozen in liquid N2 and stored at -80°C until further use. All experiments using deletion strains were performed using at least two independent biological replicates.
Before total RNA extraction, S. cerevisiae and S. pombe cells were mixed in a ratio of 3:1. Total RNAs were extracted using RiboPure yeast kit (Ambion, Life Technologies) according to the description provided by the manufacturer. Prior to biotinylation, RNA samples were heated for 10 min at 60°C and cooled down on ice for another 5 min. 200μg of total RNA were biotinylated using 200μL of 1mg/mL EZ-link HPDP- Biotin (Pierce) in 100μL of biotinylation buffer (100mM Tris- HCl pH 7.5, 10mM EDTA) and adjusted to a final volume of 1000μL with DEPC-treated RNase-free water (Sigma-Aldrich) for 3 h at room temperature. After chloroform extraction and isopropanol precipitation (1/10 vol 5M NaCl and 2.5 vol isopropanol), purified RNAs were suspended in 100μL of DEPC-treated RNase-free water (Sigma-Aldrich).
Recovered RNA samples were incubated at 65°C for 10 min and allowed to cool down on ice for 5 min. Newly-synthesized biotinylated RNAs were bound to 100μL of μMACS streptavidin microbeads (Miltenyi Biotec) for 90 min at room temperature with gentle shaking. Purification of labeled RNA was then carried out using μMACS streptavidin starting kit (Miltenyi Biotec). Columns were first equilibrated with 1mL of washing buffer (100mM Tris-HCl at pH 7.5, 10mM EDTA, 1M NaCl, 0.1% Tween20). Samples were passed through the columns twice and washed five times with incremental volumes of washing buffer (600, 700, 800, 900, and 1000μL). Ultimately, labeled RNAs were eluted twice with 200μL of 100mM DTT. Following ethanol precipitation (overnight precipitation in vol of 3M NaOAc, 3 vol of 100% ethanol and 20μg of RNA-grade glycogen), RNAs were washed in ice-cold 70% ethanol and resuspended in 20μL of DEPC-treated RNase-free water (Sigma-Aldrich). Samples were stored at -80°C until further use.
For the anchor-away strain (SPT7-FRB), 200mL of S. cerevisiae cells were grown in YPD medium at 30°C to an OD600 ≈ 0.8. At that point, the culture was divided into two equal volumes of 100mL each and to one of those cultures rapamycin was added (+RAPA) until a final concentration of 1μg/mL, for 60 min, to allow nuclear depletion of Spt7. To the other half of the culture, the control samples (-RAPA/minus rapamycin), only the vehicle (DMSO) was added. Newly synthesized RNAs were labeled for 6 min by adding 4-thiouracil (Sigma-Aldrich) until a final concentration of 5mM.
For time-course experiments, 600mL of S. cerevisiae were grown and when OD600 ≈ 0.8, 100mL of the culture were collected (timepoint 0) and rapamycin was added up to a final concentration of 1μg/mL. Labeling was performed as described above and aliquots of 100mL were collected, representing 15, 30, 60, 120 and 240 min of nuclear depletion of Spt7. In all cases, cells were collected, counted and frozen in liquid N2. In parallel, and as described above, wild-type S. pombe cells were similarly grown in YES medium, at 31°C, and labeled for 6 min to be used as a spike-in across all samples. All experiments with anchor-away strains were performed using three independent biological replicates.
RT-qPCR
When performing RT-qPCR, cDNA synthesis was performed on 2μg of total RNA or 10μL of labeled RNA using random hexamers and Transcriptor reverse transcriptase (Roche) according to the manufacturer’s instructions. Real-time qPCR were performed using SYBR Green I Master (Roche). A list of all of the primers used can be found in Supplemental Table S3. All samples were run in triplicate and using at least two biological replicates. After qPCR, all raw values were corrected for the expression of S. pombe tubulin. Finally, results were represented graphically as a relative comparison between the wild-type (set to 1) and mutant samples.
Genome-wide expression analyses
RNA samples were hybridized to GeneChip Yeast Genome 2.0 microarrays following the instructions from the supplier (Affymetrix). Briefly, biotinylated cRNA _targets were prepared from 150ng of total RNA using the “MessageAmp™ Premier RNA Amplification Kit" (Ambion), according to the Instruction Manual # 4386269 Revision B, 18 september 2007. Following fragmentation, 4 μg of cRNAs were hybridized for 16 hours at 45°C, 60rpm on GeneChip® Yeast Genome 2.0 arrays (Affymetrix). The chips were washed and stained in the GeneChip® Fluidics Station 450 (Affymetrix) using the FS450_0003 script and scanned with the GeneChip® Scanner 3000 7G (Affymetrix) at a resolution of 1.56µm. Raw data (.CEL Intensity files) were extracted from the scanned images using the Affymetrix GeneChip® Command Console (AGCC) version 4.1.2. CEL files were further processed with Affymetrix Expression Console software version 1.4.1 to calculate probeset signal intensities, using the Affymetrix statistics-based algorithms MAS-5.0 with default settings and global scaling as normalization method. The trimmed mean _target intensity of each chip was arbitrarily set to 100.
All experiments were done using at least two independent biological replicates. Raw data were normalized to S. pombe signal and used to calculate fold-changes in total and newly-synthesized RNA levels, as represented in volcano plots. Further calculations of synthesis and decay rates, based on mathematical model as previously described, were performed using a pipeline and R/Bioconductor package publicly available (Schwalb et al., 2012; Sun et al., 2012). TFIID- and SAGA-dependent or TATA-containing or TATA-less genes were defined as previously mentioned (Huisinga and Pugh, 2004; Rhee and Pugh, 2012).
Growth curve and cell viability analyses
For growth curve analysis, the anchor-away (SPT7-FRB) and parental strains were grown overnight and then diluted to OD600 ≈ 0.1 in pre-heated YPD (200mL), before rapamycin addition at a final concentration of 1μg/mL (+ RAPA). To the control samples (-RAPA), only the vehicle (DMSO) was added. Every 30min, OD600 was measured until saturation of the culture. For cell viability, strains were grown until log-phase (OD600 ≈ 0.7). After 30 and 60min an aliquot was collected, stained with trypan blue and counted using a neubauer chamber.
Cell fractionation
150 ml cell culture (OD600 ≈ 0.5) were centrifuged and treated with 0.1M Tris, pH 9.4, 10mM DTT for 5 min at room temperature. Cells were pelleted and resuspended in YPD/S (YPD with 1M Sorbitol) and collected by centrifugation. Cells were resuspended in 1mL YPD/S and 750μL of 2M sorbitol. Zymolyaze 100T was added until a final concentration of 0.3mg/mL. Cells were incubated at 30°C for 30min (progress of spheroplasting was checked every 10min). Spheroplasts were washed and resuspended in YPD/S for 30 min at 30°C. Cells were quickly cooled down in ice, centrifuged, washed and resuspended in 1.5 ml lysis buffer (20 mM K-phosphate pH 6.5, 0.5 mM MgCl2, 18% Ficoll, 1xPIC). Spheroplasts were lysed three times with a B dounce and shortly centrifuged at 4500g to remove unbroken cells. The precleared whole cell lysate (Total lysate fraction) was centrifuged at 21000g for 45 min resulting into a crude nuclear pellet (nuclear fraction) and post-nuclear supernatant cytosol fraction (cytoplasmic fraction). 200µl of each fraction was TCA precipitated, washed with ice-cold acetone and resuspended in SDS-sample buffer.
RNA half-life quantification
Wild-type and mutant S. cerevisiae cells (two independent biological replicates for each strain) were grown in 300mL YPD medium at 30°C to an OD600≈0.8. A 50mL aliquot was collected and used as timepoint = 0. Immediately after, thiolutin was added to the cells until a final concentration of 3μg/mL. Aliquots of 50mL were collected after 10, 20, 30, 45 and 60 min exposure to thiolutin. Cells were collected by centrifugation and frozen in liquid N2. RNA was extracted using RiboPure yeast kit (Ambion, Life Technologies) according to the description provided by the manufacturer. cDNA preparations and RT-qPCR analyses were performed as previously described. Results were represented graphically as a relative comparison between the wild-type (set to 1) and mutant samples.
Whole cell extract and immunoprecipitation
150mL of cultures were grown in YPD at 30°C until OD600 ≈ 0.8. Cells were pelleted, washed in ice-cold 1x PBS, centrifuged and the pellet was frozen at 80°C. 500μL of buffer I (40 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.1% Tween, 10% glycerol, 1x PIC) were added to the pellet, and cells were broken by addition of acid-washed glass beads (0.5 mm diameter). Whole-cell extracts were clarified by centrifugation for 5 min at 4°C, and the protein concentration was determined.
Immunoprecipitations were carried out using protein A sepharose beads (Sigma) equilibrated in buffer I (40 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Tween, 10% glycerol, 1x PIC). 2mg of whole cell extract were precleared with 100μL of protein A sepharose beads, and immunoprecipitated using 5μL of antibody (anti-Taf10 and anti-Ada1) and 100μL of protein A sepharose beads. Beads were washed three times with 1mL of buffer II (40 mM Tris-HCl pH 8.0, 350mM NaCl, 0.1% Tween, 10% glycerol, 1x PIC) and three times with 1mL of buffer I. Bound protein complexes were eluted twice with 50μL 3M glycine pH 2.8 and then neutralized with 6.73μL of Tris-HCl pH9.8. From these eluates, equal volumes of 10μL were separated in a SDS-PAGE and analyzed by Western blot.
Chromatin immunoprecipitation
ChiP was performed as previously reported using three independent biological replicates (Bonnet et al., 2014). Briefly, cell cultures in log-phase (OD≈0.8) were cross-linked with 1% formaldehyde for 15 min (at room temperature, with gentle agitation) and then quenched with 0.125M glycine for 5 min (4°C, with gentle agitation). Cells were harvested, washed twice with ice-cold 1xPBS and resuspended in FA lysis buffer (50mM HEPES pH 7.5, 140mM NaCl, 1mM EDTA, 0.1% sodium deoxycholate, 1% Triton X-100, 0.4mM DTT, protease inhibitor cocktail (Roche)). Cells were lysed by mechanical shearing using acid-washed glass beads for 45 mins at 4°C, with intervals where samples were cooled-down on ice to avoid over-heating. The pellets were collected, washed once with FA lysis buffer and then sonicated in FA lysis buffer using a Covaris E220 machine to achieve an average DNA fragment size of 200bp. The Covaris E220 settings were as following: peak incident power of 140 Watts, duty factor of 5%, 200 bursts per cycle and 600 seconds (10 min) duration. The sonication was performed at 4°C and for six cycles of the described protocol. After each 10 min cycle, the samples would be left in the bath while other samples would be sonicating, to avoid overheating.
For each ChIP reaction, 250µg of chromatin extract were used. The equivalent to 1% of total sonicated chromatin was used as input. Input samples were kept at 4°C until the elution stage, when it was treated the same way the immunoprecipitated samples. After pre-clearing with protein-G sepharose, chromatin extracts were incubated overnight with 50µL EZview Red anti-HA affinity gel beads (Sigma). The resins were washed once with the following buffers at room temperature for 10min, except for the wash with Tris-EDTA buffer that was performed twice under the same conditions: (i) FA lysis buffer, (ii) FA lysis buffer containing 0.5 M NaCl, (iii) LiCl-containing buffer (10mM Tris-HCl pH 7.5, 1mM EDTA, 0.25M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) and finally (iv) Tris-EDTA buffer (10mM Tris-HCl pH 7.5 and 1mM EDTA). All buffers were also complemented with protease inhibitor cocktail (Roche). Bound chromatins were eluted with 250µL of elution buffer (50mM Tris-HCl pH7.5, 10mM EDTA, 0.1M NaOAc, 1% SDS) for 30 min at 65°C with shaking and then reverse cross-linked with RNase A treatment at 65°C overnight. Immunopurified material was incubated with 20 µg proteinase K for 1 h at 55°C. DNA was extracted once with phenol:chloroform:isoamyl alcohol (25:24:1) and extracted a second time with chloroform only. DNA was precipitated overnight at -20°C (1/10 vol 3M NaOAc, 3 vol 100% ethanol and 20µg of glycogen). DNA were then recovered and resuspended in 60µL of Tris-EDTA buffer (10mM Tris-HCl pH 7.5 and 1mM EDTA).
ChIP-qPCR
For ChIP-qPCR experiments, three independent sets were performed and analyzed by qPCR. qPCR were performed using SYBR Green I Master (Roche) as described above. A list of all of the primers used can be found in Supplemental Table S3. Each sample was run in triplicate and using three biological replicates. Normalization was conducted using the input percent method and results were presented as % of input.
Western blotting
For histone marks analyses, whole-cell extracts were prepared as described (Gardner et al., 2005) with minor changes. Cells from log-phase yeast cultures were harvested by centrifugation and lysed in 400 mL of SUME buffer (8 M urea, 1% SDS, 10 mM MOPS at pH 6.8, 10 mM EDTA, 0.01% bromophenol blue) by mechanical shearing. For RNA Pol II phosphoSer2 and phosphoSer5 analyses, extracts were prepared by treating cell pellets with 0.2M NaOH for 5min and boiling the samples in 2x Laemmli buffer for 5min. Extracts were cleared from debris through centrifugation. Antibodies used in this study were as follows: anti-H3K9ac (Abcam, ab4441), anti-Flag (Sigma, M2), anti-H3K4me3 (Abcam, ab8580), anti-H3K36me3 (Abcam, ab9050), anti-RNA Pol II phosphoSer2 (3E10, Active Motif), anti-RNA Pol II phosphoSer5 (3E8, Active Motif) and anti-Taf4 (gift from P. Anthony Weil).
Data and Software Availability
Data Resources
The accession numbers for the data reported in this paper are GEO: GSE96849 and GSE97379.
Supplementary Material
Acknowledgements
We are grateful to Violaine Alunni and Christelle Thibault (IGBMC) for array hybridization. We thank Vincent Géli and Pierre Luciano (CRCM, Marseille, France) for advices and discussion, Gabe Zentner (Indiana University, USA) for advice on ChEC-seq data analysis and P. Anthony Weil for antibodies. We thank Farrah El Saafin for critically reading the manuscript. T.B. was supported by a Marie Curie-ITN fellowship (EU-FP7 PEOPLE-2013 program, PITN-GA-2013-606806, NR-NET) and ARC fellowship. This work was supported by NIH grant GM053451 and GM075114 (to S.H.), funds from the Agence Nationale de la Recherche (ANR-15-CE11-0022 SAGA2 to D.D.; ANR-13-BSV8-0021-03 DiscoverIID, to L.T.) and the European Research Council (ERC) Advanced grant (ERC-2013-340551, Birtoaction, to L.T.). This study was also supported by ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02. N.M. is a student in the Magistère de Génétique Graduate Program at Université Paris Diderot, Sorbonne, Paris Cité.
Footnotes
Author contributions
D.D., T.B., S.G., S.H. and L.T. designed the study; T.B. performed all experiments except ChEC-seq analyses; S.B. and N.M. performed ChEC-seq experiments; M.J.E.K. and H.T.M.T. provided anchor-away strains; T.B., D.D., S.G. analyzed data; D.D., T.B., S.G., S.H. and L.T. wrote the manuscript with input from all authors.
References
- Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci U S A. 2009;106:16734–16739. doi: 10.1073/pnas.0905103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene _targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. SAGA is an essential in vivo _target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- de Jonge WJ, O'Duibhir E, Lijnzaad P, van Leenen D, Groot Koerkamp MJ, Kemmeren P, Holstege FC. Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters. EMBO J. 2017;36:274–290. doi: 10.15252/embj.201695621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, Berger SL, Workman JL. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyboulet F, Wydau-Dematteis S, Eychenne T, Alibert O, Neil H, Boschiero C, Nevers MC, Volland H, Cornu D, Redeker V, et al. Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo. Nucleic Acids Res. 2015;43:9214–9231. doi: 10.1093/nar/gkv782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, Struhl K. Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell. 2014;156:812–824. doi: 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grunberg S, Henikoff S, Hahn S, Zentner GE. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J. 2016;35:2435–2446. doi: 10.15252/embj.201695020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg S, Zentner GE. Genome-wide Mapping of Protein-DNA Interactions with ChEC-seq in Saccharomyces cerevisiae. J Vis Exp. 2017 doi: 10.3791/55836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, Causse SZ, Garber M, Millan-Zambrano G, Barkai O, Chavez S, Perez-Ortin JE, Darzacq X, Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Han Y, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 2014;33:2534–2546. doi: 10.15252/embj.201488638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Bortvin AL, Ricupero-Hovasse SL, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubik S, Bruzzone MJ, Jacquet P, Falcone JL, Rougemont J, Shore D. Nucleosome Stability Distinguishes Two Different Promoter Types at All Protein-Coding Genes in Yeast. Mol Cell. 2015;60:422–434. doi: 10.1016/j.molcel.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Laprade L, Rose D, Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–2017. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Holstege FC. The discrepancy between chromatin factor location and effect. Nucleus. 2012;3:213–219. doi: 10.4161/nucl.19513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Tora L. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim Biophys Acta. 2014;1839:118–128. doi: 10.1016/j.bbagrm.2013.11.003. [DOI] [PubMed] [Google Scholar]
- O'Duibhir E, Lijnzaad P, Benschop JJ, Lenstra TL, van Leenen D, Groot Koerkamp MJ, Margaritis T, Brok MO, Kemmeren P, Holstege FC. Cell cycle population effects in perturbation studies. Mol Syst Biol. 2014;10:732. doi: 10.15252/msb.20145172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Larivière L, Wenzeck L, Seizl M, Hemann M, Tegunov D, Petrotchenko EV, Borchers CH, Baumeister W, Herzog F, et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Winston F. Chromatin and Transcription in Yeast. Genetics. 2012;190:351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H, Pugh FB. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SM, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Molina JB, Tseng SC, Simonett SP, Taunton J, Ansari AZ. Engineered Covalent Inactivation of TFIIH-Kinase Reveals an Elongation Checkpoint and Results in Widespread mRNA Stabilization. Mol Cell. 2016;63:433–444. doi: 10.1016/j.molcel.2016.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Schulz D, Sun M, Zacher B, Dumcke S, Martin DE, Cramer P, Tresch A. Measurement of genome-wide RNA synthesis and decay rates with Dynamic Transcriptome Analysis (DTA) Bioinformatics. 2012;28:884–885. doi: 10.1093/bioinformatics/bts052. [DOI] [PubMed] [Google Scholar]
- Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK. Conformational flexibility and subunit arrangement of the modular yeast Spt-Ada-Gcn5 acetyltransferase complex. J Biol Chem. 2015;290:10057–10070. doi: 10.1074/jbc.M114.624684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell. 2013;52:52–62. doi: 10.1016/j.molcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Lariviere L, Maier KC, Seizl M, Tresch A, Cramer P. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci U S A. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Berman J, Barkai N. The pattern and evolution of yeast promoter bendability. Trends Genet. 2007;23:318–321. doi: 10.1016/j.tig.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Tora L, Timmers HT. The TATA box regulates TATA-binding protein (TBP) dynamics in vivo. Trends Biochem Sci. 2010;35:309–314. doi: 10.1016/j.tibs.2010.01.007. [DOI] [PubMed] [Google Scholar]
- van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008;22:2359–2369. doi: 10.1101/gad.1682308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Wachi S, Mavrich TN, Andersen BE, Jena P, Sinnamon AJ, Jain P, Rolleri NS, Jiang C, Hemeryck-Walsh C, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanyi Z, Collart MA. Ccr4-Not is at the core of the eukaryotic gene expression circuitry. Biochem Soc Trans. 2015;43:1253–1258. doi: 10.1042/BST20150167. [DOI] [PubMed] [Google Scholar]
- Vosnakis N, Koch M, Scheer E, Kessler P, Mely Y, Didier P, Tora L. Coactivators and general transcription factors have two distinct dynamic populations dependent on transcription. EMBO J. 2017 Jul 19; doi: 10.15252/embj.201696035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. SAGA function in tissue-specific gene expression. Trends Cell Biol. 2012;22:177–184. doi: 10.1016/j.tcb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Kasinathan S, Xin B, Rohs R, Henikoff S. ChEC-seq kinetics discriminates transcription factor binding sites by DNA sequence and shape in vivo. Nat Commun. 2015;6:8733. doi: 10.1038/ncomms9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.