Abstract
Human Cytomegalovirus (HCMV) is an opportunistic pathogen that causes substantial disease in neonates and immunocompromised individuals. Reverse genetic analysis of the HCMV genome is a powerful tool to dissect the roles that various viral genes play during infection. However, genetic engineering of HCMV is hampered by both the large size of the HCMV genome and HCMV’s slow replication cycle. Currently, most laboratories that genetically engineer HCMV employ Bacterial Artificial Chromosome (BAC) mediated recombineering, which is a relatively lengthy process. We explored an alternative method of producing recombinant HCMV using the CRISPR/Cas9 system. We employed both homologous recombination (HR) and Non-homologous end-joining (NHEJ)-based methods, and find that each approach is capable of efficiently mutating the HCMV genome, with optimal efficiencies of 42% and 81% respectively. Our results suggest that CRISPR-mediated genomic engineering of HCMV is competitive with BAC-mediated recombineering and provide a framework for using CRISPR/Cas9 for mutational analysis of the HCMV genome.
Keywords: CRISPR, Cytomegalovirus, HCMV, Cas9, Recombineering
Introduction
Human Cytomegalovirus (HCMV) is a double-stranded DNA virus that infects more than half of adults in the US by age 40 (Staras et al., 2006). While typically asymptomatic in immunocompetent adults, HCMV can be a significant cause of morbidity in immunocompromised individuals including those with HIV/AIDS, organ transplant recipients, and newborns (Azevedo et al., 2015; Ramanan and Razonable, 2013; Revello et al., 2006; Wallace and Hannah, 1987). In particular, congenital HCMV infection is a major cause of birth defects in neonates and is among the leading causes of deafness, blindness, and intellectual disability in children (Damato and Winnen, 2002). Clinical treatment of HCMV currently depends in part on nucleoside analogues, which can result in toxicity and the emergence of drug resistant strains (Biron et al., 1986; Janoly-Dumenil et al., 2012). As such, there is a significant need for less toxic _targeted therapeutics.
BAC-mediated HCMV mutagenesis has become a common tool to elucidate HCMV genomic features that are important for successful viral replication. Various HCMV genome strains have been cloned as BACs, transformed into bacteria, and manipulated using recombineering techniques (Paredes and Yu, 2012). However, the BAC recombineering process presents some methodological issues. Perhaps the most notable is the length of time that BAC-mediated recombineering takes. It can frequently take up to six weeks in order to produce a useable viral stock. Another issue is that BAC recombineering requires multiple rounds of clonal selection of BAC-containing colonies resulting in a monoclonal population that fixes a single HCMV genotype, which could include any newly arising secondary mutations.
Recent developments in CRISPR/Cas9 gene editing have made it possible to introduce precise modifications into a wide variety of genomes, including those of dsDNA viruses (Lin et al., 2016). CRISPR/Cas9 _targeting relies on the co-expression of a prokaryotic Cas9 nuclease and an associated guide RNA (gRNA) sequence. Once gRNA-_targeted Cas9 creates a double-stranded break (DSB) in the genome, two main repair processes compete to repair the damage, leading to modifications of the original sequence. Non-homologous end-joining (NHEJ) is an error prone process in which base pairs are often added or subtracted to the damaged sequence before the ends are ligated together, reliably producing imprecise insertion and deletion (INDEL) mutations. Homologous Recombination (HR) repairs the sequence using sections of homologous DNA as a template, which when paired with an exogenous DNA construct allows for the introduction of precise changes to the _targeted genome (Ran et al., 2013). HR gives the ability to make specific modifications to the viral genome without relying on BAC constructs.
Here, we assessed the feasibility and efficiency of utilizing CRISPR-based techniques to engineer the HCMV genome. We find that CRISPR-based methods can be employed to efficiently _target HCMV sequences via both INDEL mutations and homologous recombination. After optimizing various experimental parameters, we find that NHEJ-mediated INDEL mutations can result in >75% gene knockout efficiency, whereas HR-_targeted site-directed mutations can reach >40% recombination efficiency. Taken together, these results provide a platform for using the CRISPR/Cas9 system in primary fibroblasts to make specific modifications to the HCMV genome, streamlining mutant virion production and facilitating the investigation of processes important to HCMV replication.
Results
Establishment of CRISPR-mediated homology-directed HCMV mutagenesis.
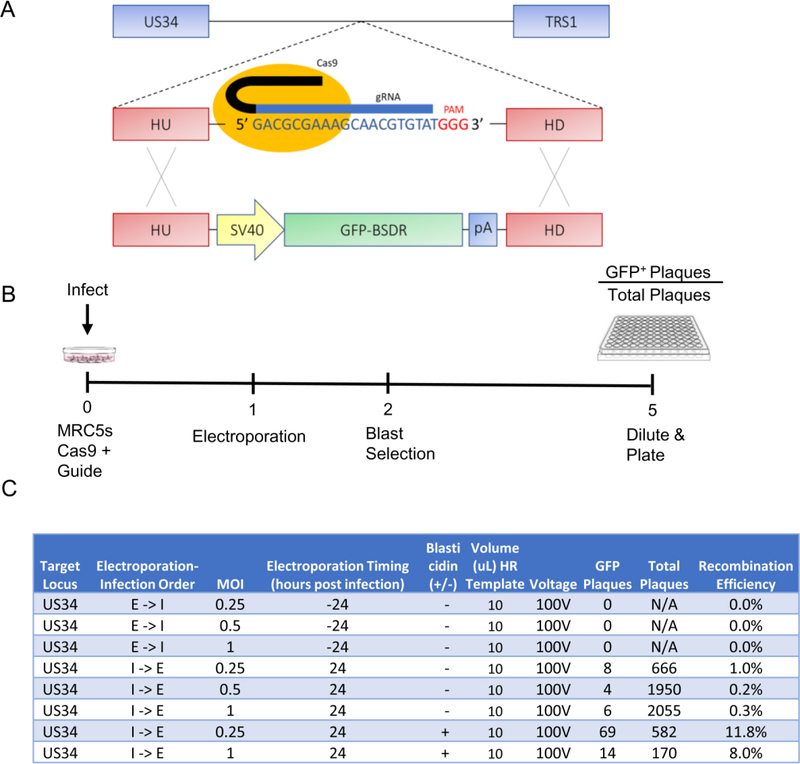
To determine the feasibility of using CRISPR/Cas9 to make large (~1.5kb) changes to the viral genome, we created a construct encoding a GFP-blasticidin deaminase fusion protein (GFP-BSDR) flanked by homologous HCMV sequences (Figure 1A), which enables homologous recombination (HR) into the HCMV genome. A non-essential locus between US34 and TRS1 was selected to assess HR efficiencies. Briefly, MRC5 fibroblasts were transduced with a lentiviral construct expressing Cas9 and a gRNA _targeting this region. These cells were then infected with wild type virus at varying multiplicities of infection (MOIs) and transfected with the GFP-BSDR template under a variety of electroporation, infection, and culture conditions to elucidate whether HR was possible, and identify the treatments resulting in the highest recombination efficiency. After five days of infection, supernatants containing viral progeny were harvested, diluted, and plated (Figure 1B). Subsequent plaque formation and monitoring of GFP expression indicated successful HR. GFP positive plaques emerged with varying efficiencies, with the results summarized in Figure 1C. No recombination was evident when electroporation was performed prior to infection (Figure 1C). Electroporation at 24h post infection gave rise to successful recombination events (Figure 1C), which were subsequently verified by sequencing several isolates to confirm GFP-recombination in the proper locus. Of the various MOIs tested, the maximum recombination efficiency was observed at an MOI = 0.25 (Figure 1C). Blasticidin selection reduced the total amount of WT virus produced (data not shown); however, it substantially increased the HR efficiency, e.g., by 12- and 26-fold at MOIs of 0.25 and 1.0 respectively, with the maximum recombination efficiency observed under these conditions being 11.8%.
Figure 1: Homology-directed recombineering of the HCMV genome in fibroblasts.
(a) Schematic of Cas9 _targeting and GFP incorporation. Homology arms are 750bp. (b) Timeline of cell treatment, infection, transfection and viral isolation over 120 hours. Per well, 10ul of HR template (10ug of GFP-containing HR template in 30uL) was transfected as a linearized plasmid either 24 hours pre- or post-infection. (c) Viral recombination efficiencies. Where indicated, cells were treated with 10ug/mL blasticidin from days 2–5. I-> E denotes infection, then electroporation. E -> I denotes electroporation, then infection.
High efficiency mutation of the HCMV genome via CRISPR-mediated non-homologous end-joining (NHEJ).
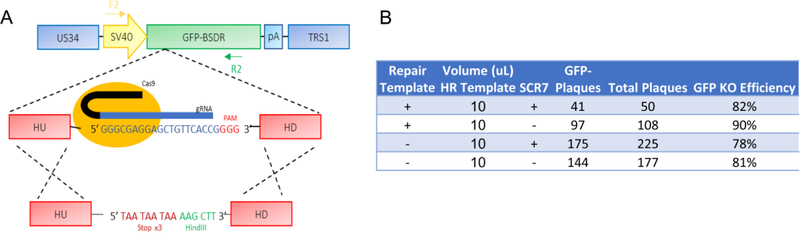
Repair of CRISPR-mediated DNA cleavage can take place via homologous recombination. Alternatively, CRISPR-mediated cleavage can occur via the more imprecise non-homologous end-joining pathway (NHEJ), which introduces insertions or deletions (INDELs) that can disrupt amino acid coding. NHEJ repair requires the activity of DNA Ligase IV, and it has been reported that the efficiency of HR can be increased via pharmaceutical inhibition of DNA Ligase IV (Chu et al., 2015). Studies have also shown that the size of the repair template can influence HR efficiencies, with smaller single stranded oligonucleotide (ssODN) templates leading to higher efficiency (Song and Stieger, 2017). We therefore assessed the effect of a DNA Ligase IV inhibitor, SCR7, and smaller HR repair templates on HR efficiency. Using the GFP+ virus generated in Figure 1, we analyzed HR-mediated and NHEJ-mediated mutation of the GFP open reading frame (Figure 2A). Primary fibroblasts that co-express Cas9 and a GFP-_targeting gRNA, were infected in the presence or absence SCR7, and a small 200bp ssODN repair template, homologous to GFP, but containing a triple stop codon (Figure 2A) to ablate GFP expression. The efficiency of GFP KO was assessed via monitoring GFP+ plaque formation. High efficiency GFP knockout was observed under all conditions (~80–90% loss of GFP). This loss of functional GFP expression occurred irrespective of the presence of repair template (Figure 2B), suggesting that HR was not responsible for the loss of GFP, and that the SCR7 treatment was not effective at inhibiting NHEJ. Subsequent cloning and sequencing of 25 different clones confirmed that the loss of GFP was primarily due to INDELs and not due to recombination with the repair template (data not shown). These results suggest that neither treatment with SCR7 nor the utilization of a small ssODN repair template improved HR efficiencies. However, our results with INDEL-mediated inactivation of GFP expression suggest that NHEJ-mediated INDEL mutation of the HCMV genome occurs at very high efficiency, and could therefore be a helpful tool for INDEL-mediated _targeting of the HCMV genome.
Figure 2: High efficiency CRISPR-mediated INDEL inactivation in the HCMV genome.
(a) Schematic of _targeting viral GFP function using CRISPR/Cas9 editing in primary fibroblasts. 24 hours post-infection, 10uL of 10uM ssODN solution was transfected per well. Where indicated, cells were treated with 10uM SCR7 from days 1–5 (b) GFP KO efficiencies were calculated by plaque assay at 10dpi.
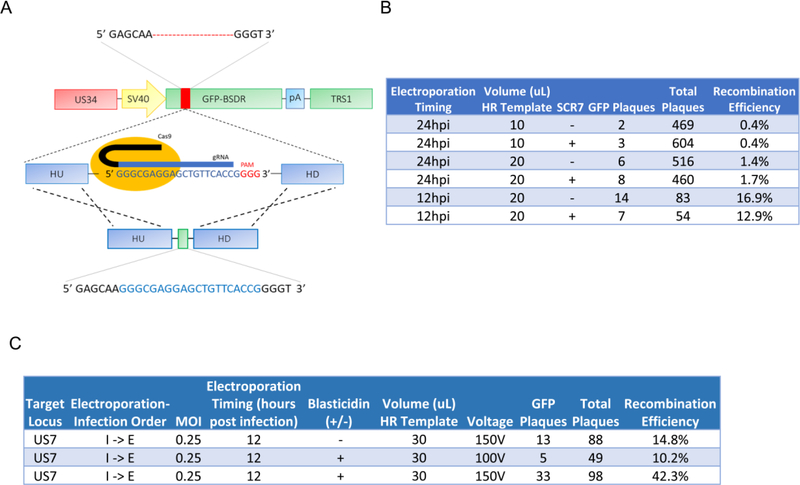
In the experiments presented in Figure 2, measurement of HR efficiency required extensive clonal sequencing. To more quickly determine HR efficiency, we assessed the HR-dependent repair of a mutant GFP allele. We isolated a viral clone created by the experiment described in Figure 2 in which GFP was inactivated by an INDEL. The clone selected possessed a 21 base-pair deletion near the N-terminus of GFP. This mutation can subsequently be repaired by a newly designed gRNA, and repair template to restore GFP function, thereby facilitating better estimates of HR efficiencies (Figure 3A). For these experiments, we assessed the effect of SCR7 treatment, repair template volume, and electroporation timing, on HR efficiency. We found that electroporation earlier during infection (12hpi vs. 24hpi) increased HR efficiencies by approximately 10-fold (Figure 3B). Further, increasing the amount of HR template (10uL vs. 20uL of a 10uM oligo solution) also increased HR efficiencies by approximately 3–4-fold. In contrast, treatment with 10uM SCR7 solution during the viral incubation cycle did not appear to significantly increase recombination efficiency. For ssODN-mediated repair maximum efficiency (14/83 GFP+ plaques or ~17%) was achieved with 20uL of oligo solution, electroporation at 12 hours post infection, and no SCR7 treatment (Figure 3B).
Figure 3: Optimization of homology-directed recombineering of the HCMV genome in fibroblasts.
(a) Schematic for repairing a 21-base pair deletion in viral GFP using CRISPR/Cas9 Homologous Recombination. HR template volumes given represent ul from a solution containing 10ug of GFP-containing HR template in 30uL. (b) Recombination efficiencies for repairing GFP function using a small ssODN repair template, counted at 10 days post infection. Volumes given represent ul of a 10 uM oligonucleotide-containing solution. (c) Recombination efficiencies of optimized homologous recombination/insertion of GFP-BSD into the US7 locus. HR template volumes given represent ul from a solution containing 10ug of GFP-containing HR template in 30uL
In Figure 1, we found the recombination templates encoding blasticidin resistance increased HR efficiencies by approximately 10-fold (Figure 1C). We therefore assessed whether blasticidin selection could improve HR efficiencies with the optimized parameters employed in Figure 3B, i.e., electroporation at 12 hpi, and increased HR template volume. We also assessed the impact of increased electroporation voltage, i.e. 150V versus 100V, and examined the frequency of recombination at a different locus, in between US6 and US7. As shown in Figure 3C, these modifications resulted in a maximum HR efficiency of 42.3%, i.e. with 30uL HR electroporation at 150 volts, 12 hours after infection at MOI = 0.25, and with blasticidin selection. These results suggest that CRISPR-mediated HR could be an effective method for homologous site-directed mutation of the HCMV genome.
Discussion
Our goal was to investigate the potential of using CAS9 for HCMV-mediated mutagenesis. Moreover, we wanted to identify key parameters that would optimize the efficiency of _targeted HR-directed and INDEL-associated mutation, while minimizing the time required to produce recombinant virus. The optimum conditions we observed generated HR-associated site-directed mutations with efficiencies of ~40% (Figure 3C), and INDEL-associated mutations with efficiencies of ~80%. (Figure 2B). Our optimized protocol is outlined in the Materials and Methods below. Collectively, our data indicate that Cas9-directed mutation of the HCMV genome is a viable strategy for HCMV genomic engineering.
We found that the timing of electroporation was a key parameter that impacted HR efficiencies. Electroporation at 12 hpi increased the HR efficiency substantially relative to 24 hpi (Figure 3B). A likely explanation is that Cas9-mediated cleavage/NHEJ repair is active soon after infection, and that a longer delay in HR template delivery increases the likelihood that the _target sequence has already been mutated with an INDEL thereby preventing HR (Mao et al., 2008). Another possibility, would be that the later times of electroporation could negatively impact HCMV replication. In addition to electroporation timing, there was also an observed difference in HR efficiency associated with MOI. An MOI of 0.25 produced the highest amount of recombinant virus relative to background. This likely indicates that that the amount of electroporated template or Cas9-gRNA complexes that make it to the nucleus are limiting at higher MOIs, thus resulting in a higher background of non-recombineered virus. It is possible that further reductions in the MOI or in the time in between infection and electroporation could result in higher efficiencies. Further, we speculate that increases in the efficiency of nuclear electroporation of HDR template would increase HDR efficiencies.
In the above studies, we used the presence or absence of GFP expression to assess recombination and INDEL efficiencies. Most site-directed mutagenesis strategies involve engineering mutations that do not include reporter insertion, i.e. without GFP as a readout for HR success. In the absence of GFP/blasticidin selection, we obtained HR efficiencies of ~15–17%. At these HR efficiencies, approximately 10–20 isolated infections/plaques would have to be screened for the introduction of a silent molecular marker, e.g. a restriction site, to obtain a successful recombinant. We found that this could readily be achieved via dilution and plating in 96-well dishes. However, as indicated above, further optimization of electroporation timing, and efficiency, as well as the MOI of infection could increase HR efficiency and thus enable screening fewer plaques.
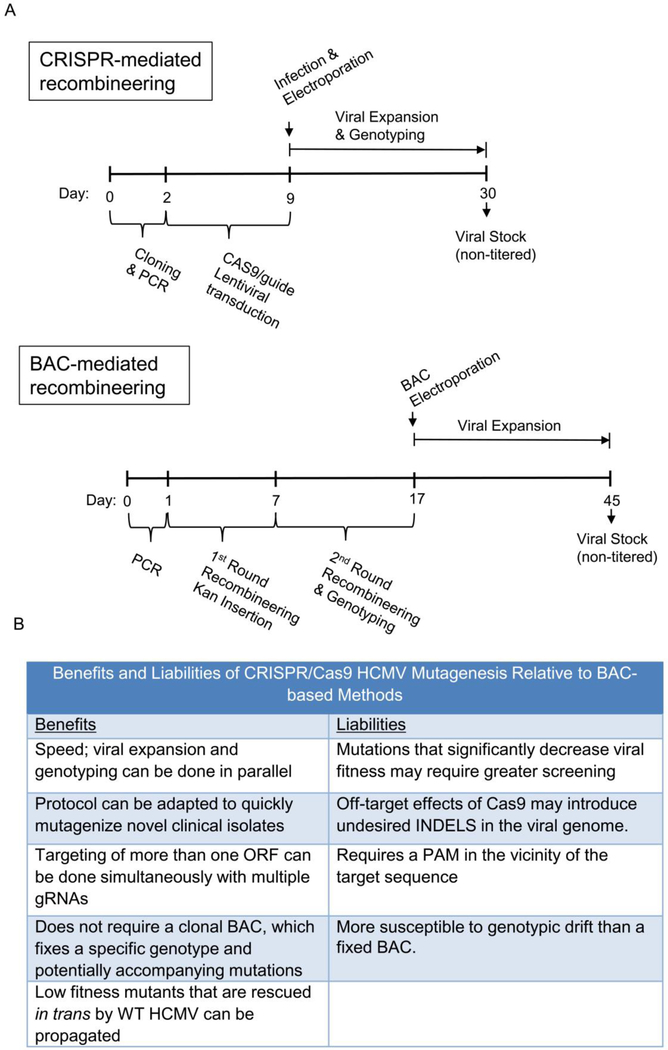
The observed HR and INDEL-associated efficiencies suggest that CRISPR-mediated recombineering is competitive with, and in some cases may be preferred to BAC-mediated recombineering. In figure 4, we illustrate the timing of CRISPR-mediated recombineering of the current protocol, against a typical BAC-mediated recombineering protocol. In general, the current CRISPR protocol compares favorably with respect to the length of time necessary to produce a viral stock (Figure 4). Given the slow HCMV replication cycle, the shorter time frame largely reflects two factors. First, with the current CRISPR protocol, viral expansion starts with an infectious virus as opposed to transfection with a naked DNA BAC clone, from which the emergence of infectious virus is relatively slow and inefficient. Secondly, with the CRISPR protocol, viral genotyping and expansion can occur simultaneously, whereas BAC-mediated recombineering requires genotyping prior to BAC transfection (Figure 4A).
Figure 4:
Timeline and Methodological Comparison of BAC recombineering and CRISPR-mediated homologous recombination.
Another potential benefit of the current CRISPR-based method is that a novel clinical isolate could be relatively quickly mutagenized without the need for cloning/insertion of BAC sequences and subsequent recombineering. Alternatively, should BAC clones of novel clinical isolates be desired, the current CRISPR-mediated HR could be employed easily to insert BAC sequences into new HCMV clinical strains. Further, in both scenarios, the polyclonal nature of the original viral population could be largely preserved as opposed to monoclonally fixing a single sampled genotype, which may not accurately reflect the infectious population.
One advantage of the current CRISPR-technique would be the relative ease of making simultaneous inactivating mutations in multiple HCMV ORFs. Such a strategy would require multiple rounds of BAC-mediated recombineering, and therefore a significant investment of time. In contrast, cells expressing guides to multiple HCMV-guide sequences would not be difficult to create and would enable inactivation of multiple HCMV ORFs in a single round, which would therefore facilitate analysis of synthetic or epistatic relationships between HCMV genes.
Viral mutants with defective replication kinetics are challenging to obtain, and we see potential advantages and disadvantages associated with both BAC-mediated and CRISPR-mediated mutagenesis in various contexts. In the case of a completely replication incompetent virus, i.e. one not capable of initiating a plaque on its own, both BAC and CRISPR-mediated recombineering will fail. However, BAC-mediated recombineering would be the preferred technique, as the lack of plaques would be apparent sooner relative to the CRISPR method, in which one would have to screen many plaques without success to come to the same conclusion. However, in another scenario, CRISPR-mediated recombineering would be predicted to represent an improvement over BAC-mediated recombineering. Specifically, the CRISPR method could enable the replication of low fitness mutants capable of being rescued in trans by the presence of wildtype HCMV. The timing of our protocol allows for a single round of co-replication, prior to plaquing and clonal expansion. During this initial single round of infection, wildtype HCMV genes may help rescue the replication of the mutant, thus enabling the generation of the infectious mutant virus. In this regard, the currently described CRISPR methodology may facilitate the generation mutants that are otherwise incapable of initiating an infection from a BAC-clone in the absence of wildtype infection. Figure 4B summarizes some of the benefits and potential liabilities associated with CRISPR-Cas9 mutagenesis in comparison to BAC-based methods.
Collectively, our results provide a blueprint for employing CRISPR-Cas9 to make modifications to the HCMV genome. The presented techniques are competitive with current BAC-recombineering techniques in terms of efficiency and the time necessary to obtain a recombineered virus. In short, CRISPR-mediated HCMV recombineering is another tool that will assist in the elucidation of how HCMV gene function contributes to HCMV biology and pathogenesis.
Materials and Methods
Cell culture and infection.
HEK293T cancer cells and immortalized MRC5-hTERT fibroblasts (referred to as MRC5 cells in the text) were cultured in Dulbecco’s modified Eagle medium (DMEM, Invitrogen), supplemented with 10% fetal bovine serum, 4.5g/L glucose, and 1% penicillin-streptomycin. MRC5s were seeded at 3.4 × 105 in a 10cm dish 72 hours prior to viral infection with the HCMV strain AD169. Cells were exposed to a 4mL viral inoculum at the specified MOI for 2 hours, after which, the virus was aspirated and replaced with fresh media.
Pharmacological Inhibitors.
SCR7 Pyrazine and Blasticidin S Hydrochloride were purchased from Sigma Aldrich and solubilized in DMSO. Cells were treated with 10μM SCR7 in DMEM for the full 120 hours of initial infection. Cells were treated with 10ug/mL Blasticidin from 48 to 120 hours post infection.
Homologous recombination templates.
For GFP-BSD insertion, upstream and downstream homology arms (see homology arm sequences and PCR primers below) were PCR amplified from 750 base-pair gene blocks (IDT). The SV40 promoter and polyadenylation sequences were PCR amplified from pEPkan-S, and EGFP/BSDR was amplified from LeGO/BSD (Addgene). These individual components were then assembled through Gibson Assembly into the PstI site of pBluescript KS (−) (Addgene). Correct clones were identified by sequencing using the T7F and M13R promoter primers. For the Stopx3-HindIII and GFP-deletion repair templates (see sequences below), oligos were ordered as 200 base-pair Ultramers from IDT and dissolved to 100uM in water.
Guide RNA Cloning of US34/TRS1 _targeted CRISPR/Cas9 lentiviral vector.
Oligos with the following sequences: 5’CACCG GAC GCG AAA GCA ACG TGT AT 3’ and 5’AAAC AT ACA CGT TGC TTT CGC GTC C 3’ were phosphorylated and annealed in a thermocycler (Applied Biosystems 2720 Thermo Cycler) under the following conditions: 37°C for 30 min, 95°C for 5 min then ramp down to 25°C at 5°C/min. Oligos were then cloned into the BsmB1 site of pLentiCRISPRv2 (Addgene), amplified via transformation into stbl3 E. coli, and confirmed by sequencing using a h-U6F promoter primer. Similar cloning approaches where employed for guide RNA cloning constructs _targeting US6/7, the Stopx3-HindIII insertion, and the GFP deletion repair: US6/7 oligos: 5’ CACCG CCC CGA TGA ACG CTC TCG TC 3’; 5’ AAAC GA CGA GAG CGT TCA TCG GGG 3’;, Stopx3-HindIII insertions oligos: 5’ CACCG GGG CGA GGA GCT GTT CAC CG 3’, 5’ AAAC CG GTG AAC AGC TCC TCG CCC C 3’; GFP-deletion repair oligos:5’CACCG GAG CAA GGT GGT GCC CAT CC 3’, 5’ AAAC GG ATG GGC ACC ACC TTG CTC C 3’.
Lentiviral transfection/transduction to introduce viral-_targeted Cas9.
293T cells were seeded at 6.8 × 105 cells in a 10 cm dish and grown for 24 hours. Using the Fugene 6 transfection reagent (Promega), each 10 cm dish was transfected with 2.6 μg pLentiCRISPRv2 (Addgene), 0.25 μg pVSVg, and 2.5μg PAX2. After 24 hours, the media were replaced by 4 mL fresh media. After another 24 hours, lentiviral-containing media were taken from each plate, filtered through a 0.45μm filter, and applied to MRC5 fibroblasts with polybrene over the course of 24 hours. The media were then replaced with 10 mL fresh media and, after another 48 hours, cells were placed under 1μg/mL puromycin selection for 4 days.
Electroporation of GFP-BSD HR template into Fibroblasts.
Ten μg of plasmid HR template were linearized with HindIII in a 30uL reaction and purified using a Qiaquick PCR Purification Kit (Qiagen). A 10 cm dish of 90% confluent MRC5 fibroblasts (~1×106 cells) was trypsinized, resuspended in 100 μL Nucleofection Solution #6 (140mM sodium phosphate buffer, 5 mM MgCl2, 5 mM KCl, pH 7.2) and electroporated with 30uL linearized HR template using the exponential decay pulse setting at 100V or 150V, and 960 μF. Similar results were obtained when electroporating cells from a single well of a six well dish (~3×105 cells) using identical electroporation conditions as indicated above. Cells were then re-plated into a single well of a 6-well plate. For the STOPx3HindIII insertion and GFP-deletion repair, cells were treated as described above, except 10uL of 100uM HR template solution was added per electroporation.
Plaque Assays.
Forty-eight hours prior to infection, 5 × 104 MRC5-hTERT cells were plated into each well of a 12-well plate. Prior to infection, using a deep (1mL) 96-well plate, each viral stock was serially diluted from 10−1 to 10−6 to a volume of 1mL per well. For a given viral stock, 450 ul of each dilution was added to 1 well of cells and incubated overnight. Twenty-four hours post infection, viral-inocula were aspirated and replaced with pre-warmed gel overlay solution (50% 2X DMEM with 1% PenStrep, 40% of 2.5% low melting NuSieve agarose in H20, 10% FBS). Plaques were typically counted 10 days post infection, and viral clones isolated 15 days post infection.
Viral/Cellular DNA Extraction.
Seventy-two hours post infection with viral supernatant, cells were scraped in their media, pelleted by centrifugation at 2000 rpm for 5 min, washed with PBS, and centrifuged once more. Cells were resuspended in 200–500uL lysis buffer (100mM NaCl, 100mM Tris at pH 8, 25mM EDTA, 0.5% SDS, 0.1 mg/mL Proteinase K, 40ug/mL RNAse A) and incubated at 55°C overnight. Samples were extracted once with an equal volume amount of Phenol/Chloroform/Isoamyl Alcohol (25:24:1). A second extraction was performed with Chloroform/Isoamyl Alcohol (24:1). DNA was precipitated by addition of 0.1 volume of 3M sodium acetate and 2 volumes of 95% ethanol at −20 degrees. DNA was pelleted by centrifugation for 15 min at 14,000 rpm, and washed once with 70% ethanol. The DNA pellet was resuspended in 60uL H20.
Homology Arm Sequences.
US34/TRS1 upstream (left) homology arm:
5’CGGACCGAGCCAGAGGGACCTTCGCTTGGCGTTCAGGCCTTAATACTAGCCGCA TCTTACCTGTCGGCAGCATGTATCGGGGCTCCGACGCCTTACCCGCCGGCCTGTATCGTCCCGAAGAAGAGGTGTTCCTCCTCTTGAATCGCTGCCATGGGCCACTGTCAACGCCGAAAAATGCTTGTCTGGCTGAGGTTGGTGTCGCTAATGCCACTTTTTTGTCTCGCTTCAATGTCGGTGATTTTCACGGAGCGTCATGGGAAAACGGTACCGCTCCCGATGGAGAGCCCGGGGTATGCTGAAATTCCTCTTAAAATTTCGTAAACGACGTCGTCCAGTCGTTGTGCCGCGATTCGTACGGTTCATCGTCTACGTCGTTTTGTTCACCGTCGCTGTGCAACGCGTGAAACAAGAGCGTGATGCGCACCTTCGGCGGTATGAAGAACGATTACGGAAAAACCGCGCACGGCGTCGGCAGTCTTTTCCGTGACTTGGGGCGATGGGTCCGAGCTGCGGTATGGGTCACGGCGGCGTGTGTTTTATTGACGAAGATGCCGATGTGTGACTAAAAACGTCCCAGCCCCAGAGCGATATGTTTCAATAAAAAAAATATGTAGTATCATATTATGCGTGTCCTGGTTTTTCATTTTTGGATGTATGTATCGCATAAAGGGTGGCGAGGTGTGAGGATGAAACATATGCAGATACGCAGTGTTGTTATCCGAACGAAACCCGTGTAATGCGTACAACGGT 3’
US34/TRS1 downstream (right) homology arm:
5’ATATTGCTAATCATGCGTCTTGCAGTACAGATAGCCGCTGTATCTTACGCGTATTGTCGCAACAGTTCCACATCGGTGTAATTGGATGTCTGGTACTTATCACTGGCGTCGTTATAACATTGTAAAACAAGTTTTCGAAACATAACGACAGCTGCAAAAGAAAACCAGTTTATTGAGCATTGTAATGGTAGTGTGTGGCTATATTAGAAAACGTGACGCGTCGCATGTCGCGGCACAATCTGGCAGCGGGGTCGGGGTAGGGTACGGTGGGAGGCATGTACACAGATGGAACAAAAGCAGAAGTAACGTGAGAAGGAGCATACAGTCCAGTATCCAGCGGTTCCTGAGTAGCACCACCCATCAACTGAATGCCCTCATGAGTAAAAGTCTGCGGGCGACAGCCCTTGGGGACCGTTGGCATGGGACGATCAATCTCCAAACCACAGCGTAACACCGTTTTCTTCCAACGTCGTTGATAGACGTCGTTTTTACGGTTACTCCCAAGAACCCAGAAAGTCTCGTCCAAGTCGTACCAGGAATCTTCTCCGGGGAGACGCGACGGTTTCCAATCCTCGTCGTCTCGTCTCAAAGCACGTCCCAAACTGGCTTGAGGAGTCAACGGTGGTTCTGTGGGTCGGGTGTAGCGCGAGTGTTTTCCCTTCATGAGCGATTCATCCTCCTTGCCTTTAGGCTTTTTGGTCTTTTTGTGTATCATCTGGCCGCCGGCCTCCATAACCACCGTGGCCAAGT 3’
US6/7 upstream (left) homology arm:
5’ GCCGTGTTGAAGGAACGCGCTTTTATTGAGACGATAAAACAGCATCAGGAGCCACAACGTCGAATCCCACGTCCAGTCGATTCGTATGTTATGCTGCACAGCAATGCTAGAATAACAACCAGCAGGGTAATCCCGCAACATAAATACAAAGTCACAGCGAAGAATCCGTGTCGTTCTATCAAGCGAAACGCGTTCCAAACGGCCCCGTCACAGACGCAGTTATTCATAAGCGTTAACAACCGGTGGCTAGGATGAATATCCAAATCACAGGGCAGTAGCCGACGGACTCGTTGACAGGTCAGCCTACCCTCAAGGTTCCTATCGTTCGGACGGGATTTGTGCGTTTTAGGCCTCTTTTTCGCCGCCTGCAAGCATTGGTGCGCAAAGTCCTCACCCAGCTGTTTCCAGCTATCATCTGCATCTGTGCAGTCCCCTGTATCGTTGTAACAAACGGGTCTGTGCGACTTCGTTCTCGGAACACAAGCTTGTTGTCGCGGAGACAGAGAGAGAAGGGTTTTCGGGTCACGCGAAGACCGCTCACCGGGGGTCGGCAACGCACACATCAACAGAAAACCGAGACGAATCAAGAGATCCATAGTGAAGGAGTGATATCGACGTGCTTACGAAACGGCGATTATATATGTTCTCAACAATACCGCCCTACGTTGTATGATGTAACGTGTGACGTGAGTCTGATCCAACACTGAACGCTTTCGTCGTGTTTTTCATGCAGCTTTTACAGACCATGAC 3’
US6/7 downstream (right) homology arm:
5’ TACACAAACTCCATATATTTGTTACGATAGAATACGGAACGGAGGAGGCTTTCGC CACACCTATCCTGAAAGCGTTGCATTCTTTATGATAGGTGTGACGATGTCTTTACCATTCCCACGGCTGCTTTGCGTGATGATGACATTCATCATGTATTTCCATTCACACATACCTTTTGTGCATACGGTTTATATATGACCATCCACGCTTATAACGAACCTAACAGTTTATTAGCCCTTGACAGGATAGGTCAAAAGATTATATGTAGGTTTTCCGGTAAACCGAATTGTGATATTTCTCTGCAGGAAATAGAACAGCCTGGTACCTATAAAACGGACAATGCAGTACTGTAGCAGCGTAACCAAGTAGGTCCACATGAACACGTACAAAATTATGGTAAGCCATCGTTTTTCATACCACAGCCTGTAGCTGTCGTACATGAATGAGGACGGTCGAGGAACCCAGGGTAGTTGTAATTGGGGGCGACATTCGTACTGTCCAGAAGACAATTGCACGGGTTTCAGTGAGATGAGTACTTTAGCGATGTCGGCGGGGGCGCTACGTTTCACCGTGACGGTGAGAACTTGACCGTCGTTTTGTATTTCATGAGGCACGTTATACAAGCCACTGGTATCATGAAGGATGACCTCTGATGCGATGTGAGGATTAAATTGTCCCTCAAACCGCCAAACGCTGGTCATGTTTCCACCGTCAATTACGCAGCTGACGGTGTGAGATACCACGATGTTGGA
GFP-Blasticidin Cassette:
5’GTGTGTCAGTTAGGGTGTGGAAAGTCCCCAGGCTCCCCAGGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCTGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCCTGGCCCACCCTCGTGACCACCCTGACCTACGGCGTGCAGTGCTTCAGCCGCTACCCCGACCACATGAAGCAGCACGACTTCTTCAAGTCCGCCATGCCCGAAGGCTACGTCCAGGAGCGCACCATCTTCTTCAAGGACGACGGCAACTACAAGACCCGCGCCGAGGTGAAGTTCGAGGGCGACACCCTGGTGAACCGCATCGAGCTGAAGGGCATCGACTTCAAGGAGGACGGCAACATCCTGGGGCACAAGCTGGAGTACAACTACAACAGCCACAACGTCTATATCATGGCCGACAAGCAGAAGAACGGCATCAAGGTGAACTTCAAGATCCGCCACAACATCGAGGACGGCAGCGTGCAGCTCGCCGACCACTACCAGCAGAACACCCCCATCGGCGACGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGTCCGCCCTGAGCAAAGACCCCAACGAGAAGCGCGATCACATGGTCCTGCTGGAGTTCGTGACCGCCGCCGGGATCACTCTCGGCATGGACGAGCTGTACAAGGCCAAGCCTTTGTCTCAAGAAGAATCCACCCTCATTGAAAGAGCAACGGCTACAATCAACAGCATCCCCATCTCTGAAGACTACAGCGTCGCCAGCGCAGCTCTCTCTAGCGACGGCCGCATCTTCACTGGTGTCAATGTATATCATTTTACTGGGGGACCTTGTGCAGAACTCGTGGTGCTGGGCACTGCTGCTGCTGCGGCAGCTGGCAACCTGACTTGTATCGTCGCGATCGGAAATGAGAACAGGGGCATCTTGAGCCCCTGCGGACGGTGCCGACAGGTGCTTCTCGATCTGCATCCTGGGATCAAAGCCATAGTGAAGGACAGTGATGGACAGCCGACGGCAGTTGGGATTCGTGAATTGCTGCCCTCTGGTTATGTGTGGGAGGGCTAAAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTA 3’
HR-template for STOPx3-HindIII Insertion into GFP:
5’ CTCTGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAATGGTGAGCAAGGGCGAGGAGCTGTTAATAATAAAAGCTTTGGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAGGGCGATGCCACCTACGGCA 3’
HR template for repairing GFP-deletion virus:
5’ TCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAATGGTGAGCAAGGGCGAGGAGCTGTTCACCGGGGTAGTGCCCATCCTGGTCGAGCTGGACGGCGACGTAAACGGCCACAAGTTCAGCGTGTCCGGCGAGGGCGAG 3’
PCR Primers.
US34/TRS1 left homology forward primer:
5’ CTC TAG AAC TAG TGG ATC CCC CGG GCT GCA CGG ACC GAG CCA GAG GGA CCT TCG CTT GGC 3’.
US34/TRS1 left homology reverse primer:
5’ TGG GGA CTT TCC ACA CCC TAA CTG ACA CAC ACC GTT GTA CGC ATT ACA CGG GTT TCG TTC 3’ US34/TRS1
EGFP/BSDR forward primer:
5’ GAA CGA AAC CCG TGT AAT GCG TAC AAC GGT GTG TGT CAG TTA GGG TGT GGA AAG TCC CCA 3’
US34/TRS1 EGFP/BSDR reverse primer:
5’ CTG TAC TGC AAG ACG CAT GAT TAG CAA TAT TAA GAT ACA TTG ATG AGT TTG GAC AAA CCA 3’
US34/TRS1 right homology forward primer:
5’ CTG TAC TGC AAG ACG CAT GAT TAG CAA TAT TAA GAT ACA TTG ATG AGT TTG GAC AAA CCA 3’
US34/TRS1 right homology reverse primer:
5’ GAC GGT ATC GAT AAG CTT GAT ATC GAA TTC ACT TGG CCA CGG TGG TTA TGG AGG CCG GCG 3’
US6/7 left homology forward primer:
5’CTC TAG AAC TAG TGG ATC CCC CGG GCT GCA GCC GTG TTG AAG GAA CGC GCT TTT ATT GAG 3’
US6/7 left homology reverse primer:
5’ TGG GGA CTT TCC ACA CCC TAA CTG ACA CAC GTC ATG GTC TGT AAA AGC TGC ATG AAA AAC 3’
US6/7 EGFP/BSDR forward primer:
5’ GTT TTT CAT GCA GCT TTT ACA GAC CAT GAC GTG TGT CAG TTA GGG TGT GGA AAG TCC CCA 3’
US6/7 EGFP/BSDR reverse primer:
5’ CTA TCG TAA CAA ATA TAT GGA GTT TGT GTA TAA GAT ACA TTG ATG AGT TTG GAC AAA CCA 3’
US6/7 right homology forward primer:
5’ TGG TTT GTC CAA ACT CAT CAA TGT ATC TTA TAC ACA AAC TCC ATA TAT TTG TTA CGA TAG 3’
US6/7 right homology reverse primer:
5’ GAC GGT ATC GAT AAG CTT GAT ATC GAA TTC TCC AAC ATC GTG GTA TCT CAC ACC GTC AGC 3’
Optimized CRISPR-mediated recombineering protocol Preparation of sgRNA-Cas9 pLentiCRISPRv2 Expression Vector
Adapted from (Sanjana et al., 2014; Shalem et al., 2014).
-
For _targeting a single locus, we recommend designing three sets of guide RNAs using the online webtool at https://zlab.bio/guide-design-resources. Guide RNAs can be ordered from IDT as oligos with the following template:
- 5’ CACCG------------------−−3’
- 3’ C--------------------CAAA 5’
where dashes represent the _target sequence, and underlined portions the BsmB1 cut sites for clonal insertion.
-
Five micrograms of pLentiCRISPRv2, which enables constitutive co-expression of Cas9 and sgRNA, are digested with BsmB1, as outlined below.
5ug pLentiCRISPRv2 3uL 10X Buffer 3.1 (NEB) 3uL BsmB1 (NEB) To 30uL ddH2O Digest at 55 degrees for 2 hours, then add 3 uL Calf Intestinal Phosphatase (CIP, NEB) and incubate for another 1–2 hours (at 55C?). Purify 10–12kb band on 2% agarose gel.
-
Dilute each pair of oligos to 100uM in water, and phosphorylate and anneal in a thermocycler as follows:
1uL 100uM Oligo 1 1uL 100uM Oligo 2 1uL 10X T4 Ligation Buffer 6.5uL ddH2O 0.5uL T4 PNK Incubate at 37 degrees Celsius for 30 min, 95 degrees for 5 min, then ramp down to 25 degrees at 5 degrees/min.
Dilute annealed reaction mixture 1:200 in H2O or EB Buffer (Qiagen).
-
Prepare a ligation reaction with the following conditions:
50ng digested pLentiCRISPRv2 plasmid from step 2 1uL Diluted oligo duplex reaction mix 5uL 2X Quick Ligase Buffer To 10uL ddH2O 1uL Quick Ligase (NEB) It is also recommended that a negative control ligation reaction be performed in which no oligo duplex is added. Incubate ligations at room temperature for 10 minutes, and transform into competent stbl3 E. coli. We’ve also had success with transforming other E. coli strains, including DH10B.
Lentiviral Transfection and Transduction of HCMV Host Cell Line (MRC5s)
Day 1:
One day prior to transfection, split a confluent 10 cm dish of 293Ts 1:8 in 10 mL DMEM. Ideally cells should be 30–40% confluent before transfection. Three 10 cm dishes of 293Ts will be required for each 10 cm dish of MRC5s to be transduced.
Day 2:
For each plate of 293Ts to be transfected, combine the following in a 15 mL conical tube:
600 uL PBS
2.4 ug psPax2
0.25 ug pVSVg
2.6 ug pLentiCRISPRv2 / gRNA ligation mixture
Incubate mixture at room temperature for 5 minutes.
Add 18 ul FuGene (Promega) to each conical and incubate at room temperature for 20–30 minutes.
Finally, at the edge of each plate, add 600uL of Fugene/DNA/PBS complex dropwise to avoid dislodging cells from dish. Gently rock the plates back and forth, and incubate overnight at 37C, 5% CO2.
Day 3 (BSL2+):
Aspirate media from each plate of transfected 293Ts, and replace with 4 mL fresh DMEM.
Additionally, seed 3–4e5 MRC5s in the desired number of 10 cm plates to be transduced. Be sure to include an extra dish of MRC5 for a non-transduced control.
Day 4 (BSL2+):
For each 10 cm plate of MRC5s to be transduced, aspirate medium and immediately add lentivirus collected from one dish of 293Ts that had been previously filtered through a 0.45 um filter. Then, add polybrene to 10 ug/mL. Incubate MRC5s with lentivirus at at 37C, 5% CO2 for 3–4 hours, aspirate media and repeat incubation with filtered lentivirus collected from the second dish of transfected 293Ts, omitting polybrene this time. Allow the plate to sit for an additional 3–4 hours, then repeat transduction a third time with filtered lentivirus collected from the last plate of 293Ts. Incubate transduced cells with lentivirus overnight.
Day 5 (BSL2+):
From each dish of transduced MRC5s, aspirate lentiviral inoculum, and add fresh DMEM.
Day 7:
If sufficiently confluent (>80%), passage cells 1:3 into three 10 cm dishes with DMEM and 1ug/mL puromycin. The non-transduced control cells should be passaged and placed under selection at this step as well. Puromycin selection is usually complete after 3–4 days, as assessed by complete toxicity of non- transduced MRC5s.
If cells are insufficiently confluent, they can be split at lower dilutions or be allowed to fill in for several days before splitting.
Day 10+:
Once selected for, transduced cells can be used immediately or frozen and cryogenic stored.
WT HCMV Infection and Electroporation of Homologous Recombination (HR) Template
To maximize the amount of HR template delivery per cell and reduce the amount of virus used, we recommend that host cells be grown in a 6-well plate prior to infection and electroporation.
Day 1:
Cells should ideally be 50–80% confluent prior to infection. Incubate cells with WT HCMV in DMEM for 2 hours at an MOI of 0.25 (approximately 6e4 pfu in a confluent dish). Replace viral inoculum with fresh medium and incubate overnight.
Day 2:
Approximately 12 hours after infection, trypsinize, inactivate serum and resuspend cells as usual. Centrifuge cells at 1500 RPM for 5 min to pellet. Resuspend cells in 80 uL of the following solution: 140mM sodium phosphate buffer, 5 mM MgCl2, 5 mM KCl, pH 7.2. Transfer cells to a 4mm electroporation cuvette and add 30uL 10uM ssODN or 5–10ug plasmid HR template. Tap to mix the DNA/cell suspension.
Electroporate with the following settings using an exponential decay pulse: 150V, 960uF. We’ve found 150 volts to be an effective middle ground between maximizing transfection and minimizing cell death, although specific electroporation conditions may need to be empirically optimized.
Immediately transfer the electroporated cells into 1 well of a 6-well dish with 2mL fresh pre-warmed DMEM. Swirl the plate to disperse the cells, and incubate overnight.
Day 3:
Change medium on the electroporated cells with fresh warmed DMEM.
Day 6:
To harvest virus, scrape cells in their medium and transfer to a 15 ml conical. Sonicate 3 times, for 30 seconds each time, vortex gently, then centrifuge at 2500 rpm for 5 minutes. Transfer the viral supernatants to cryovials and discard the pellet. For storage, viral supernatants should be first frozen on dry ice, then transferred to liquid nitrogen (not −80C?).
Plaque Assays and Isolating Viral Mutants
Per stock of virus, plate and grow to at least 70% confluence a single 12-well plate of MRC5s.
-
Using one deep-well 96-well plate for every six stocks, add 900 uL DMEM to each well in rows A-F. Each stock will be serially diluted in duplicate according to the chart below.
Add 100uL of viral stock to the first well containing 900 uL (corresponds to row A in the layout above). This represents the 10−1 dilution.
Using a multichannel pipettor, pipette the first row up and down to mix well then add 100uL of the first dilution to the second row. This represents the 10−2 dilution. Repeat these steps for the remaining dilutions making sure to changed tips in between each dilution.
Working with one 12 well plate of MRC5 at a time, aspirate media from cells. For a given viral stock, immediately add 500uL of each dilution to the cells Work quickly to avoid letting cells dry out during this step. Incubate cells with viral inocula overnight at 37C, 5% CO2.
-
Twenty four hours later, make gel overlay solution:
- In a 50 mL concical, combine the following:
- Prewarmed to 37 degrees C:
- 25 mL 2× DMEM
- 5 mL FBS
- 500 uL Penicillin / streptomycin stock
- Prewarmed to 55 degrees C:
- 20 mL 2.5% Low Melting Nusieve Agarose prepared in dH2O
Allow mixture to cool slightly before adding to cells. This makes enough overlay for two 12 well plates. Scale overlay recipe up or down as needed.
Aspirate viral inoculum from cells. Add 2 mL of gel overlay to each well. Allow gel overlay to solidify for 30 min at room temperature then return plates to the incubator.
Plaques can be counted starting at 8 days post infection. We recommend taking two counts, at 10 and 12 days.
For isolating individual plaques, we recommend waiting until days 12–14 – when individual plaques in the higher dilutions are visible with the naked eye. At this point, single plaques can be picked by piercing the gel with a small pipette tip and aspirating the plaque of interest after carefully dislodging it with the tip. Dispense the pipette contents into a microcentrifuge tube containing 500 uL DMEM, sonicate for 30 seconds, and incubate overnight with MRC5s previously seeded in a 12 well plate. Virus from single plaques can sun subsequently be amplified and genotyped via PCR (see Materials and Methods – Viral/Cellular DNA extraction).
Highlights.
CRISPR-mediated recombination can be utilized to efficiently mutate the Human Cytomegalovirus genome
The speed of mutant generation is an advantage of the current CRISPR-based technique
Acknowledgements
The authors would like to thank all of the members of the Munger Lab for their guidance and support. We would also like to thank Xenia Schafer for critical reading of the manuscript and Luis Martinez-Sobrido for the GFP-BSD sequences. This work was supported by grants to JM including an NIH R01 (AI081773) and a Research Scholar Grant from the American Cancer Society (RSG-15–049-01-MPC). MK was supported by the URMC PREP program funded by an R25 award (GM064133).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azevedo LS, Pierrotti LC, Abdala E, Costa SF, Strabelli TMV, Campos SV, Ramos JF, Latif AZA, Litvinov N, Maluf NZ, Caiaffa Filho HH, Pannuti CS, Lopes MH, dos Santos VA, Linardi C. da C.G., Yasuda MAS, Marques H.H. de S., 2015. Cytomegalovirus infection in transplant recipients. Clin. São Paulo, Brazil) 70, 515–23. 10.6061/clinics/2015(07)09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron KK, Fyfe JA, Stanat SC, Leslie LK, Sorrell JB, Lambe CU, Coen DM, 1986. A human cytomegalovirus mutant resistant to the nucleoside analog 9-{[2-hydroxy-1-(hydroxymethyl)ethoxy]methyl}guanine (BW B759U) induces reduced levels of BW B759U triphosphate. Proc. Natl. Acad. Sci. U. S. A 83, 8769–8773. 10.1073/pnas.83.22.8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R, 2015. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol 33, 543–548. 10.1038/nbt.3198 [DOI] [PubMed] [Google Scholar]
- Damato EG, Winnen CW, 2002. Cytomegalovirus Infection: Perinatal Implications. J. Obstet. Gynecol. Neonatal Nurs 31, 86–92. [DOI] [PubMed] [Google Scholar]
- Janoly-Dumenil A, Rouvet I, Bleyzac N, Morfin F, Zabot MT, Tod M, 2012. A pharmacodynamic model of ganciclovir antiviral effect and toxicity for lymphoblastoid cells suggests a new dosing regimen to treat cytomegalovirus infection. Antimicrob. Agents Chemother 56, 3732–3738. 10.1128/AAC.06423-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Li H, Hao M, Xiong D, Luo Y, Huang C, Yuan Q, Zhang J, Xia N, 2016. Increasing the Efficiency of CRISPR/Cas9-mediated Precise Genome Editing of HSV-1 Virus in Human Cells 6, 34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bozzella M, Seluanov A, Gorbunova V, 2008. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst). 7, 1765–1771. 10.1016/j.dnarep.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes A, Yu D, 2012. Human Cytomegalovirus: Bacterial Artificial Chromosome (BAC) Cloning and Genetic Manipulation. Curr. Protoc. Microbiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan P, Razonable RR, 2013. Cytomegalovirus infections in solid organ transplantation: A review. Infect. Chemother 45, 260–271. 10.3947/ic.2013.45.3.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revello MG, Zavattoni M, Furione M, Fabbri E, Gerna G, 2006. Preconceptional primary human cytomegalovirus infection and risk of congenital infection. J. Infect. Dis 193, 783–7. 10.1086/500509 [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F, 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F, 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (80-. ). 343, 84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song F, Stieger K, 2017. Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks. Mol. Ther. - Nucleic Acids 7, 53–60. 10.1016/j.omtn.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staras SAS, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ, 2006. Seroprevalence of cytomegalovirus infection in the United States, 1988–1994. Clin. Infect. Dis 43, 1143–51. 10.1086/508173 [DOI] [PubMed] [Google Scholar]
- Studies of Ocular Complications of AIDS Research Group, in C. with the A.C.T.G., 1992. Mortality in Patients with the Acquired Immunodeficiency Syndrome Treated with either Foscarnet or Ganciclovir for Cytomegalovirus Retinitis. N. Engl. J. Med 326, 213–220. 10.1056/NEJM199201233260401 [DOI] [PubMed] [Google Scholar]
- Wallace JM, Hannah J, 1987. Cytomegalovirus pneumonitis in patients with AIDS. Findings in an autopsy series. Chest 92, 198–203. 10.1378/chest.92.2.198 [DOI] [PubMed] [Google Scholar]