Abstract
Mutations in genes encoding related proteins, termed presenilin 1 (PS1) and presenilin 2 (PS2), are linked to the majority of cases with early-onset familial Alzheimer’s disease (FAD). To clarify potential function(s) of presenilins and relationships of presenilin expression to pathogenesis of AD, we examined the expression of PS1 and PS2 mRNA and PS1 protein in human and mouse. Semi-quantitative PCR of reverse-transcribed RNA (RT-PCR) analysis revealed that PS1 and PS2 mRNA are expressed ubiquitously and at comparable levels in most human and mouse tissues, including adult brain. However, PS1 mRNA is expressed at significantly higher levels in developing brain. In situ hybridization studies of mouse embryos revealed widespread expression of PS1 mRNA with a neural expression pattern that, in part, overlaps that reported for mRNA encoding specific Notch homologs.In situ hybridization analysis in adult mouse brain revealed that PS1 and PS2 mRNAs are enriched in neurons of the hippocampal formation and entorhinal cortex. Although PS1 and PS2 mRNA are expressed most prominently in neurons, lower but significant levels of PS1 and PS2 transcripts are also detected in white matter glial cells. Moreover, cultured neurons and astrocytes express PS1 and PS2 mRNAs. Using PS1-specific antibodies in immunoblot analysis, we demonstrate that PS1 accumulates as ∼28 kDa N-terminal and ∼18 kDa C-terminal fragments in brain. Immunocytochemical studies of mouse brain reveal that PS1 protein accumulates in a variety of neuronal populations with enrichment in somatodendritic and neuropil compartments.
Keywords: Alzheimer’s disease, presenilins, development, mRNA expression, protein accumulation, immunocytochemistry
Alzheimer’s disease (AD), the most common progressive type of dementia occurring in adult life, is characterized neuropathologically by the presence of numerous senile plaques and neurofibrillary tangles in cerebral cortex and hippocampus (Wisniewski and Terry, 1973). AD is a genetically heterogeneous disorder (St. George-Hyslop et al., 1992; Schellenberg, 1995). Missense mutations in the presenilin 1 (PS1) gene and a highly related presenilin 2 (PS2) segregate with the vast majority (∼40-50%) of early-onset cases of familial Alzheimer’s disease (FAD; Alzheimer’s Disease Collaborative Group, 1995; Campion et al., 1995; Chapman et al., 1995; Levy-Lahad et al., 1995a,b; Pereztur et al., 1995; Rogaev et al., 1995; Sherrington et al., 1995; Wasco et al., 1995; Boteva et al., 1996).
Presenilins 1 and 2 are transmembrane proteins (Rogaev et al., 1995;Sherrington et al., 1995) that exhibit significant homology to theCaenorhabditis elegans spe-4 (∼25% identity; L’Hernault and Arduengo, 1992) and sel-12 (∼50% identity; Levitan and Greenwald, 1995) polypeptides. Neither the normal biological function(s) of presenilins nor the mechanism(s) by which FAD-associated mutations in presenilins cause disease are known. In view of the significant homology between presenilins and sel-12, a molecule that functions in signaling mediated by lin-12 (Levitan and Greenwald, 1995), it has been suggested that presenilins play a role in mammalian cell-fate decisions. Additional functional roles for presenilins in ion gating or membrane organization have been suggested solely on the basis of predicted structures of the molecules (Alzheimer’s Disease Collaborative Group, 1995; Levy-Lahad et al., 1995b; Selkoe, 1995;Sherrington et al., 1995; Slunt et al., 1995; Kovacs et al., 1996). Although the structural similarities between PS1 and PS2 suggest that the molecules play highly related functional roles, highly divergent hydrophilic regions at the N-terminal “head” and the central “loop” domains are likely to mediate cell- or PS-specific functions via differential interaction(s) with other ligands.
Previous studies demonstrated that both PS1 and PS2 are expressed ubiquitously in a variety of human tissues and brain regions (Rogaev et al., 1995; Sherrington et al., 1995). More recently, in situhybridization analysis revealed that both PS1 and PS2 are expressed at highest levels in hippocampus and in cerebellum (Kovacs et al., 1996;Suzuki et al., 1996). However, these previous studies did not provide information regarding the quantitative levels of each presenilin transcript in brain or in systemic organs, and there are apparent disagreements in the cellular specificity of presenilin expression in brain (Kovacs et al., 1996; Suzuki et al., 1996).
To clarify potential function(s) of presenilins and relationships of presenilin expression in the pathogenesis of AD, we examined the expression of PS1 and PS2 mRNA and PS1 protein in human and mouse. Our semi-quantitative RT-PCR studies reveal that PS1 and PS2 mRNAs are expressed at similar levels in most tissues, with minor exceptions.In situ hybridization analysis of mouse embryos at various developmental stages and adult mouse brain indicate that PS1 and PS2 transcripts are widely expressed. In brain, PS1 and PS2 mRNAs are expressed at highest levels in neurons, with somewhat lower levels in glial cells. Immunoblot analysis using two anti-PS1 antibodies raised against nonoverlapping epitopes revealed that PS1 accumulates as proteolytically processed fragments in brain and systemic tissues. Finally, we performed immunocytochemical investigations to identify the cellular and subcellular distributions of PS1 in mouse brain. We document that PS1 is expressed in somatodendritic and neuropil compartments of neurons in the neocortex and hippocampal formation.
MATERIALS AND METHODS
PS1 and PS2 mRNA analysis. Total RNA was isolated by homogenization of selected tissue in 4 m guanidium thiocyanate and centrifugation of the lysates over a 5.7 mcesium chloride cushion (Chirgwin et al., 1979).
The levels of PS1 and PS2 mRNA in mouse and human brain, peripheral tissues, and developmental stages were assayed by PCR of reverse-transcribed RNA (RT-PCR; Chelly et al., 1988; Golde et al., 1990; Slunt et al., 1994). Reverse-transcribed RNA was added to a PCR reaction mixture containing degenerate primers (1 μmeach) and 32P-end-labeled sense primer (10 pm), reaction buffer, and Taq DNA polymerase. For mouse, we used a sense primer, 5′-(A/G)ACGG(G/T)CAGCT(A/C)ATCTACAC-3′, and an antisense primer, 5′-GAT(A/G)AA(C/T)ACCAGGGCCATGAG-3′, to amplify a 386 bp fragment encoding amino acids 110–238 of PS1 and the homologous region of mouse PS2. For human, we used a sense primer, 5′-ATCATGCT(G/C)TTTGT(G/C)CCTG-3′, and an antisense primer, 5′-TTCTCTCCTG(A/G)GC(A/T)GTTTC-3′, to amplify a 588 bp fragment encoding amino acids 83–278 of human PS1 and the homologous region of human PS2. The resulting PCR products were double-digested withPflMI, which cuts only the PS1 cDNA, and NcoI, which cuts only the PS2 cDNA, to generate PS1- and PS2-specific fragments of different length. The fragments were fractionated on 2% Metaphore-agarose gels (FMC Bioproducts, Rockford, ME) and dried. The radioactivity in the PS1 and PS2 cDNAs was quantified by phosphorimaging. To determine the linear range of amplification for both PS1 and PS2 templates, we terminated RT-PCR reactions at 22, 24, 26, 28, and 30 cycles. The linearity of amplification over the cycling range for either PS1 or PS1 cDNAs was analyzed by semilog plots of radioactivity versus cycle numbers (Chelly et al., 1988; Golde et al., 1990; Slunt et al., 1994).
In situ hybridization. Mouse PS1 cDNA corresponding to nucleotides 1758–1945 of mouse PS1 mRNA (Sherrington et al., 1995) was generated by RT-PCR and subcloned into pBluescript KS+ plasmid (Stratagene, La Jolla, CA). The cDNA encoding the loop region of mouse PS2 (homologous to codon 275–334 of human PS2) was generated by PCR, using a partial mouse PS2 as template and subcloned into pBluescript KS+ plasmid (Stratagene). Sense and antisense 33P-labeled riboprobes were generated from each template with a riboprobe kit (Promega, Madison, WI). A partial mouse PS2 cDNA was obtained with a sense primer (5′-CTCTGTGCATGATCGTGGTGG-3′) encoding amino acids 96–102 of human PS2 and an antisense (5′-GAAGAGGAGGAAAGGGGCGT-3′) primer complementary to sequences encoding amino acids 334–340 of human PS2.
Sections (10 μm) cut from either fresh-frozen or paraffin-embedded tissues were mounted onto slides and reacted with either sense or antisense probes (1 ng/slide, ∼106 cpm/ng). Sections were hybridized overnight at 55°C in a buffer containing 50% formamide, 10% dextran sulfate, 0.3 m NaCl, 20 mm Tris, pH 7.5, 10 mm DTT, 5 mm EDTA, 1× Denhardt’s solution and 0.4 mg/ml yeast tRNA, and 1 ng/slide of labeled probe (∼2 × 106 cpm/ng). Slides were washed as follows: 5× SSC (1× SSC = 0.15 m NaCl and 0.015 msodium citrate) and 1 mm DTT at 55°C for 20 min; 50% formamide, 2× SSC, and 1 mm DTT at 60°C for 30 min; 10 mm Tris, pH 7.5, 0.5 m NaCl, and 5 mm EDTA (TNE buffer) at 37°C three times for 10 min each; TNE buffer containing 20 μg/ml RNase A at 37°C for 45 min; TNE buffer at 37°C for 15 min; 50% formamide, 2× SSC, and 1 mm DTT at 60°C for 30 min; 2× SSC at 37°C for 20 min; and 0.1× SSC at 37°C for 20 min. After dehydration, slides were dipped into Kodak emulsion NTB-2 from 3 d to 3 weeks and developed in Kodak D-19. The sections were counterstained lightly with hematoxylin and eosin and coverslipped with Permount.
Primary neuronal and glial cultures. Primary neuronal and glial cultures from mouse neocortex were obtained as described by Hertz et al. (1985) with minor modifications. For neuronal cultures, 16 d mouse embryos were removed asceptically from timed pregnant female mice (129/BL6) and rinsed in 70% ethanol; neocortices were dissected into Hank’s Buffered Saline Solution (HBSS). The cortices were minced into ∼1 mm pieces and treated with 0.2% trypsin in HBSS for 5 min at room temperature. Trypsin was inactivated by addition of Neurobasal medium (Life Technologies, Bethesda, MD) containing 10% heat-inactivated horse serum. The tissue was pelleted by centrifugation, resuspended in B27-Neurobasal medium supplemented with B27 additive (Life Technologies), and cells were dissociated by trituration. Neuronal cells were diluted further in B27-supplemented Neurobasal media and plated at 2 × 105cells/cm2 on poly-d-lysine-coated tissue culture plates. The media was changed every 3 d.
The glial culture, enriched in astrocytes, was established from cortices of 2-d-old mouse pups. The cortices were removed asceptically into HBSS, minced into ∼1 mm pieces, and treated with trypsin as above. Cells were dissociated and plated on plastic tissue culture dishes at 2 × 105 cells/cm2 in DMEM containing 10% fetal calf serum. Total cellular RNA from 7 d neuronal and glial cultures was isolated with Trizol reagent (Life Technologies).
Immunoblot analysis of PS1 protein. Tissue and cell homogenates were prepared in TNE buffer (10 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 5 mm EDTA) containing protease inhibitors (5 mm PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin) and detergents (1% SDS, 0.25% deoxycholate, and 0.25% NP-40). Protein concentrations of each homogenate were determined by bichichonic acid protein assay (Pierce, Rockford, IL). The homogenates were diluted further with the Laemmli sample buffer (Laemmli, 1970).
Protein in the above homogenates was fractioned by SDS-PAGE (Laemmli, 1970) and transferred to nitrocellulose filter membranes (Towbin et al., 1979). PS1-related peptides were detected using Ab14, a rabbit polyclonal antibody generated against amino acids 1–25 of PS1, or αPS1Loop, a rabbit polyclonal antibody generated against amino acids 263–407 of PS1 (the “loop” region between putative transmembrane domains 6 and 7; Sherrington et al., 1995; Thinakaran et al., 1996). To establish the purity of neuronal and glial cultures, we analyzed the homogenates from primary neuronal and glial cultures by Western blotting with an anti-neuron-specific β-tubulin mouse monoclonal antibody (TuJ1; Lee et al., 1990) or anti-glial fibrillary acidic protein (GFAP) rabbit polyclonal antisera (ICN, Costa Mesa, CA). Bound primary antibodies were visualized via125I-conjugated protein A, followed by autoradiography or appropriate horseradish peroxidase-conjugated secondary antibodies and then followed by chemiluminescence detection (Pierce).
Immunocytochemistry. Mice were perfused intra-aortically with PBS followed by 4% paraformaldehyde. Brains were cryoprotected in 30% sucrose. Sections were cut (40 μm) on a sliding microtome and processed immunocytochemically by using the peroxidase/antiperoxidase method with diaminobenzidine as chromogen (Martin et al., 1991;Rothstein et al., 1994). To enhance immunocytochemical staining, we pretreated sections by boiling them in distilled water for 3 min before permeabilization in 0.4% Triton-X 100 and blocking them in 4% normal goat serum. PS1 immunoreactivity was visualized with affinity-purified αPS1Loop antibodies or Ab14 antiserum raised against a GST fusion protein containing human PS1 amino acids 263–407 or a synthetic peptide corresponding to human PS1 amino acids 1–25, respectively (Thinakaran et al., 1996). αPS1Loop antibodies were used at a concentration of 0.82 μg/ml, whereas Ab14 antisera was used at a dilution of 1:1000. For preadsorption controls, αLoopPS1 or Ab14 antibodies were incubated with 25 μg/ml GST fusion proteins or synthetic peptide antigen, respectively, for 16 hr before immunocytochemistry.
RESULTS
Levels of PS1 and PS2 mRNAs in human and murine tissues
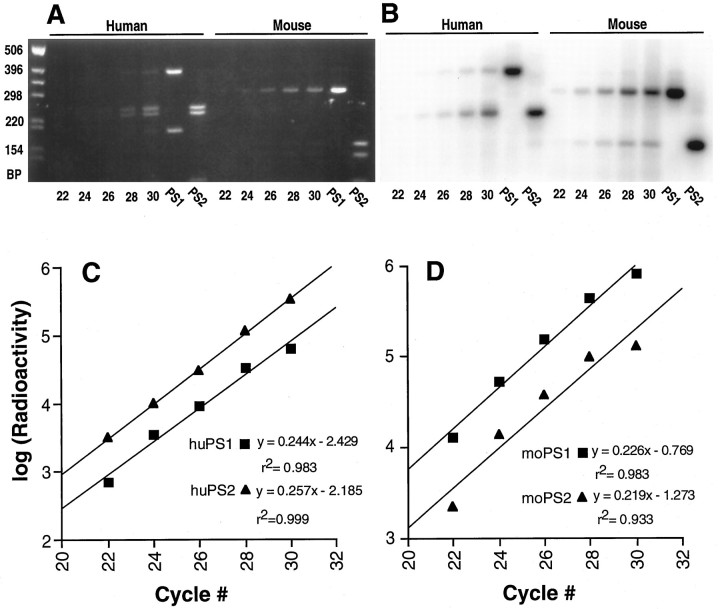
Earlier Northern blotting studies revealed that, although PS1 and PS2 transcripts are expressed in a variety of human tissues, PS1 mRNA seemed to be over-represented considerably, relative to PS2 mRNA (Rogaev et al., 1995). However, differences in probe-specific activities and hybridization stringencies potentially could confound the interpretation of these studies. To estimate the relative steady-state levels of PS1 and PS2 mRNA in various human and mouse tissues, we used a semi-quantitative RT-PCR strategy used previously to examine the relative levels of dystrophin and aldolase mRNA in human muscle and nonmuscle tissue (Chelly et al., 1988), the relative levels of alternatively spliced amyloid precursor protein (APP) transcripts in human tissues (Golde et al., 1990), the relative levels of alternatively spliced APP transcripts in rat dorsal root ganglia (Sisodia et al., 1993), and the relative levels of APP and APLP2 mRNA in mouse tissues (Slunt et al., 1994). To establish optimal conditions for RT-PCR analysis, we reverse-transcribed human brain or mouse spinal cord mRNA with the random hexamer primers and subjected the resulting cDNA products to PCR amplification with degenerate primer pairs that hybridize to highly conserved sequences within PS1 and PS2 cDNAs. The degenerate primers were chosen to generate identical-length products from either PS1 or PS2 templates. Double digestion of resulting products with PflMI, which cuts only PS1 cDNA, andNcoI, which cuts only PS2 cDNA, generates a PS1- or a PS2-specific restriction fragment pattern (Fig.1A,B). Because only the sense primer is 32P-end-labeled, a single PS1- or PS2-specific fragment is detected by autoradiography (Fig. 1B). Linear regression analysis of the amount of RT-PCR amplified product versus the number of PCR cycles (Fig. 1C,D) defines the linear range and demonstrates that the degenerate primers amplify PS1 and PS2 cDNAs with similar efficiencies. The ratio of PS1/PS2 mRNAs in each tissue sample then can be determined by direct comparison of the levels of cDNA generated at a given cycle within the linear range of amplification. We demonstrate that, for ∼100 ng of RNA subject to reverse transcription, a linear range of PCR amplification of PS1/PS2 cDNA is obtained between 20 and 28 cycles. Notably, analysis of human fetal brain RNA revealed that, although PS1 transcripts predominate (because the y-axis is logarithmic, we estimate PS1 mRNA to be ∼4- to 5-fold higher than PS2 mRNA), the relative levels of PS1 or PS2 cDNA generated from respective mRNA templates across the cycling range were identical. Hence, we demonstrate that two mRNA populations within a one-half order of magnitude in abundance can be compared by the semi-quantitative method used herein. Similarly, the relative levels of cDNA obtained by amplification of PS1 and PS2 mRNA from mouse spinal cord, each at cycles 24, 26, and 28, are comparable despite an approximately eightfold difference in transcript abundance.
Fig. 1.
Quantitative PCR amplification of PS1 and PS2 cDNAs from human brain and mouse spinal cord. A, EtBr-stained gel of PflMI/NcoI double-digested PCR products from human fetal brain and adult mouse spinal cord cDNA after 22, 24, 26, 28, and 30 cycles of amplification. Also shown are PCR products obtained by using PS1 and PS2 cDNA as templates. The marker DNA fragments are indicated in bp.B, Autoradiogram of the gel shown in A.C, D, Semilog plots of radioactivity (density) versus cycle number for the human (C) and mouse (D) PS1 and PS2 fragments shown in B. The linear regression equation for each plot is shown in theinset.
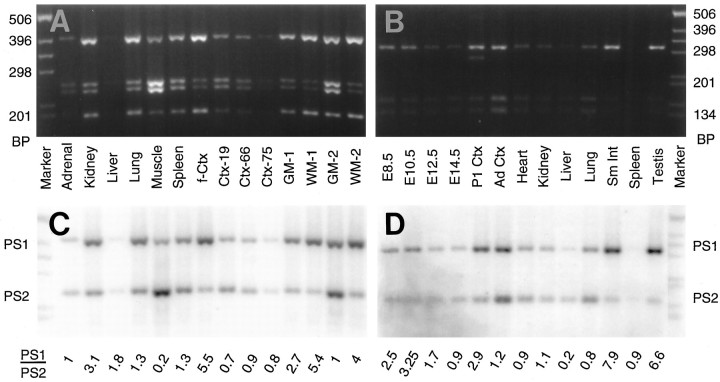
Using optimized RT-PCR conditions (100 ng of reverse-transcribed RNA and 26 cycles), we examined the relative levels of PS1 and PS2 mRNAs in human fetal tissues. Our studies, consistent with earlier RNA blot studies (Rogaev et al., 1995; Sherrington et al., 1995), reveal that both PS1 and PS2 are expressed widely in a variety of tissues (Fig. 2A,C); mRNAs encoding PS1 and PS2 accumulated to similar levels in the adrenal, liver, lung, and spleen. However, PS1 mRNA is considerably higher than PS2 mRNA in fetal brain and kidney, whereas PS2 transcripts are enriched relative to PS1 transcripts in the skeletal muscle.
Fig. 2.
RT-PCR analysis of PS1 and PS2 mRNA in human and mouse tissues. A, EtBr-stained gel ofPflMI/NcoI double-digested PCR product generated by RT-PCR amplification of RNAs from human fetal tissues (adrenal, kidney, liver, lung, skeletal muscle, and spleen), brains from fetus and adults [cortex from a fetus (f-Ctx), a 19 yr old (Ctx-19), a 66 yr old (Ctx-66), and a 75 yr old (Ctx-75)], cortical white matter (WM-1and WM-2), and cortical gray matter (GM-1and GM-2). B, EtBr-stained gel ofPflMI/NcoI double-digested PCR products generated by RT-PCR amplification of RNAs from mouse embryos [embryonic days 8.5 (E8.5), 10.5 (E10.5), 12.5 (E12.5), and 14.5 (E14.5)], brain [neonatal cortex (P1 Ctx) and adult cortex (Ad Ctx)], and adult tissues [heart, kidney, liver, lung, small intestine (Sm Int), spleen, and testis]. C, D, Autoradiogram of the gels shown in A and B, respectively. The ratio of PS1 to PS2 cDNA products, averaged from two independent PCR reactions, is shown at the bottom. The amount of template (reverse-transcribed RNA) was equated for its actin mRNA content (data not shown).
Analysis of PS1 and PS2 mRNA levels in human cortex from a late second trimester fetus, 19 yr old, 66 yr old, and 75 yr old reveals that the relative expression levels of PS1 and PS2 mRNAs change significantly as a function of age. In fetal cortex, the expression level of PS1 mRNA is approximately fivefold higher than that of PS2 mRNA, consistent with studies shown in Figure 1, A andC. However, in young adult and aged brain, PS1 and PS2 mRNAs are expressed at similar levels. Finally, analysis of cortical gray and white matter from a 21 yr old (Fig. 2A,C, lanes GM-1, WM-1) or an 80 yr old (Fig.2A,C, lanes GM-2, WM-2) revealed that PS1 mRNA is enriched in the white matter, as compared with PS2 mRNA, suggesting that glial cells (mostly oligodendrocytes) selectively express more PS1 mRNA than PS2 mRNA. This observation is consistent with earlier Northern blot studies that showed high levels of PS1 mRNA expression in corpus collosum (Rogaev et al., 1995;Sherrington et al., 1995). In gray matter, PS1 and PS2 mRNAs seem to be expressed at similar levels.
Both PS1 and PS2 mRNAs also are expressed widely in adult mouse tissues (Fig. 2B,D). PS1 mRNA levels are significantly higher than PS2 mRNA in small intestine and in testis, whereas PS2 mRNA is four times more abundant than PS1 mRNA in liver. In the remaining adult mouse tissues, PS1 and PS2 mRNAs accumulate to comparable levels. However, during mouse embryonic development, PS1 mRNA expression occurs at earlier developmental stages, with subsequent increases in the relative level of PS2 mRNA (Fig. 2B,D). In day 8.5 and 10.5 whole-mouse embryos, PS1 mRNA levels are approximately twice that of PS2 mRNA. By day 12.5, both PS1 and PS2 transcripts are expressed at equivalent levels, and PS2 mRNA may be expressed at slightly higher levels in 14.5 d embryos. In the cortex of newborn mice, PS1 is expressed at approximately three times the level of PS2 mRNA. Consistent with the results obtained using postnatal human brain (Fig. 2A,C), the level of PS2 mRNA is similar to the level of PS1 mRNA in adult mouse cortex.
These studies demonstrate that, although both PS1 and PS2 transcripts are expressed in many tissues, the relative levels of PS1/PS2 mRNAs are variable between tissues and during brain development. Specifically, the level of PS2 mRNA increases relative to PS1 mRNA during postnatal development of both human and mouse brain, suggesting that the expression of PS1 and PS2 transcripts is regulated differentially during brain maturation. Clearly, analysis of total RNA samples fails to score differences in mRNA accumulation within specific cell types/substructures; hence, our studies are only a first approximation. Nevertheless, although the structural conservation and relatively ubiquitous expression pattern of PS1 and PS2 mRNAs suggest some degree of functional redundancy, differences in relative levels of expression suggest that the biological roles of PS1 and PS2 may differ during development and in adult tissues.
Distribution of PS1 and PS2 transcripts in mouse embryos
The high degree of homology between presenilins and sel-12, aC. elegans protein that mediates cell-fate decisions elicited by Notch/lin-12, has led to the suggestion that PS1/PS2 may function in Notch signaling pathway(s) in mammals. To determine whether PS1 transcripts are expressed in a distinctive pattern overlapping that known for Notch mRNA during development, we examined the expression of PS1 mRNA in mouse embryos by in situ hybridization analysis.
In 10 and 12 d mouse embryos, PS1 mRNA is expressed throughout the neuraxis and developing organs and at comparable levels (Fig. 3A,B,E,F). Notably, in the neuroepithelium, PS1 mRNA is expressed at nearly indistinguishable levels in cells within the ventricular and the marginal zones and may have slightly higher levels of expression in terminally differentiated neurons located in ventral horn and dorsal root ganglia (Fig.3C,D). These observations contrast with several reports demonstrating that the highest levels of Notch mRNA expression in the neuroepithelium are in cells located within the ventricular zone (Reaume et al., 1992; Weinmaster et al., 1992; Lardelli et al., 1994;Lindsell et al., 1995; Williams et al., 1995). Interestingly, analysis of day 16 embryos revealed that PS1 mRNA is expressed by most organs with significantly higher levels of PS1 transcripts in the epithelial cells of the embryonic intestine and skin (Fig.3G,H). Our findings may be significant in view of earlier studies showing high levels of Notch 1 mRNA expression in the latter tissues (Weinmaster et al., 1992).
Fig. 3.
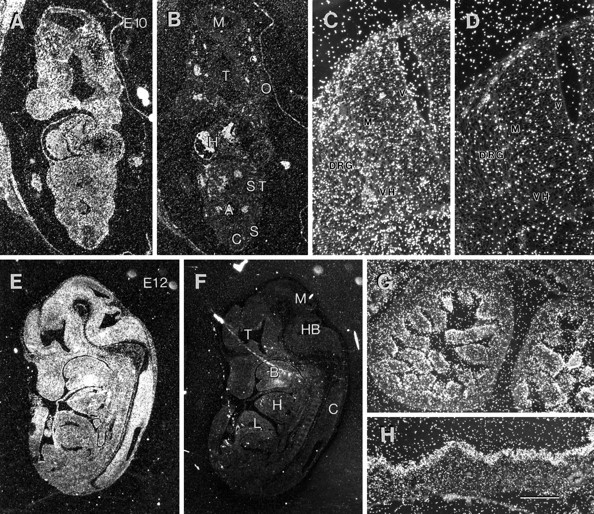
In situ localization of PS1 mRNA in mouse embryos. Paraffin-embedded sections of mouse embryos were processed for in situ hybridization by PS1-specific riboprobes. The silver grains over the sections, representing specific hybridization, were visualized by dark-field microscopy. The sections hybridized with antisense and control sense probes were photographed and reproduced using identical conditions. A, B, PS1 mRNA expression in E10 mouse embryos. Sections from E10 mouse embryos probed with antisense (A) and sense (B) mouse PS1 riboprobes reveal the presence of PS1 mRNA throughout the embryo. A, Aorta; C, spinal cord;H, heart; M, mesencephalon;O, optic vesicle; S, somite;ST, septum transversum; T, telencephalon.C, D, Detailed view of the spinal neural tube from sections shown in A and B. The antisense-probed section (C) shows higher grain density throughout the neural epithelium, as compared with the section probed with sense probe (D). DRG, Dorsal root ganglion; M, marginal zone; V, ventricular zone; VH, ventral horn. E, F, Sagittal sections of E12.5 mouse embryo hybridized with antisense (E) and sense (F) mouse PS1 riboprobe show PS1 mRNA expression throughout the nervous system and peripheral tissues. B, Brachial arch; C, spinal cord; H, heart; HB, hindbrain;L, liver; M, mesencephalon;T, telencephalon. G, H, Sections of E16.5 mouse embryo show high levels of PS1 mRNA expression in epithelial cells of small intestine (G) and skin (H). Scale bars: C, D, 150 μm;G, H, 300 μm.
Comparison of PS1 and PS2 mRNA expression with APP mRNA expression in adult mouse brain
Although earlier reports show that both PS1 and PS2 mRNAs are expressed in a variety of brain regions (Rogaev et al., 1995;Sherrington et al., 1995), information regarding the regional/cellular specificity of PS1 and PS2 mRNA expression in brain is limited (Kovacs et al., 1996; Suzuki et al., 1996). We have used in situhybridization to define the expression of PS1 and PS2 mRNA in mouse brain. In parallel, the expression of APP mRNA was analyzed in adjacent brain sections to compare and contrast directly the distribution of mRNA of the three major genes implicated in the pathogenesis of FAD.
Consistent with several earlier reports, our analysis of coronal sections through mouse brain at the level of medial dorsal hippocampus revealed that APP mRNA is expressed widely at high levels in most brain regions (Fig. 4A). Adjacent sections hybridized with 33P-labeled RNA probes complementary to PS1 or PS2 mRNA show that presenilin transcripts also are expressed in a variety of brain regions (Fig. 4C,E). Consecutive sections hybridized with sense RNA probes did not show significant hybridization (Fig. 4B,D,F), demonstrating the specificity of the antisense RNA probes. Neurons seem to express PS1 transcripts at the highest levels (Fig. 5A); PS1 mRNA expression is also evident in oligodendrocytes and astrocytes in white matter tracts, although at somewhat lower levels (Fig. 5C). The expression of PS1 or PS2 mRNAs in glial cells in the neuropil could not be determined conclusively using in situ hybridization because of the paucity of the cytoplasm in these cells. Nevertheless, expression of presenilin mRNA and protein has been confirmed in primary cultures of purified astrocytes (see below, Fig.9).
Fig. 4.
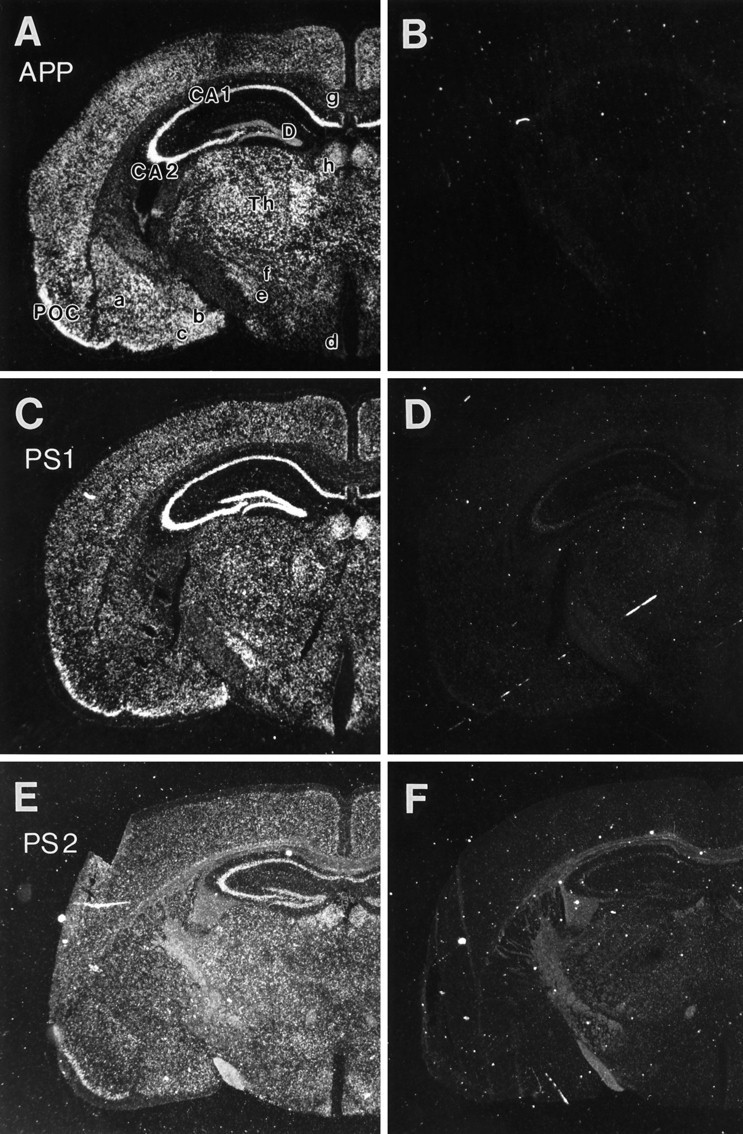
Expression of APP, PS1, and PS2 transcripts in mouse brain. Coronal sections of adult mouse brain at the level of dorsal hippocampus hybridized with antisense riboprobes specific for APP (A), PS1 (C), and PS2 (E). Adjacent sections were hybridized with sense probe (B, D, F) to show the specificity of antisense probes. The silver grains associated with specific transcripts were visualized with dark-field microscopy. The antisense and respective sense control sections were photographed and reproduced under identical conditions. A, B, APP mRNA is highly expressed in most brain regions, with particularly high levels in hippocampal CA fields (CA1, CA2) and primary olfactory cortex (POC). The APP sections were exposed to emulsion for 3 d. PS1 (C, D) and PS2 (E, F) mRNA are widely distributed, but the distribution of cells expressing high (Figure legend continues)levels of each transcript is restricted. The PS1 and PS2 sections were exposed on emulsion for 3 weeks. Longer exposure times were required for photography of sections hybridized with PS2 probes (E, F) because of lower signal levels of the section shown in E. The longer exposure times under dark-field illumination led to higher levels of nonspecific signal associated with the white matter tracts in both antisense (E) and sense (F)-probed sections.CA1, CA2, D, Hippocampal CA-fields and dentate gyrus; POC, primary olfactory cortex;Th, thalamus; a, b, c, lateral, medial, and cortical amygdala; d, arcuate nucleus; e, subthalamic nucleus; f, zona inserta; g, corpus callosum; h, habenulae.
Fig. 5.
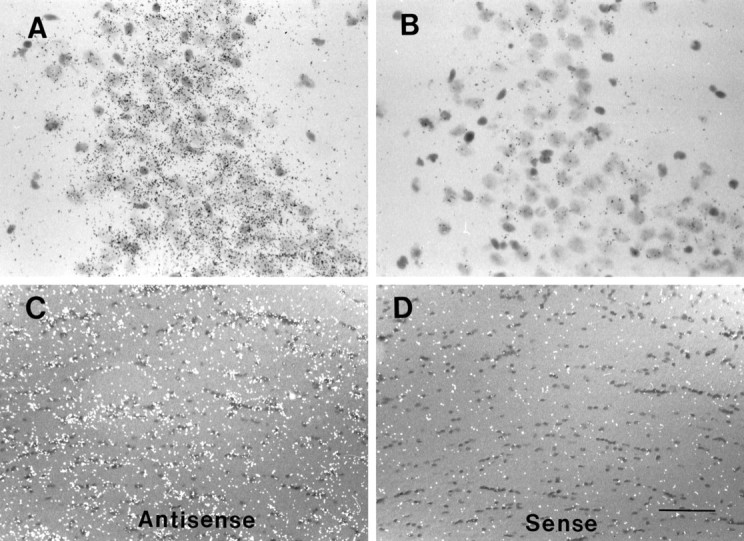
Neuronal and glial expression of PS1 mRNA in brain. A, B, Bright-field image of CA2 region of hippocampus from sections hybridized with antisense (A) or sense (B) PS1 riboprobes. High levels of PS1 mRNA-associated silver grains are most evident in pyramidal neurons.C, D, Corpus callosum from brain sections hybridized with antisense (C) or sense (D) riboprobes shows expression of PS1 mRNA in glial cells of white matter tracts. The silver grains were visualized by differential interference contrast microscopy and are shown as white dots. All of the sections were lightly counterstained with hematoxylin/eosin. Scale bars: A, B, 75 μm; C, D, 150 μm.
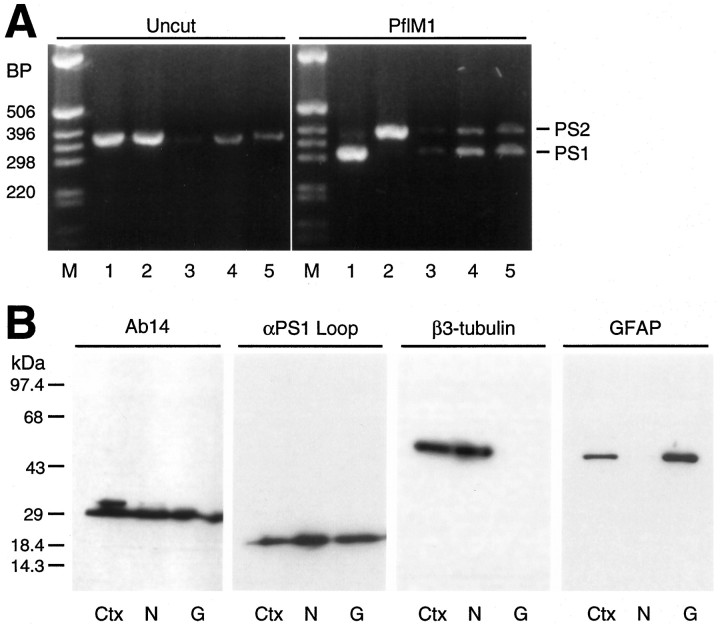
Fig. 9.
Expression of PS1 and PS2 in mouse neurons and glia in culture. A, EtBr-stained gels of PS1 and PS2 products amplified from mouse neuronal or glial RNA. PS1 was distinguished from PS2 by digestion with PflMI.Lane 1, PS1 plasmid; lane 2, PS2 plasmid;lane 3, mouse neocortex; lane 4, primary neuronal culture; lane 5, primary glial culture. Marker fragments (M) are indicated in bps.B, Immunoblot analysis of total SDS-extracts (50 μg) from mouse cortex (Ctx), cultured neurons (N), and cultured glia (G) using Ab14 and αPS1Loop antibodies; for analysis of β3-tubulin and GFAP, 5 μg of total SDS-extracts was assayed. Blots probed with antibodies to PS1 N-terminal fragment (Ab14) or PS1 C-terminal fragment (αPS1Loop) show that expressed PS1 protein accumulates as processed fragments in cultured neurons and glia. Immunoblot analysis of parallel extracts using antibodies to either neuron-specific β-tubulin (β3-tubulin) or GFAP (GFAP) demonstrates the purity of each culture. Molecular weight standards are indicated in kDa.
Despite the widely distributed expression pattern of PS1, PS2, and APP transcripts, not all neuronal populations express all of these transcripts at high levels. In general, APP mRNA is abundant in most neurons, as previously reported (Bendotti et al., 1988; Neve et al., 1988; Spillantini et al., 1989; Hyman et al., 1993; Sola et al., 1993;Tanzi et al., 1993). Similar to APP, hippocampal pyramidal neurons and the neurons of primary olfactory cortex express both PS1 and PS2 at high levels (Fig. 4A–F). However, PS1 and PS2 mRNAs are expressed in dentate granule neurons at levels comparable with neurons in the CA-fields (Fig. 4C–F), a pattern distinct from that observed for APP mRNA (Fig. 4A). In addition, PS1 mRNA also seems to be expressed (albeit at significantly lower levels than in the CA-fields) in the habenula, the medial and cortical amygdala, subthalamic nucleus, and the arcuate nucleus. In contrast, APP mRNAs seem to be expressed in these latter areas at levels comparable to hippocampal and cortical regions (Fig.4A,B).
Coronal sections through the caudate at the level anterior to hippocampus and sections through caudal hippocampus at the level of entorhinal cortex and substantia nigra show high levels of APP mRNA expression in all cortical and subcortical regions (Fig.6A,D), including neurons in the thalamus, lateral amygdyla, and substantia nigra as well as cortical and hippocampal neurons. Examination of PS1 and PS2 mRNA in adjacent sections shows that, in addition to cells in the hippocampus, presenilin transcripts are expressed prominently in neurons of subiculum and entorhinal cortex (Fig. 6E,F). However, unlike APP mRNA, presenilin mRNA expression in nigral neurons and in the lateral amygdala was unremarkable (Fig.6D–F). With the possible exception of the caudate putamen, the general distribution of PS2 mRNA is similar to that observed for PS1 mRNA (Fig. 6B,C).
Fig. 6.
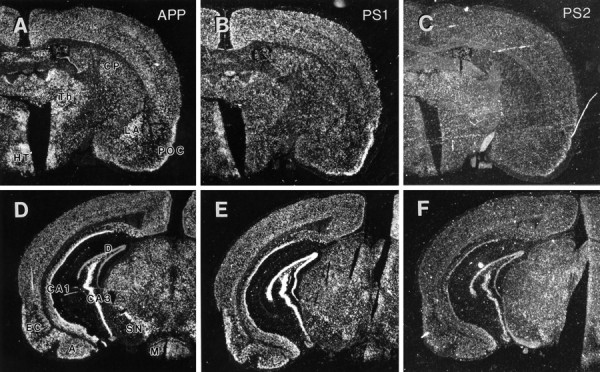
Distribution of APP, PS1, and PS2 mRNA expression at the level of caudate and entorhinal cortex. A–C, Coronal sections through the caudate putamen at the level anterior to hippocampus hybridized with antisense riboprobes specific for APP (A), PS1 (B), or PS2 (C) mRNA. In lateral amygdyla, APP mRNA is expressed at high levels relative to PS1 and PS2 transcripts. Also note that, whereas the level of PS1 mRNA in caudate putamen is qualitatively similar to other cortical regions in this section, PS2 mRNA is much more prominent in the CP relative to neighboring cortical regions. CP, Caudate putamen; HT, hypothalamus; LA, lateral amygdyla, POC, primary olfactory cortex;Th, thalamus. D–F, Coronal section through caudal hippocampus at the level of entorhinal cortex (EC) and substantia nigra (SN) hybridized with antisense riboprobes specific for APP (D), PS1 (E), and PS2 (F). All three transcripts are distributed throughout this coronal section with significant expression of APP, PS1, and PS2 mRNAs in hippocampus, entorhinal cortex, and amygdyla. APP mRNA level is very high in SN neurons, whereas PS1 and PS2 mRNAs are expressed at similar levels in SN and regions adjacent to it.A, Amygdyla; CA1, CA3, the CA-fields of hippocampus; D, dentate gyrus;M, mamillary body.
Finally, analysis of PS1 and PS2 transcripts in the cerebellum shows high levels of presenilin expression in granule cells, Purkinje cells, and neurons in the pons (Fig. 7). Grains over smaller nuclei located in the Purkinje cell layer suggest that PS1 and PS2 mRNAs also may be expressed in Bergmann glia or other supporting cells (Fig. 7). In contrast to the general expression pattern seen with PS1/PS2, the highest level of APP expression in cerebellum is limited to the Purkinje cells and the deep cerebellar nuclei (Sola et al., 1993).
Fig. 7.
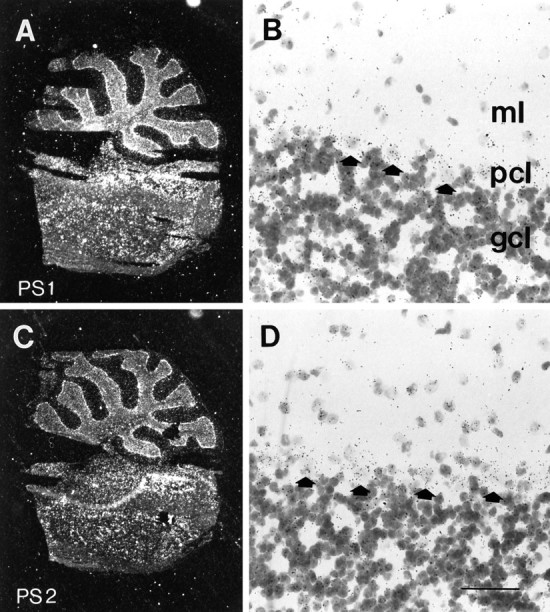
Expression of PS1 and PS2 mRNAs in cerebellum.A, C, Dark-field microscopy images of sagittal section through cerebellum and pons hybridized with antisense riboprobe for PS1 (A) and PS2 (B). B, D, High power view of cerebellar cortex showing PS1 (B) and PS2 (D) mRNA-associated silver grains over granule cell layer (gcl) and Purkinje cells (arrows) in Purkinje cell layer (pcl). Note that grains inpcl are not restricted to Purkinje cells, and the cells in the molecular layer (ml) also show specific hybridization. Scale bar, B, D, 75 μm.
Accumulation of proteolytically processed PS1 derivatives in mouse brain and tissues
To complement our analysis of PS1 mRNA expression, we examined the expression of PS1 protein by immunoblot analysis. We detected PS1 protein in whole SDS-soluble extracts of mouse brain regions and selected non-neural tissues by using a rabbit polyclonal antibody, Ab14, generated against the N-terminal 25 amino acids of PS1 and a rabbit polyclonal antibody, αPS1Loop, generated against a bacterially synthesized glutathione-S-transferase (GST) fusion protein containing amino acids 263–407 of human PS1 (Thinakaran et al., 1996). We previously demonstrated that, in human, monkey, and mouse brain, PS1 accumulates as an N-terminal, Ab14-reactive ∼28 kDa fragment and a C-terminal, αPS1Loop-reactive, ∼18 kDa fragment, supporting the idea that PS1 is subject to processing endoproteolytically in vivo (Thinakaran et al., 1996).
Immunoblot analysis of mouse brain regions, cranial nerves, and peripheral tissues reveals that PS1 accumulates as two principal fragments (Fig. 8). Notably, although the Ab14-reactive ∼28 kDa N-terminal fragment resolves as a doublet with possibly a third minor species, only a single C-terminal fragment is detected in most non-neural tissues. The apparent heterogeneity of the N-terminal fragment suggests that PS1 may be subject to tissue-specific post-translational modifications or, alternatively, be encoded by alternatively spliced transcripts. In support of the latter idea, alternatively spliced human PS1 mRNA has been identified in lymphoblasts, liver, spleen, and kidney (Rogaev et al., 1995; Sahara et al., 1996). In contrast, we detected a single major C-terminal fragment of ∼18 kDa in all mouse tissues examined. On longer exposures of Western blots of brain extracts with both Ab14 and αPS1Loop antibodies, we also observed an ∼43-45 kDa polypeptide that presumably represents full-length PS1 (data not shown).
Fig. 8.
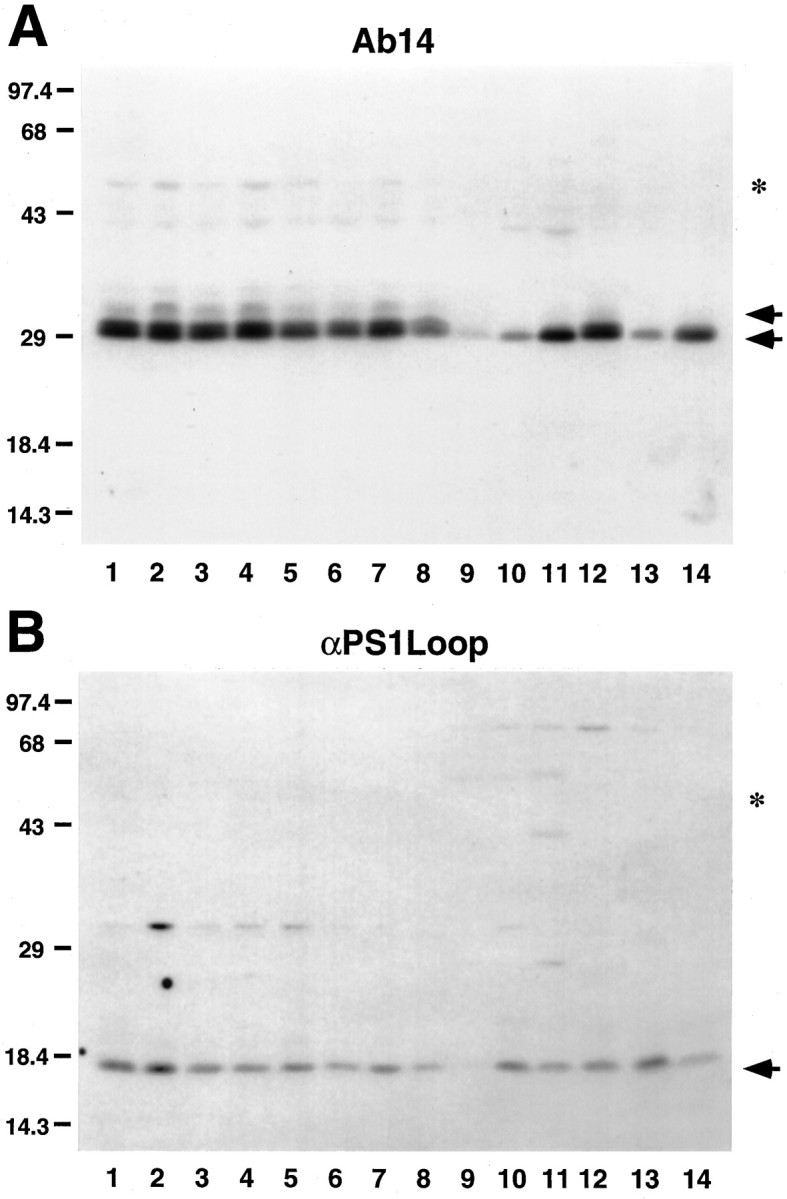
Accumulation of processed PS1 fragments in mouse tissues. SDS-soluble extracts from mouse neural (olfactory bulb,lane 1; caudate putamen, lane 2; thalamus, lane 3; neocortex, lane 4; hippocampus, lane 5; cerebellum, lane 6; brain stem, lane 7; optic nerve, lane 8) and non-neural (heart, lane 9; kidney, lane 10; liver, lane 11; lung, lane 12; small intestine, lane 13; spleen,lane 14) tissues were immunoblotted and probed with PS1-specific Ab14 (A) or αPS1Loop (B) antibodies. Arrows denote specific immunoreactive species, which were competible with peptide (Ab14) or GST fusion protein (PS1Loop; Thinakaran et al., 1996) (data not shown). The predicted mobility of the full-length PS1 is indicated by the asterisk. The molecular mass standards are indicated at left in kDa.A, Ab14 reacts with the N-terminal PS1 fragment at ∼28–30 kDa. Note that, in neural tissues (lanes 1–8), the PS1 N-terminal fragment resolves as a major doublet and a minor species with retarded migration (arrows). With exception of lung (lane 12), only a single Ab14-reactive PS1 fragment is detected in non-neural tissues (lanes 9–14). B, αPS1Loop antibody specifically reacts with an ∼18 kDa PS1 fragment in all tissues.
Our semi-quantitative analysis of total levels of PS1 in mouse neocortex revealed that the PS1 accumulates to <0.001% of total brain protein (Thinakaran et al., 1996). The present analysis clearly shows that, relative to neocortex, PS1 protein accumulates to even lower levels in most brain regions and non-neural tissues. Moreover, very low levels of PS1 protein accumulate in cranial nerves consisting of myelinated axons and oligodendrocytes (Fig. 8).
Expression of PS1 and PS2 in cultured mouse neurons and glia
To confirm the cellular specificities of PS1 and PS2 expression in neural tissues, we examined the expression of PS1 and PS2 mRNAs in primary cultures of mouse cortical neurons and glia by using optimized RT-PCR conditions (Fig. 9A). Our studies clearly show that cultured mouse neurons and glia express similar levels of PS1 and PS2 mRNA (Fig. 9A). To examine the relative levels and the nature of the accumulated PS1 protein SDS-extracts prepared from neurons and glia, we performed immunoblotting with the two anti-PS1 antibodies described above (Fig.8). Immunoblot analysis demonstrated that both neurons and glia accumulate similar levels of N-terminal and C-terminal fragments (Fig.9B). To establish the purity of neuronal and glial cultures, we subjected extracts from each culture to immunoblot analysis with antibodies specific for neuron-specific β-tubulin isotype (Lee et al., 1991) and glial fibrillary acidic protein (GFAP). Our studies confirm that neuronal culture is free of glial cells, and the glial culture is free of neurons (Fig. 9B). Hence, PS1 and PS2 are expressed in neurons and glial cells.
Immunocytochemical localization of PS1 in mouse brain
To date, only a single report has attempted to localize PS1 in brain (Moussaoui et al., 1996). The authors demonstrated that an antibody raised against a synthetic peptide corresponding to a region within the PS1 “loop” showed PS1-immunoreactive (PS1-IR) in a wide variety of CNS neurons. However, we have several concerns regarding the specificity of the antibody: first, successful immunoprecipitation of PS1 from an in vitro translation reaction programmed with synthetic PS1 mRNA was the only biochemical assay for antibody specificity; second, immunocytochemical staining was only competible with extremely high levels of peptide (1 mg/2 μl antiserum); and third, Western blot analyses of brain homogenates were not presented. With these latter reservations, we initiated immunocytochemical studies to define the cellular and subcellular distribution of PS1 in rodent brain with Ab14 and αPS1Loop antibodies (see above, Figs. 8, 9). Using affinity-purified αPS1Loop antibodies, we demonstrate that PS1 is widely expressed and enriched in neurons; in neocortex, pyramidal and nonpyramidal neuronal cell bodies are highly PS1-IR. Moreover, dendrites in all layers and the neuropil were diffusely positive for PS1 (Fig. 10E). Consistent with ourin situ hybridization studies (Figs. 4, 5, 6), pyramidal neurons in all hippocampal CA subfields and granule cells in the dentate gyrus were strongly PS1-immunoreactive (Fig.10D–F). In addition, the stratum lucidum of CA3 (corresponding to the distribution of mossy fiber terminals) was intensely immunoreactive (Fig. 10D,F), consistent with the localization of PS1 in granule cells of the dentate gyrus. Preadsorption of αPS1Loop antibody with a GST fusion protein containing the PS1 “loop” region fully competed neuronal, somatodendritic, and neuropil PS-IR (Fig. 1C). Using Ab14 antibodies, we observed a pattern of staining nearly indistinguishable to that observed with αLoopPS1 antibody (Fig.10G). Hence, we conclude that the PS1 N- and C-terminal fragments accumulate in similar, if not identical, subcellular compartments in CNS neurons.
Fig. 10.
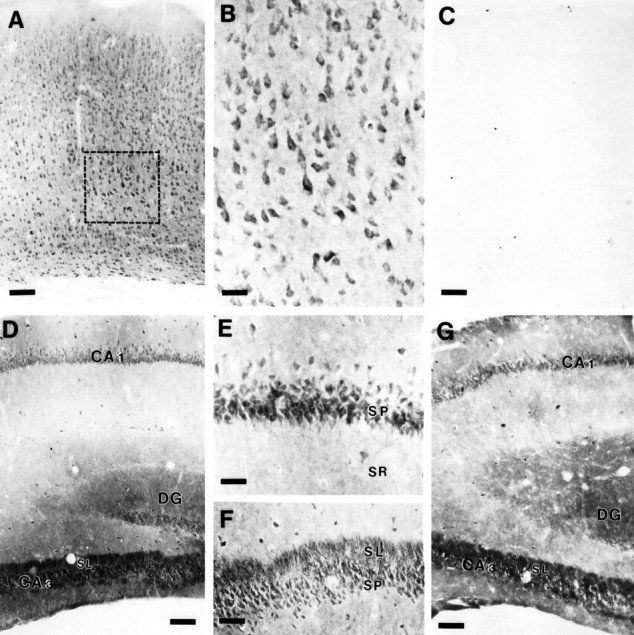
Immunohistochemical localization of PS1 in mouse brain. A, αPS1Loop staining in neocortex. PS1-IR is present in all layers of neocortex. Box delineates area shown at higher magnification in B. Scale bar, 160 μm.B, High power view of αPS1Loop staining in neocortex. PS1-IR is enriched in somatodendritic compartments in neurons, with lighter staining in the neuropil. Scale bar, 53 μm.C, Somatodendritic and neuropil PS1-IR are fully competed by preadsorption of αPS1Loop antibody with GST-PS1 loop fusion protein. Scale bar, 160 μm. D, αPS1Loop staining in the hippocampal formation. PS1-IR is present inCA1, CA3, and dentate gyrus (DG). The cell bodies of pyramidal neurons inCA1 and CA3 and those of granule cells of the DG are intensely PS1-IR. In addition, the stratum lucidum (SL) of CA3, corresponding to the terminal fields of mossy fibers originating from granule cells in theDG, is also intensely PS1-IR. Scale bar, 320 μm.E, Higher magnification of αPS1Loop staining inCA1. PS1-IR is prominent in neuronal cell bodies in the stratum pyramidale (SP) and to proximal dendrites in the stratum radiatum (SR). Scale bar, 64 μm.F, Higher magnification of αPS1Loop staining inCA3. PS1-IR is present in neuronal cell bodies of theSP and prominent in the SL. Scale bar, 160 μm. G, Ab14 staining in the hippocampal formation. PS1-IR is similar to that using αPS1Loop antibody. Scale bar, 280 μm.
DISCUSSION
To address the potential functional role(s) of presenilins during development and to provide a conceptual framework that relates presenilin expression and selective neuronal vulnerability in AD, we examined the expression of presenilins 1 and 2 during embryonic development and in mature brain. The present studies provide new information concerning the expression of presenilin transcripts and protein, the potential biological function(s) of PS, and the roles of PS in the pathogenesis of AD.
First, and in contrast with earlier Northern blotting studies concluding that PS2 mRNA is expressed at low levels relative to PS1 mRNA, our quantitative RT-PCR studies revealed that, although both PS1 and PS2 transcripts are expressed in many tissues, the relative levels of PS1/PS2 expression can vary significantly between tissues and during brain development. Thus, although the structural conservation and relatively ubiquitous expression pattern of PS1 and PS2 mRNAs suggest some degree of functional redundancy, differences in relative levels of expression suggest that PS1 and PS2 may play different biological roles in tissue- or development-specific processes.
Second, in view of the demonstration that sel-12, a C. elegans homolog of presenilins, mediates developmental cell-fate decisions elicited by lin-12 (Levitan and Greenwald, 1995), we examined the expression of presenilins during embryonic development. Although elevated levels of PS1 mRNA in intestine and in skin of E16 mouse embryos overlap the expression pattern reported for Notch 1 mRNA (Weinmaster et al., 1992), the general spatial and temporal expression patterns of presenilins mRNAs do not coincide directly with the expression patterns of any specific member of the known mammalian Notch homologs (Reaume et al., 1992; Weinmaster et al., 1992; Lardelli et al., 1994; Lindsell et al., 1995; Williams et al., 1995). For example, although Notch mRNA expression in the developing neural tube is limited to cells near the ventricular zone, PS1 mRNA is expressed in neuroblasts in the ventricular zones as well as in terminally differentiated neurons. Our demonstration that PS1 mRNA is expressed in neuroblasts suggests that PS1 can play a role in mammalian Notch signaling. In support of this view, we have demonstrated that human PS1 can substitute functionally for C. elegans sel-12 proteinin vivo (D. Levitan, S. Sisodia, and I. Greenwald, unpublished observation). Nevertheless, the ubiquitous expression of PS1 in terminally differentiated populations of cells indicates that PS1 function is not limited to Notch signaling alone.
Third, our in situ hybridization analysis of adult mouse brain reveals that both PS1 and PS2 transcripts are expressed in a variety of neuronal populations. Moreover, prominent expression of presenilin transcripts in neurons supports the view that mutations in presenilin genes cause FAD by directly compromising neuronal function. However, in view of significant levels of presenilin mRNA expression in glial cells, pathogenic mechanisms involving glial cells cannot be excluded. Most notable is our finding that neuronal populations known to be at risk in AD coexpress high levels of APP and presenilin transcripts. For example, neurons in the hippocampal CA-fields, neurons of the medial and cortical amygdala, and neocortical neurons express presenilins and APP transcripts at high levels. In contrast, neurons in regions less prone to AD-associated pathology express these transcripts at more variable levels. For example, granule cells in the dentate gyrus and cerebellum express high levels of presenilins mRNA but lower levels of APP mRNA; in contrast, neurons in lateral amygdala and substantia nigra express high levels of APP mRNA but low levels of presenilin transcripts. Thus, our demonstration of high-level presenilin mRNA expression in neurons known to be at risk in AD further supports the view, proposed earlier by Kovacs et al. (1996), that mutations in PS compromise neuronal function and contribute to the pathology of presenilin-linked FAD. At a conceptual level, coexpression of APP and presenilins at high levels in selected neuronal populations raises the intriguing possibility that mutations in presenilins enhance pathogenic processing of APP, hence rendering specific neurons to be more vulnerable in AD. In this regard, elevated levels of highly amyloidogenic β-amyloid 1-42(43) have been reported in the plasma and conditioned medium of fibroblasts from individuals harboring PS1 or PS2 mutations (Scheuner et al., 1996). Notably, recent immunocytochemical studies documented overwhelming amyloid deposition in the brain parenchyma and blood vessels of patients with an FAD-linked PS1 E280A mutation and revealed, for the first time, the presence of ubiquitin-positive neurites surrounding compact amyloid plaques in the cerebellum, particularly in the Purkinje cell layer (Lemere et al., 1996).
Fourth, our studies are the first to examine the expression and distribution of PS1 by using two highly specific antibodies generated against independent epitopes of PS1. We document that PS1 protein accumulates as two proteolytically processed fragments in all tissues examined, similar to our earlier description of PS1 derivatives in human, primate, and rodent brain and brains of transgenic mice expressing human PS1 (Thinakaran et al., 1996). At present, we do not understand the functional significance of proteolytic processing of PS1. Nevertheless, and in view of the paucity of accumulated full-length PS1 and the generality of PS1 processing across tissues, we suggest that PS1 fragments are the relevant functional units.
Finally, our immunocytochemical studies of mouse brain provide strong evidence for the ubiquitous and prominent accumulation of PS1 polypeptides in neurons. PS1-IR in neurons is localized primarily to somatodendritic compartments, but strong neuropil PS1-IR is also evident. The neuropil-specific PS1-IR may correspond to PS1 within axon terminals, a view supported by our demonstration of prominent PS1-IR in the stratum lucidum of CA3, an area enriched in mossy fiber terminals. Alternatively, neuropil-IR may correspond to distal dendrites or to glial processes. In this regard, and despite our demonstration that low levels of PS1 protein accumulate in cranial nerves (Fig.8A,B), PS1 mRNA expression in glial cells (Figs.5B, 9B), and PS1 protein accumulation in cultured glial cells (Fig. 9B), we rarely detected PS1-IR in glial cell bodies in the neuropil. However, in view of findings that certain glial proteins (e.g., glial glutamate transporters; Rothstein et al., 1994) are enriched in the neuropil and do not show appreciable accumulation in cell bodies, it is not unreasonable that PS1 is localized to glial processes. Future immunoelectron microscopy studies are warranted to identify unambiguously the localization of PS1 within neuropil compartments in brain.
Footnotes
This work was supported by National Institutes of Health Grants AG05146 and NS20471 and by grants from the Adler Foundation and the Develbiss Fund. S.S.S. is the recipient of an Alzheimer’s Association Zenith Award. We thank Cornelia Von Koch [Johns Hopkins Medical Institutes (JHMI), Baltimore, MD] for providing mouse embryos, Donna Suresch for technical help with tissue sections, and Dr. Juan Troncoso (JHMI) for helpful discussions.
Correspondence should be addressed to Dr. Sangram S. Sisodia, The Johns Hopkins University School of Medicine, Neuropathology Laboratory, 558 Ross Research Building, 720 Rutland Avenue, Baltimore, MD 21205-2196.
REFERENCES
- 1.Alzheimer’s Disease Collaborative Group. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat Genet. 1995;11:219–222. doi: 10.1038/ng1095-219. [DOI] [PubMed] [Google Scholar]
- 2.Bendotti C, Forloni GL, Morgan RA, O’Hara BF, Oster-Granite ML, Reeves RH, Gearhart JD, Coyle JT. Neuroanatomical localization and quantification of amyloid precursor protein mRNA by in situ hybridization in the brains of normal, aneuploid, and lesioned mice. Proc Natl Acad Sci USA. 1988;85:3628–3632. doi: 10.1073/pnas.85.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boteva K, Vitek M, Mitsuda H, de Silva H, Xu P-T, Small G, Gilbert JR. Mutation analysis of presenilin 1 gene in Alzheimer’s disease. Lancet. 1996;347:130–131. doi: 10.1016/s0140-6736(96)90261-5. [DOI] [PubMed] [Google Scholar]
- 4.Campion D, Flaman JM, Brice A, Hannequin D, Dubois B, Martin C, Moreau V, Charbonnier F, Didierjean O, Tardieu S, Penet C, Puel M, Pasquier F, Ledoze F, Bellis G, Calenda A, Heilig R, Martinez M, Mallet J, Bellis M, Clergetdarpoux F, Agid Y, Frebourg T. Mutations of the presenilin 1 gene in families with early-onset Alzheimer’s disease. Hum Mol Genet. 1995;4:2373–2377. doi: 10.1093/hmg/4.12.2373. [DOI] [PubMed] [Google Scholar]
- 5.Chapman J, Asherov A, Wang N, Treves TA, Korczyn AD, Goldfarb LG. Familial Alzheimer’s disease associated with S182 codon 286 mutation. Lancet. 1995;346:1040. doi: 10.1016/s0140-6736(95)91723-3. [DOI] [PubMed] [Google Scholar]
- 6.Chelly J, Kaplan JC, Maire P, Gautron S, Kahn A. Transcription of the dystrophin gene in human muscle and non-muscle tissues. Nature. 1988;333:958–960. doi: 10.1038/333858a0. [DOI] [PubMed] [Google Scholar]
- 7.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 8.Golde TE, Estus SE, Usiak M, Yunkin LH, Younkin SG. Expression of β amyloid protein precursor mRNAs: recognition of a novel alternatively spliced form and quantitation in Alzheimer’s disease using PCR. Neuron. 1990;4:253–267. doi: 10.1016/0896-6273(90)90100-t. [DOI] [PubMed] [Google Scholar]
- 9.Hertz L, Juurlink H, Szuchet S, Walz W. Cell and tissue cultures. In: Boulton AA, Baker GB, editors. Neuromethods 1. Humana; Clifton, NJ: 1985. pp. 117–167. [Google Scholar]
- 10.Hyman BT, Wenniger JJ, Tanzi RE. Nonisotopic in situ hybridization of amyloid beta protein precursor in Alzheimer’s disease: expression in neurofibrillary tangle-bearing neurons and in the microenvironment surrounding senile plaques. Mol Brain Res. 1993;18:253–258. doi: 10.1016/0169-328x(93)90197-w. [DOI] [PubMed] [Google Scholar]
- 11.Kovacs DM, Fausett HJ, Page KJ, Kim T-W, Moir RD, Merriam DE, Hollister RD, Hallmark OG, Mancini R, Felsenstein KM, Hyman BT, Tanzi RE, Wasco W. Alzheimer-associated presenilins 1 and 2: neuronal expression in brain and localization to intracellular membranes in mammalian cells. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 12.Laemmlie UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Lardelli M, Dahlstrand J, Lendahl U. The novel notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 14.Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter AF. Expression and post-translational modification of a neuron-specific beta-tubulin isotope during chick embryogenesis. Cell Motil Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- 15.Lemere CA, Lopera F, Kosik KS, Ossa J, Saido TC, Ruiz A, Martinez A, Madrigol L, Hincapie L, Arango JC, Selkoe DJ, Arango JC. Immunocytochemical characterization of plaques, tangles, and vascular amyloid in the brains of 2 FAD patients with an identified PS1 mutation. Neurobiol Aging [Suppl] 1996;17:S17. [Google Scholar]
- 16.Levitan D, Greenwald I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer’s disease gene. Nature. 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 17.Levy-Lahad E, Wijsman EM, Nemens E, Anderson L, Goddard KAB, Weber JL, Bird TD, Schellenberg GD. A familial Alzheimer’s disease locus on chromosome 1. Science. 1995a;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 18.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu C-E, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu Y-H, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995b;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 19.L’Hernault SW, Arduengo PM. Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J Cell Biol. 1992;119:55–68. doi: 10.1083/jcb.119.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch 1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 21.Martin LJ, Sisodia SS, Koo EH, Cork LC, Dellovade TL, Weidmann A, Beyreuther K, Masters C, Price DL. Amyloid precursor protein in aged nonhuman primates. Proc Natl Acad Sci USA. 1991;88:1461–1465. doi: 10.1073/pnas.88.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moussaoui S, Czech C, Pradier L, Blanchard V, Boniei B, Gohin M, Imperato A, Revah F. Immunohistochemical analysis of presenilin-1 expression in the mouse brain. FEBS Lett. 1996;383:219–222. doi: 10.1016/0014-5793(96)00250-5. [DOI] [PubMed] [Google Scholar]
- 23.Neve RL, Finch EA, Dawes LR. Expression of the Alzheimer amyloid precursor gene transcripts in the human brain. Neuron. 1988;1:667–677. doi: 10.1016/0896-6273(88)90166-3. [DOI] [PubMed] [Google Scholar]
- 24.Pereztur J, Froelich S, Prihar G, Crook R, Baker M, Duff K, Wragg M, Busfield F, Lendon C, Clark RF, Roques P, Fuldner RA, Johnston J, Cowburn R, Forsell C, Axelman K, Lilius L, Houlden H, Karran E, Roberts GW, Rossor M, Adams MD, Hardy J, Goate A, Lannfelt L, Hutton M. A mutation in Alzheimer’s disease destroying a splice acceptor site in the presenilin-1 gene. NeuroReport. 1995;7:297–301. [PubMed] [Google Scholar]
- 25.Reaume AG, Conlon RA, Zirngibl R, Yamaguchi TP, Rossant J. Expression analysis of a Notch homologue in the mouse embryo. Dev Biol. 1992;154:377–387. doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- 26.Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, Mar L, Sorbi S, Nacmias B, Piacentini S, Amaducci L, Chumakov I, Cohen D, Lannfelt L, Fraser PE, Rommens JM, St. George-Hyslop PH. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 27.Rothstein JD, Martin L, Levey A, Dykes-Hoberg M, Jin L, Wu D, Nash N, Kuncl R. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 28.Sahara N, Yahagi Y, Takagi H, Kondo T, Okochi M, Usami M, Shirasawa T, Mori H. Identification and characterization of presenilin I-467, I-463, and I374. FEBS Lett. 1996;381:7–11. doi: 10.1016/0014-5793(96)00054-3. [DOI] [PubMed] [Google Scholar]
- 29.Schellenberg GD. Genetic dissection of Alzheimer disease, a heterogeneous disorder. Proc Natl Acad Sci USA. 1995;92:8552–8559. doi: 10.1073/pnas.92.19.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. The amyloid β protein deposited in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations lined to familial Alzheimer’s disease. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 31.Selkoe DJ. Missense on the membrane. Nature. 1995;375:734. doi: 10.1038/375734a0. [DOI] [PubMed] [Google Scholar]
- 32.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, Tsuda T, Mar L, Foncin J-F, Bruni AC, Montesi MP, Sorbi S, Rainero I, Pinessi L, Nee L, Chumakov I, Pollen D, Brookes A, Sanseau P, Polinsky RJ, Wasco W, Da Silva HAR, Haines JL, Pericak-Vance MA, Tanzi RE, Roses AD, Fraser PE, Rommens JM, St. George-Hyslop PH. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 33.Sisodia SS, Koo EH, Hoffman PH, Perry G, Price DL. Identification and transport of full-length amyloid precursor proteins in rat peripheral nervous system. J Neurosci. 1993;13:3136–3142. doi: 10.1523/JNEUROSCI.13-07-03136.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slunt HH, Thinakaran G, Von Koch C, Lo ACY, Tanzi RE, Sisodia SS. Expression of a ubiquitous, cross-reactive homologue of the mouse β-amyloid precursor protein (APP). J Biol Chem. 1994;269:2637–2644. [PubMed] [Google Scholar]
- 35.Slunt HH, Thinakaran G, Lee MK, Sisodia SS. Nucleotide sequence of the chromosome 14-encoded S182 cDNA and revised secondary structure prediction. Amyloid. Int J Exp Clin Invest. 1995;2:188–190. [Google Scholar]
- 36.Sola C, Mengod G, Ghetti D, Palacios JM, Triarhou LC. Regional distribution of the alternatively spliced isoforms of βAPP RNA transcript in the brain of normal, heterozygous and homozygous weaver mutant mice as revealed by in situ hybridization histochemistry. Mol Brain Res. 1993;17:340–346. doi: 10.1016/0169-328x(93)90020-p. [DOI] [PubMed] [Google Scholar]
- 37.Spillantini MG, Hunt SP, Ulrich J, Goedert M. Expression and cellular localization of amyloid β-protein precursor transcripts in normal human brain and in Alzheimer’s disease. Mol Brain Res. 1989;6:143–150. doi: 10.1016/0169-328x(89)90048-x. [DOI] [PubMed] [Google Scholar]
- 38.St. George-Hyslop PH, Haines J, Rogaev E, Mortilla M, Vaula G, Pericak-Vance M, Foncin J-F, Montesi M, Bruni A, Sorbi S, Rainero I, Pinessi L, Pollen D, Polinsky R, Nee L, Kennedy J, Macciardi F, Rogaeva E, Liang Y, Alexandrova N, Lukiw W, Schlumpf K, Tanzi R, Tsuda T, Farrer L, Cantu J-M, Duara R, Amaducci L, Bergamini L, Gusella J, Roses A, Crapper McLachlan D. Genetic evidence for a novel familial Alzheimer’s disease locus on chromosome 14. Nat Genet. 1992;2:330–334. doi: 10.1038/ng1292-330. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki T, Nishiyama K, Murayama S, Yamamoto A, Sato S, Kanazawa I, Sakaki Y. Regional and cellular presenilin 1 gene expression in human and rat tissues. Biochem Biophys Res Commun. 1996;219:708–713. doi: 10.1006/bbrc.1996.0299. [DOI] [PubMed] [Google Scholar]
- 40.Tanzi RE, Wenniger JJ, Hyman BT. Cellular specificity and regional distribution of amyloid β protein precursor alternative transcripts are unaltered in Alzheimer hippocampal formation. Mol Brain Res. 1993;18:246–252. doi: 10.1016/0169-328x(93)90196-v. [DOI] [PubMed] [Google Scholar]
- 41.Thinakaran G, Borchelt DR, Lee MK, Slunt H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Norstedt C, Seeger M, Hardy J, Levey A, Gandy SE, Jenkins NA, Copeland NG, Price DL, Sisodia SS. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 42.Towbin H, Staehlin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wasco W, Pettingell WP, Jondro PD, Schmidt SD, Gurubhagavatula S, Rodes L, DiBlasi T, Romano DM, Guenette SY, Kovacs DM, Growdon JH, Tanzi RE. Familial Alzheimer’s chromosome 14 mutations. Nat Med. 1995;1:848. doi: 10.1038/nm0995-848a. [DOI] [PubMed] [Google Scholar]
- 44.Weinmaster G, Roberts VJ, Lemke G. Notch 2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 45.Williams R, Lendahl U, Lardelli M. Complementary and combinatorial pattern of Notch gene family expression during early mouse development. Mech Dev. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- 46.Wisniewski HM, Terry RD. Reexamination of the pathogenesis of the senile plaque. In: Zimmerman HM, editor. Progress in neuropathology, Vol 2. Grune and Stratton; New York: 1973. pp. 1–26. [Google Scholar]