Abstract
Four Dlx homeobox genes, Dlx1,Dlx2, Dlx5, and Dlx6 are expressed in the same primordia of the mouse forebrain with temporally overlapping patterns. The four genes are organized as two tail-to-tail pairs, Dlx1/Dlx2 and Dlx5/Dlx6, a genomic arrangement conserved in distantly related vertebrates like zebrafish. The Dlx5/Dlx6 intergenic region contains two sequences of a few hundred base pairs, remarkably well conserved between mouse and zebrafish. Reporter transgenes containing these two sequences are expressed in the forebrain of transgenic mice and zebrafish with patterns highly similar to endogenous Dlx5 andDlx6 expression. The activity of the transgene is drastically reduced in mouse mutants lacking both Dlx1and Dlx2, consistent with the decrease in endogenousDlx5 and Dlx6 expression. These results suggest that cross-regulation by Dlx proteins, mediated by the intergenic sequences, is essential for Dlx5 andDlx6 expression in the forebrain. This hypothesis is supported by cotransfection and DNA-protein binding experiments. We propose that the Dlx genes are part of a highly conserved developmental pathway that regulates forebrain development.
Keywords: diencephalon, evolution, homeobox, mouse, striatum, telencephalon, zebrafish
The Dlx family of vertebrate homeobox genes comprises six members in mammals and at least eight in the zebrafish (Stock et al., 1996). Four Dlx genes,Dlx1, Dlx2, Dlx5, and Dlx6are involved in development of the ventral telencephalon and diencephalon of mammals (Porteus et al., 1991; Price et al., 1991;Robinson et al., 1991; Simeone et al., 1994; Liu et al., 1997), and the expression patterns of these four genes, although distinct overall, overlap significantly. Mice lacking either Dlx1 orDlx2 function show normal or nearly normal development of the subcortical telencephalon. However, mice lacking bothDlx1 and Dlx2 functions show stronger abnormalities in the development of the striatal subventricular zone, in the differentiation of striatal matrix neurons, and in the migration of neocortical interneurons from the subcortical telencephalon (Anderson et al., 1997a,b). Interestingly, expression ofDlx5 and Dlx6 is reduced in the subventricular zone, but not in the mantle of the double mutants, suggesting thatDlx1 and/or Dlx2 might be required for the maintenance of Dlx5/Dlx6 expression in subventricular zone cells. Mice lacking Dlx5 function show defects in the branchial arches and in epithelium derived from the olfactory and otic placodes, but not in the forebrain (Acampora et al., 1999; Depew et al., 1999). Mutants lacking both Dlx5 and Dlx6functions have yet to be reported.
The zebrafish dlx1, dlx2, dlx4, anddlx6 genes are the orthologs of the mammalianDlx1, Dlx2, Dlx5, and Dlx6genes, respectively (Akimenko et al., 1994; Stock et al., 1996). These four zebrafish genes are also expressed in the ventral forebrain with patterns very similar to those of their murine counterparts (Ellies et al., 1997). The similarities between the mouse and zebrafishDlx orthologs also extend to their genomic organization. In both species, the four genes are organized as two pairs of convergently transcribed genes, the Dlx1/Dlx2 pair and theDlx5/Dlx6 pair (dlx4/dlx6 in zebrafish; Simeone et al., 1994; McGuinness et al., 1996; Ellies et al., 1997). The relatively short distances (2.5–10 kb) that separate the two genes in such pairs and the similarities in the expression patterns of the two genes that constitute a pair (Ellies et al., 1997) suggest the presence, in the intergenic region, of shared cis-acting regulatory elements.
In the present study, we have examined the molecular basis for the overlapping expression of Dlx genes in the ventral forebrain of vertebrates. We have identified highly conserved sequences in the intergenic region between Dlx5/Dlx6 (dlx4/dlx6). These sequences extend over a few hundred base pairs and are the potential site of action of a vast number of regulatory factors. We present evidence that the Dlx proteins themselves constitute some of these factors. Taken together, these results suggest that cross-regulatory mechanisms between Dlx genes and enhancer-sharing are important aspects of Dlx regulation in the forebrain.
MATERIALS AND METHODS
Identification of conserved sequences in the zebrafishdlx4/dlx6 and mouse Dlx5/Dlx6 intergenic regions. Restriction fragments of a genomic clone containing the zebrafish dlx4/dlx6 locus (Ellies et al., 1997) were radiolabeled and hybridized to a Southern blot of various restriction digests of a mouse genomic clone containing the orthologousDlx5/Dlx6 locus (Liu et al., 1997). Of the zebrafish restriction fragments from the dlx4/dlx6 locus, only a 1.4 kb XhoI–EcoRI fragment from the intergenic region hybridized to the mouse genomic fragments (Fig.1). This zebrafish fragment and the hybridizing mouse fragments were sequenced using the dideoxy procedure. Sequence accession numbers are: for the zebrafish sequence (AF201695) and for the mouse sequences (AF201696 and AF201697). A search of the GenBank database with the zebrafish 1.4 kbXhoI–EcoRI fragment enabled us to identify a human BAC clone containing the DLX5/DLX6 locus (sequence accession number AC004774).
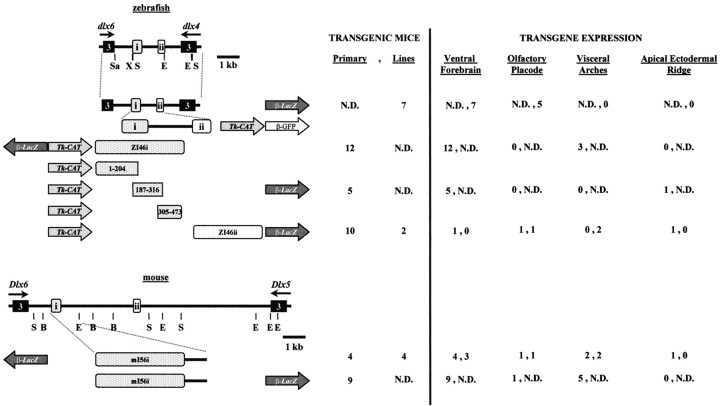
Fig. 1.
Genomic organization of the zebrafishdlx4 and dlx6 genes (top) and of the orthologous murine Dlx5 andDlx6 (bottom), indicating the location of conserved sequences with putative regulatory function. The third exons of zebrafish dlx4 and dlx6 and of mouseDlx5 and Dlx6 are represented byboxes. Direction of transcription is indicated byarrows. B, BamHI;E, EcoRI; X,XhoI; S, SacI;Sa, SalI. The constructs for the production of transgenic animals and for transfection experiments are schematized. The position and orientation of the intergenic fragments relative to the reporter genes (lacZ,CAT, or GFP) is shown.β, Minimal β-globin promoter;tk, thymidine kinase promoter. Numbers of primary transgenic embryos or embryos from transgenic lines that showlacZ expression in various sites of Dlxexpression are indicated to the right of each construct.
Nucleotide sequence comparisons were done using the GCG software package and the CLUSTAL W version 1.7 multiple sequence alignment program (Thompson, 1984).
Transgenic animals. DNA fragments from either the zebrafishdlx4/dlx6 locus or from the mouse Dlx5/Dlx6 locus were subcloned into the p1229 or p1230 vectors (Yee and Rigby, 1993). For the production of transgenic mice, the transgene was excised from the plasmid construct and injected at a concentration of 5 ng/μl in eggs from FVB/n crosses using standard procedures (Hogan et al., 1986). Transgenes were analyzed in either founder embryos or from established transgenic lines. Presence of the transgene was determined by PCR on DNA prepared from extra-embryonic tissues with the following oligonucleotide primers 5′-AGGGCAGAGCCATCTATTGC-3′ and 5′-CGCTCATCCGCCACATATCC-3′ derived, respectively, from theβ-globin promoter and lacZ sequences of the p1229/p1230 vectors. Amplification of a fetal hemoglobin gene sequence was used as a positive control (primers are x1: 5′-GATCATGACCGCCGTAGG-3′ and x2: 5′-CATGAACTTGTCCCAGGCTT-3′.
For the production of transgenic zebrafish, a 1.4 kbXhoI–EcoRI fragment of the zebrafishdlx4/dlx6 intergenic region was inserted upstream of theβ-globin promoter fragment taken from the p1230 vector and of the coding sequence of a variant of the green fluorescent protein (GFP) GM2 that emits ∼30-fold higher fluorescence than does the wild-type GFP, under standard FITC conditions (Cormack et al., 1996). Linearized plasmid DNA was injected into single-cell wild-type zebrafish embryos that were examined for GFP expression at various time points thereafter as previously described (Long et al., 1997).
Morphological analysis of transgenic animals. Founder transgenic embryos or embryos from the cross of a transgenic male with normal FVB or CD1 females were harvested at various embryonic stages. Transgene expression was also analyzed in newborn pups, young mice, and adults from established lines. Embryos were fixed in 1% formaldehyde, 0.2% glutaraldehyde, 0.02% NP-40 in PBS for 30 min at 4°C, washed in PBS for 20 min at room temperature, and stained for β-galactosidase activity overnight at 28°C in a solution of 1 mg/ml X-gal, 5 mmK3Fe(CN)6, 5 mmK4Fe(CN)6, 2 mm MgCl2, and 0.02% NP-40 in PBS.
Breeding with mouse null mutants. Mice heterozygous for thezfdlx4/6lacZ-transgene were mated to partners heterozygous for a deletion of the Dlx1 and Dlx2 genes (strain C57 Bl/6; described in Qiu et al., 1997). Offspring were genotyped by PCR, with the primers described above to detect the transgene, and with primers corresponding to the neomycin resistance gene that marks theDlx1/2 deletion. Animals that were heterozygous for both modifications (genotype Dlx1/2+/−;zfdlx4/6lacZ+/−) were mated to generate mice that were homozygous for the deletion of the Dlx1/2 locus (Dlx1/2−/−; zfdlx4/6lacZ). Mutant embryos were identified by either diagnosing a cleft palate (E15 and later) or by the absence of a PCR product, with primers that recognize the deleted portion of Dlx1.
Constructs for transient cotransfection experiments. An effector plasmid that expresses the zebrafish dlx2 gene under control of the SV40 early promoter was constructed by inserting an 845 bp PCR-amplified EcoRI fragment of the zebrafishdlx2 cDNA (Akimenko et al., 1994) encompassing the full coding sequence into the EcoRI site of the pTL2 expression vector (M. Petkovich, unpublished observations). Reporter plasmids were constructed by inserting fragments of the zebrafishdlx4/dlx6 intergenic region into the pBLCAT2 vector (Luckow and Schütz, 1987), which contains thethymidine kinase (tk) minimal promoter driving expression of the chloramphenicol acetyltransferase (CAT) gene. The 1.4 kb XhoI–EcoRI fragment from the zebrafish dlx4/dlx6 intergenic region (I4/6; Fig.1) was subcloned into pBLCAT2 directly upstream of the tkpromoter. The zI46i and zI46ii fragments and deletions of zI46i were prepared by PCR from a pBluescript clone containing the 1.4 kbXhoI–EcoRI I4/6 fragment. The zI46ii fragment was inserted in pBLCAT2 directly upstream of the tkpromoter, and the zI46i fragment was inserted immediately downstream of the CAT gene (i.e., 4.5 kb upstream of the tk promoter in the circular plasmid).
The following oligonucleotides: 1060, 5′-GCTCTAGAATTAGTTTAACGTCGAA-3′; 473, 5′-GGGGTACCGCTGGGGCATCCACGAT-3′;187,5′-GGGGTACCATTCTCATAAATGCAG-3′;204, 5′-GGGGTACCTGCATTTATGAGAATG-3′; 305, 5′-GGGGTACCATCTTTATTTGGATT-3′; and 316, 5′-GGGGTACCAAATAAAGATGCCTTT-3′ were used to prepare deletion fragments from zI46i using PCR. The numeric name of the oligonucleotide refers to the position, in the conserved intergenic sequence (Fig.2A), that borders the amplified fragment. Restriction sites (XbaI orKpnI) were introduced at the 5′ end of each oligonucleotide. PCR products: full-length zI46i (positions 1–473 in GenBank sequenceAF201695), zI46i 1–204 (positions 1–204 in the same sequence), zI46i 187–316, and zI46i 305–473, were PCR-amplified and subcloned into pBLCAT2.
Fig. 2.
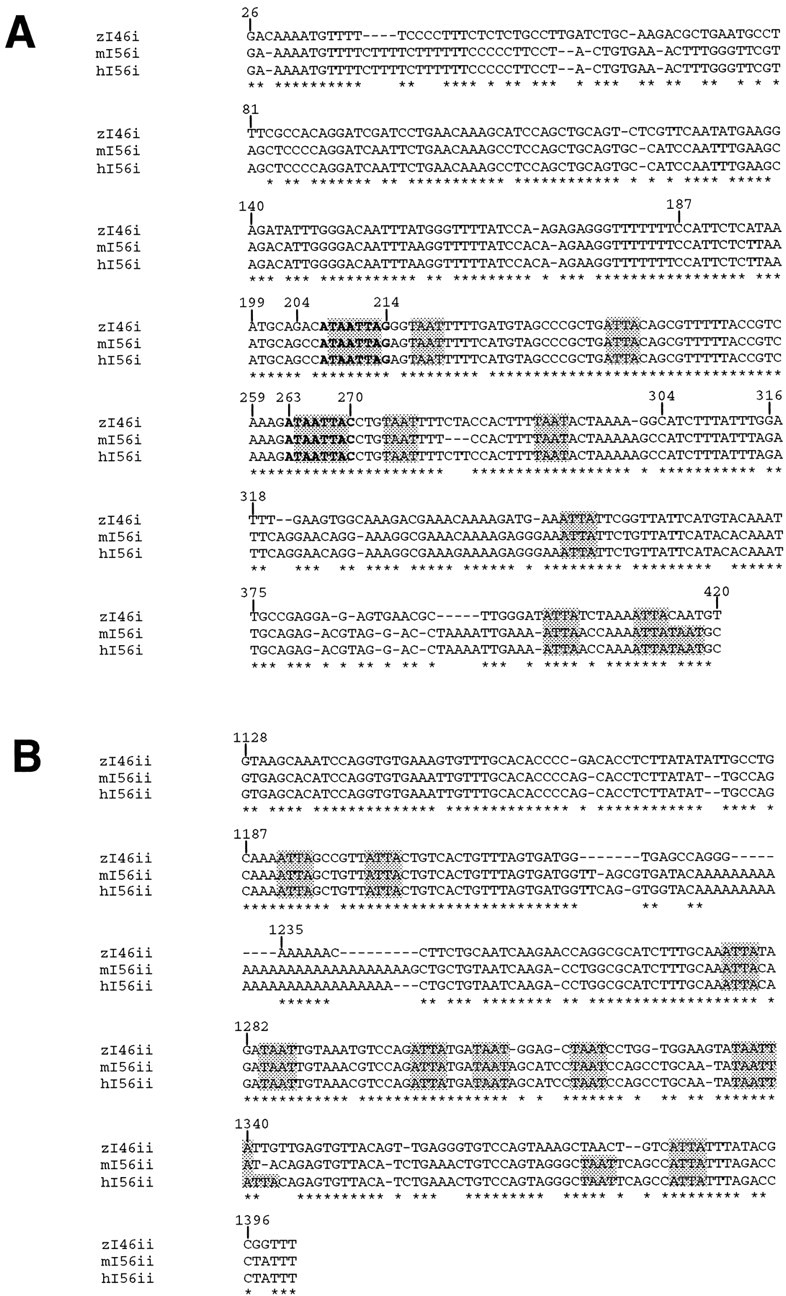
Conserved sequences in the intergenic region that separates a pair of vertebrate Dlx genes.A, Alignment of I56i sequences from human (h) and mouse (m) and zI46i from zebrafish (zf). B, Alignment of I56ii sequences from human and mouse and zI46ii from zebrafish. The human sequences were retrieved from GenBank, accession number AC004774. Complete sequence identity across the three species is indicated by anasterisk. The nucleotide positions of the intergenic sequences corresponding to putative homeodomain protein recognition sites as characterized by a TAAT/ATTA core sequence areshaded. The two putative Dlx binding sites that were mutagenized are shown in bold. Numbering is relative to the XhoI site (assigned the numerical position of 1) flanking a 1.4 kb element nearest to dlx6 in zebrafish and that contains both I4/6 elements (Fig. 1).
Mutagenesis. Two putative Dlx-binding sites (Feledy et al., 1999) in zI46i, found at sequence positions 207–214 and 263–270; were mutagenized using the Sculptor in vitro mutagenesis system (Amersham, Arlington Heights, IL). The AATTA and AATT sequences in those sites were changed to TCTAG and CTAG, respectively, to generate the Δ210 and Δ266 mutations. The double mutant Δ210/Δ266 was obtained by mutagenesis of the zI46i fragment already containing the Δ266 mutation with the same oligonucleotide used to produce the Δ210 mutation. The 187–316 fragments of zI46i containing either the Δ210 mutation, the Δ266 mutation, or both of them, were inserted into the pBLCAT2 vector for transfection experiments or in the p1230 vector to produce transgenic animals.
Transient cotransfection experiments. Transient cotransfection experiments were performed in the P19 murine embryonic carcinoma (EC) cell line essentially as described previously (Zerucha et al., 1997). Cells were seeded 24 hr before transfection at a density of 107 cells per 100 mm dish. Transfections were performed by the calcium phosphate precipitation procedure (Sambrook et al., 1989). A total of 10 μg of DNA per dish was used in each transfection. This included 2 μg pRSV-βgal as an internal control for transfection efficiency, 2 μg of reporter plasmid, 2 μg of effector plasmid, and sheared calf thymus DNA (Boehringer Mannheim, Indianapolis, IN) to the total of 10 μg. Precipitates were left on the cells for 16 hr, and the cells were harvested 64 hr after transfection. Cells were collected in PBS, pelleted by centrifugation, and resuspended in freeze/thaw buffer (250 mm Tris-HCl, pH 8, 10 mm DTT, and 15% glycerol). Cell extracts were prepared by repeated cycles of freezing and thawing. β-Galactosidase activity was assayed as described bySambrook et al. (1989). CAT activity was determined by thin-layer chromatography and measured as the percentage of conversion of monoacetylated and diacetylated chloramphenicol relative to unmodified plus acetylated chloramphenicol using the Bio-Rad (Hercules, CA) GS-525 Molecular Imager system. CAT activity was standardized to β-galactosidase levels to compensate for variations in transfection efficiency. Experiments were performed in duplicate and repeated a minimum of three times. Error bars in the figures represent SEMs of all replications.
Stable transfectant cell line. A PCR-amplifiedEcoRI fragment encompassing the full-length (845 bp) coding region of zebrafish dlx2 cDNA (described above) was subcloned into the pTL-MTG vector (Prefontaine et al., 1998) downstream of and in frame with six repeats of a c-myc sequence that encodes a polypeptide consisting of an epitope recognized by the 9E-10 monoclonal antibody (myc-tag; Santa Cruz Biotechnology, Santa Cruz, CA). Expression of this fusion protein is under control of the SV40 early promoter. This construct (pTL-MTG-Dlx2) was cotransfected together with pCMVneo into SF7 SCIDfibroblastic cells, as described above, using the calcium phosphate procedure with the following modifications: 8 μg of pTL-MTG-Dlx2 and 2 μg of pCMVneo made up the total DNA transfected per 100 mm dish; 40 hr after transfection 600 μg/ml G418 was added to the cells. Cells were maintained in this concentration of G418 until the formation of discernible colonies. Individual colonies of cells were isolated and grown separately. Each clone was screened by PCR for the presence of a zebrafish dlx2 sequence. MTG-Dlx2 protein was prepared from nuclear extracts of the stable transfectant cell line SF7-MTG-Dlx2 essentially as described by Andrews and Faller (1991). In brief, cells from each confluent 100 mm dish were harvested and resuspended in 1.5 ml PBS on ice. The cell suspension was pelleted and resuspended in 400 μl of a cold solution of (in mm): 10HEPES–KOH, pH 7.9, 1.5 MgCl2, 10 KCl, 0.5 DTT, and 0.2 PMSF, incubated on ice 10 min, then vortexed 10 sec. Insoluble nuclei were pelleted and the supernatant, containing cytoplasmic contents and outer membrane, was discarded. Nuclei were resuspended in 20–100 μl of a cold solution of (in mm): 20HEPES–KOH, pH 7.9, 1.5 MgCl2, 420 NaCl, 0.2 EDTA, 0.5 DTT, and 0.2 PMSF and 25% glycerol, and incubated on ice for 20 min. Cellular debris was removed by centrifugation. Protein concentration of the supernatant was determined by the Bio-Rad protein assay, and single use aliquots were stored at −80°C.
The presence of MTG-Dlx2 was determined by immunoblotting with the 9E-10 monoclonal antibody. Four individual clones were positive for MTG-Dlx2 after both rounds of screening, and one was chosen for subsequent experiments. It was thereafter maintained in 400 μg/ml G418.
Electrophoretic mobility shift assays. DNA fragments corresponding to positions 1–204, 187–316, and, 305–473 of zI46I (Fig. 2A), as well as the Δ210, Δ266, and Δ210/Δ266 mutagenized versions of the 187–316 fragment were amplified by PCR. The PCR product was inserted into the pCRII vector (Invitrogen, San Diego, CA). The fragment was excised withEcoRI before filling of the 5′ overhangs with the large fragment of DNA polymerase I (Klenow) in presence of radiolabeled [α32P]dATP. Binding reactions were performed in a total volume of 20 μl in (in mm): 12 HEPES–KOH, pH 7.9, 1 EDTA, 0.4 MgCl2, 100 NaCl, 0.6 DTT, and 0.6 PMSF, and 13% glycerol. Nuclear extract (10 μg) from the stable SF7-MTG-Dlx2 cell line or from control SF7 cells was pre-incubated with the 9E-10 anti-myc monoclonal antibody or an equivalent volume of water at room temperature for 30 min. After pre-incubation, 1 μg of bovine serum albumin, 1 μg of sheared calf-thymus DNA (Boehringer Mannheim), and 15,000 cpm of radiolabeled probe were added and incubated at room temperature for 20 min. Protein–DNA complexes were resolved on a 4% polyacrylamide (29:1 acrylamide:bis-acrylamide) gel run in 1× Tris-borate-EDTA.
RESULTS
Identification of highly conserved elements in the zebrafish dlx4/dlx6 and mouse Dlx5/Dlx6 intergenic regions
We identified two highly conserved sequences in the region between the zebrafish dlx4 and dlx6 genes and their mouse orthologs Dlx5/Dlx6. A 1.4 kbXhoI–EcoRI fragment from the zebrafishdlx4/dlx6 intergenic region (Fig. 1) was found to hybridize to a pair of restriction fragments in the mouseDlx5/Dlx6 intergenic region. Nucleotide sequence analysis revealed two sequences, of ∼400 and 300 bp, respectively, that are highly similar between the two species (Fig.2A,B). The orientation and the relative position of the two sequences relative to dlx4 (Dlx5) anddlx6 (Dlx6) are identical (Fig. 1). The 400 bp sequence, named zI46i, is closer to the dlx6 gene than is the 300 bp sequence, hereafter named zI46ii. The orthologous mammalian elements are hereafter called mI56i and mI56ii, respectively. We have identified highly similar sequences at the humanDLX5/DLX6 locus by searching the GenBank database (Fig.2A,B).
Nucleotide sequence comparisons indicate the human and mouse mI56i elements to be identical except for a 3 bp insertion in the human sequence (Fig. 2A). The zI46i sequence is 83% identical to its mammalian counterparts over 384 bp (Fig.2A), including a central 131 bp with 94% sequence identity. The human and mouse mI56ii sequences are 98% identical, and zI46ii shares ∼85% identity over 275 bp with its mammalian counterparts (Fig. 2B). The zI46ii sequence lacks a stretch of adenines, ∼20 nucleotides in length, that is found in the two mammalian sequences.
A zebrafish dlx4/dlx6 intergenic fragment _targets reporter gene expression to the forebrain and olfactory placodes in transgenic mice
A construct containing the lacZ reporter gene under the control of a β-globin minimal promoter and the entire zebrafish dlx4/dlx6 intergenic region (plus a short segment of the transcription unit of dlx4 and a few base pairs of the transcription unit of dlx6;zfdlx4/6lacZ transgene; Fig. 1) was injected into fertilized mouse eggs to produce transgenic animals. We obtained seven lines of transgenic mice. All seven lines showed lacZexpression, beginning at approximately embryonic day 10 (E10), in two groups of forebrain cells, one in the ventral thalamus/hypothalamus and one in the basal telencephalon (Fig.3A; I and II, respectively). Examination of whole-mount embryos stained for β-galactosidase activity indicated that the patterns of reporter transgene expression are strikingly similar to patterns of endogenous mouse Dlxexpression in the forebrain (Shimamura et al., 1997). Mouse embryos express Dlx5 and Dlx6 in two separate domains within the forebrain. Domain I is a longitudinal alar plate stripe that begins at the zona limitans intrathalamica and extends rostrally through the ventral thalamus (VT) and several hypothalamic areas (Hy) to the rostral midline. Domain II is a longitudinal region in the basal telencephalon that extends rostrally from part of the caudal ganglionic eminence (CGE; amygdala primordium), through the lateral ganglionic eminence (LGE) and medial ganglionic eminence (MGE), and into the septal and preoptic (POA) primordia (Bulfone et al., 1993b; L. Puelles, E. Kuwana, A. Bulfone , K. Shimamura, J. Keleher, S. Smiga, E. Puelles, and J. Rubenstein, unpublished observations).
Fig. 3.
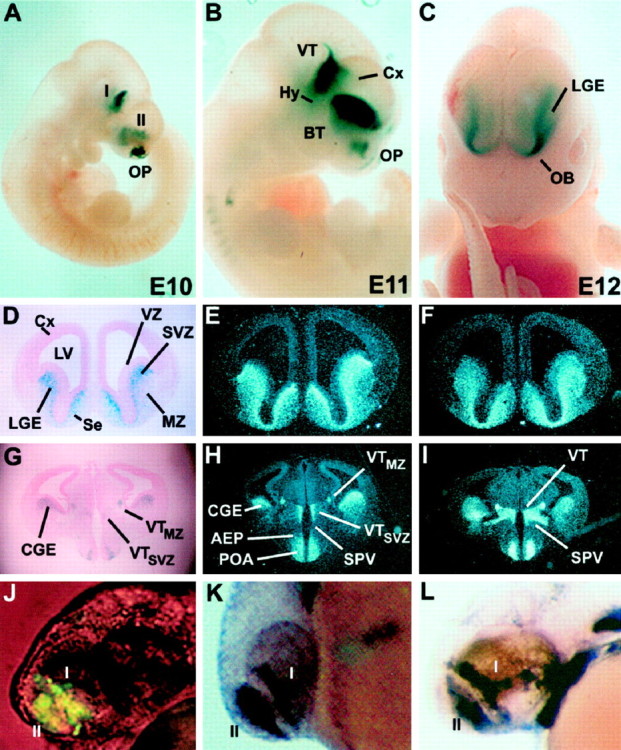
A DNA fragment encompassing the zebrafishdlx4/dlx6 intergenic region directs expression oflacZ in transgenic mouse embryos with patterns that closely recapitulate endogenous Dlx5 andDlx6 expression in the forebrain. A–C,lacZ expression in the ventral thalamus (VT), basal telencephalon (BT), and olfactory placodes (OP) in E10 (A), E11 (B), and E12 (C) whole-mount mouse embryos.D, Coronal section of an E14.5 stage mouse embryo withlacZ expression in the lateral ganglionic eminence (LGE). Higher β-galactosidase activity is seen in the subventricular zone (SVZ) compared to the mantle (MZ). E, F,In situhybridizations with Dlx5 (E) andDlx6 (F) probes on coronal sections adjacent to that seen in D. Note that the relative patterns of Dlx5 expression in the SVZ and MZ more closely resemble that seen in transgenic animals (D) than do the relative patterns ofDlx6 expression. G–I, More caudal sections of the same embryonic brain. G, Expression of β-galactosidase in the caudal ganglionic eminence (CGE), preoptic area (POA), and ventral diencephalon. H, I,In situhybridizations with Dlx5 and Dlx6 probes, respectively, on sections adjacent to those seen in G. Embryos in A and B are from line 7679, and those in C–I are from line 1469.J–L, A 1.4 kb XhoI/EcoRI fragment from the zebrafish dlx4/dlx6 intergenic region directs expression of a transgene that recapitulates endogenousdlx expression in the forebrain of zebrafish embryos.J, Expression of GFP directed by the 1.4 kb I4/6 zebrafish fragment in a 36 hr embryo. The patterns are similar to the expression of the endogenous dlx4(K) and dlx6(L) genes. The domains I and II ofdlx expression correspond, by analogy, to the diencephalic (I) and telencephalic (II) domains of Dlx expression in the mouse.I, Domain I; II, domain II;AEP, anterior entopeduncular area; BT, basal telencephalon; Cx, cortex; Hy, hypothalamus; LV, lateral ventricle; OB, prospective olfactory bulb; Se, septum;SPV, supraoptic paraventricular area; VZ, ventricular zone.
Similarly to endogenous Dlx5 and Dlx6, expression of the reporter transgene decreased after E14.5, and lacZ transcripts were virtually undetectable at postnatal day 0 (P0). Yet, β-galactosidase activity persisted much longer and, in some areas, remained strong even in the adult (P120) brain (data not shown). A higher sensitivity of the enzymatic assay for β-galactosidase activity compared to in situ hybridization or the sheltering of the β-galactosidase protein from metabolism could explain the apparent persistence of β-galactosidase compared to thelacZ transcripts. In addition to the developing forebrain, the reporter transgene under the control of zebrafish sequences was expressed in the olfactory placodes in five of seven mouse lines (Figs.1, 3A,B). There were very few additional sites of transgene expression: one line had a few labeled eye cells; one line had expression in the developing shoulder area, and one showed expression in the trunk somites (data not shown). Additional sites of endogenous mouse Dlx5 and Dlx6 expression were negative, including the branchial arches, the otic vesicle, and the limb apical ectodermal ridge (AER).
To assess the degree to which expression of thezfdlx4/6lacZ transgene matches endogenousDlx5 and Dlx6 expression in the forebrain, we have compared their expression patterns, using X-gal staining and radioactive in situ hybridization, respectively, on transverse brain sections. We analyzed sequential sections from E10.5, E12.5, E14.5, E17.5, and P0 mice.
Dlx5 and 6 are expressed in domains I and II in slightly different, but overlapping patterns: Dlx5 is expressed strongest in the subventricular zone (SVZ), whereasDlx6 is expressed strongest in the mantle zone (MZ) (Fig.3E,F). Neither gene is expressed appreciably in the ventricular zone (VZ) (Liu et al., 1997).
Remarkably, the zfdlx4/6lacZ transgene is expressed in a pattern extremely similar to that of the mouseDlx5 and Dlx6 genes. It is apparent, however, that despite the degree of overlap between thezfdlx4/6lacZ transgene and Dlx5 andDlx6 genes, there is a greater similarity between thezfdlx4/6-enhancer-driven lacZ and Dlx5expression patterns. β-Galactosidase activity and Dlx5transcripts can first be detected in the forebrain at ∼E10, as a thin layer of cells overlying parts of the ventricular zones in the basal telencephalon and diencephalon, respectively. On E10.5, E12.5, and E14.5, zebrafish dlx4/6-enhancer driven β-galactosidase expression in the mouse telencephalon matches mouseDlx5 expression more closely than that of Dlx6(Fig. 3D–I; data not shown). In domain I, thezfdlx4/6lacZ transgene has a pattern that also appears to be more similar to that of Dlx5 thanDlx6 (Fig. 3G–I).
The zebrafish dlx4/dlx6 intergenic enhancer is active in the forebrain of zebrafish embryos
To determine if the intergenic sequences that _target reporter gene expression to the forebrain of transgenic mice can reproducedlx expression in zebrafish embryos, we microinjected, into one-cell stage embryos, a construct that contained the 1.4 kb zebrafishEcoRI–XhoI intergenic fragment fromdlx4/dlx6 (Fig. 1), the same β-globin minimal promoter fragment as for the transgenic mouse experiments, and the gene coding for the GFP as a reporter. Primary transgenic zebrafish embryos carrying this construct expressed GFP specifically in forebrain cells forming two domains (Fig. 3J) with patterns strikingly similar to endogenous dlx4/dlx6 expression (Fig.3K,L). Of the 750 embryos that survived microinjection until the second day of embryonic development, four had very high levels of GFP expression in the forebrain, 25 had 5–10 GFP-positive forebrain cells, and 30 had one or two positive forebrain cells. Fifteen embryos showed one or a few GFP-positive cells in ectopic locations. The onset of GFP expression in the forebrain was ∼17–19 hr after fertilization (hpf), shortly after the onset of dlx4 expression as detected by in situ hybridization. GFP expression persisted in the forebrain until at least 36 hpf.
The two domains of dlx expression in the forebrain of zebrafish embryos (Akimenko et al., 1994) are reminiscent of the two domains observed in the mouse embryonic forebrain. To compare expression patterns of dlx genes in the zebrafish forebrain, we made sections of 48-hr-old embryos hybridized with dlxprobes. Interestingly, the patterns of expression of dlx1and dlx2 in both the telencephalon and the diencephalon indicate that the two genes are expressed in more immature cells, as reflected by their position closer to the ventricular walls than the cells that express dlx4 and dlx6 (Fig.4). A similar observation had been made previously for the mouse orthologs of these four genes (Liu et al., 1997).
Fig. 4.
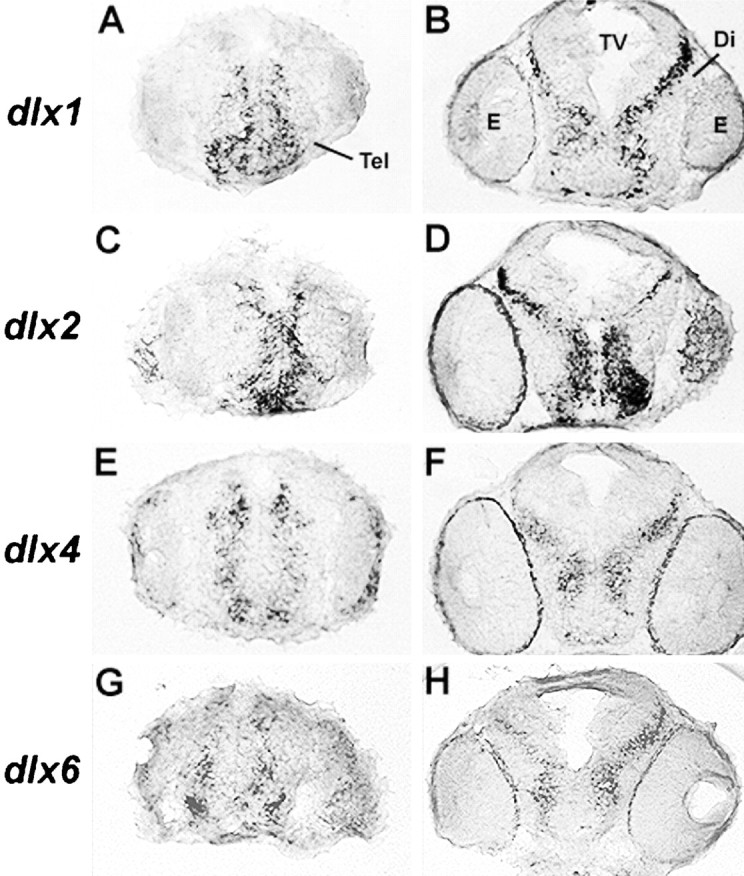
The dlx1 and dlx2genes are expressed in more immature cells of the zebrafish forebrain than their dlx4 and dlx6 paralogs. Transverse sections of 48-hr-old zebrafish embryos at the level of the telencephalon (A, C, E, G) and of the diencephalon (B, D, F, H) are shown with dorsal at thetop. Cells that express dlx1 anddlx2 are closer to the ventricle compared to those expressing dlx4 or dlx6. The expression of dlx2 closer to the ventricle compared todlx4 confirms our previous observation (Akimenko et al., 1994).
Most of the forebrain activity of the dlx4/dlx6intergenic enhancer is located in zI46i
The two conserved sequences located in the zebrafishdlx4/dlx6 intergenic region (zI46i and zI46ii) were inserted separately into reporter constructs and used to produce transgenic mouse embryos. At E11, forebrain lacZ expression _targeted by the zI46i enhancer construct was indistinguishable from that _targeted by the full-length I4/6 enhancer (compare Figs.5A, 3B), although none of the embryos showed expression in the olfactory epithelium (n = 12; Fig. 1).
Fig. 5.
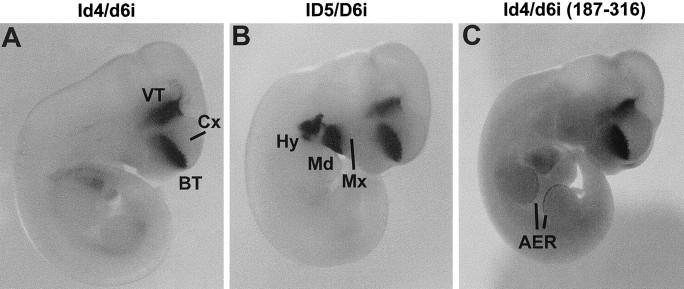
Specific intergenic sequences from either mouse or zebrafish _target gene expression to the forebrain of E11 mouse embryos with highly similar patterns. A, Zebrafish zI46i.B, Mouse mI56i. In addition to the forebrain, β-galactosidase activity was observed in the first two branchial arches in two of three lines and two of four primary transgenic embryos. C, The 187–316 fragment of zebrafish zI46i. In addition to the forebrain, β-galactosidase was also expressed in the apical ectodermal ridge (AER) of the limb buds in one of five primary transgenic embryos carrying this construct. Note that the forebrain expression patterns in A–C are highly similar to those of Figure 3B (full zebrafish intergenic fragment). Mx, Maxillary component of the first branchial arch; Md, mandibular component of the first branchial arch; Hy, hyoid arch. Other abbreviations as in Figure 3.
We also generated transgenic mice with a reporter construct that contained the second conserved sequence from the zebrafishdlx4/dlx6 intergenic region zI46ii. Two lines of transgenic mice and 10 primary transgenic mouse embryos were produced. Embryos from one transgenic line showed lacZ expression in the olfactory epithelium (data not shown) resembling that obtained with the full-length I4/6. This was also observed in one primary transgenic embryo, which, in addition, had expression in the AER of the limb buds, where Dlx genes are expressed (Dollé et al., 1992;Bulfone et al., 1993a). Finally, one primary zI46ii transgenic embryo showed correct lacZ expression in the forebrain (data not shown). Thus, zI46ii was much less efficient at _targetinglacZ to the forebrain (0 of 2 lines; 1 of 10 primary transgenic embryos) compared to full-length I4/6 (seven of seven lines) or to zI46i (12 of 12 primary transgenic embryos; Fig. 1).
We next tested whether the orthologous mouse mI56i could regulate correct Dlx expression in the forebrain. Four stable lines and four primary transgenic embryos were produced with the mI56i construct, and nearly all showed forebrain expression (Fig.5B); one transgenic line did not express lacZanywhere, possibly because of an integration effect. Reversing the orientation of mI56i had no effect on its expression (nine of nine primary transgenic embryos; Fig. 1; data not shown). Thus, both the orthologous mI56i and the zI46i fragments are enhancers that efficiently replicate the correct pattern of Dlx expression in the forebrain.
Unlike the zI46i enhancer, mI56i in either orientation frequently reproduced correct Dlx expression in the branchial arches (two of four stable lines and seven of 13 primary embryos; sum of both orientations, Figs. 1, 5B; data not shown), olfactory placode (one line and one primary embryo) and AER (one line; Fig. 1). No expression in the otic vesicle was observed in any embryos, but some expression was detected in the middle ear, which is consistent with the branchial arch expression.
To begin to identify the essential sequences within these enhancers, we used a deletion fragment of the zI46i enhancer, corresponding to positions 187–316 (Fig. 2A) and examined its activity in transgenic mouse embryos at E11. Of five primary transgenic embryos, all appeared to have correct expression in domain II in the forebrain (Fig. 5C). However, β-galactosidase expression in domain I (ventral thalamus and hypothalamus) was occasionally weaker or not detectable (data not shown). A similar construct also _targeted expression of GFP principally to the forebrain of zebrafish embryos (data not shown).
Activity of the zebrafish intergenic enhancer is reduced in mice lacking Dlx1 and Dlx2
Mutant mice that lack both Dlx1 and Dlx2function have a time-dependent block in basal telencephalon differentiation (Anderson et al., 1997b). Although early neurogenesis appears to be normal, later neurogenesis is not. This phenotype seems to be caused by a defect in the production and/or function of the subventricular zone. Accordingly, in Dlx1/2 mutantsDlx5 and Dlx6 expression is not detectable in the subventricular zone of the LGE and MGE, but is maintained in early born mantle cells at E12.5 (Anderson et al., 1997b). As described above, the zebrafish and mouse intergenic enhancers are highly active in the SVZ of the basal telencephalon. Therefore, it is possible that the Dlx1 or Dlx2 proteins might be, at least in part, responsible for the activity of this enhancer. To test this hypothesis, we bred mice containing the zebrafish dlx4/dlx6 full intergenic reporter construct with mice heterozygous for a mutation that inactivates both Dlx1and Dlx2. We then inbred mice that are heterozygous for both the mutation and the transgene to generate Dlx1/2−/− homozygotes that also had at least one zfdlx4/dlx6lacZallele.
In embryos that are homozygous for the Dlx1/Dlx2 mutation, β-galactosidase activity is strikingly reduced in the subventricular zone of the developing striatum (Fig.6A–D). These results parallel the changes in endogenous Dlx5 and Dlx6expression in the Dlx1/2 mutant mice (Fig.6E–L). Based on these results, we propose thatDlx1 and/or Dlx2 function is required, directly or indirectly, to regulate Dlx5 and Dlx6expression via their intergenic enhancer.
Fig. 6.
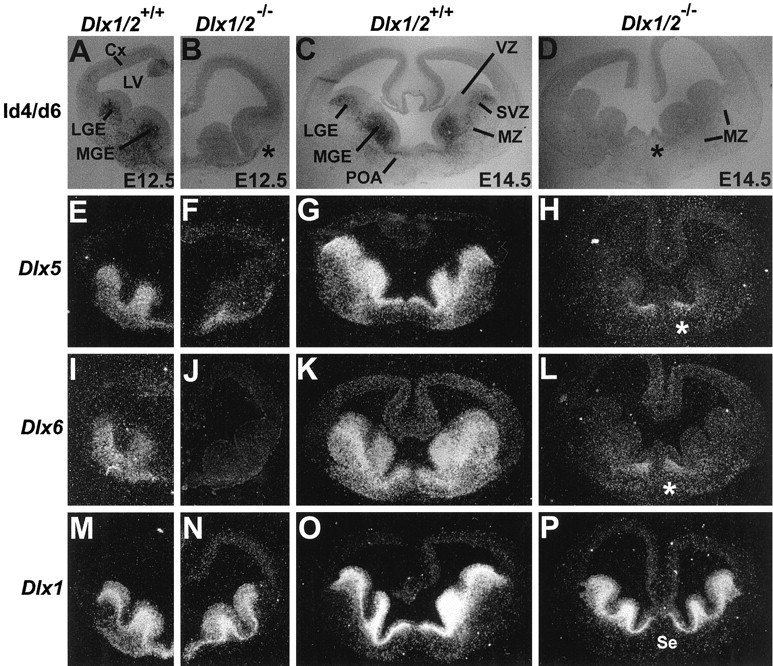
Forebrain expression of a reporter gene driven by I4/6 is drastically reduced in mice with a _targeted null mutation of the Dlx1 and Dlx2 genes. A, B, Coronal sections through the telencephalon of wild-type (A) and Dlx1/Dlx2 mutant (B) E12.5 embryos that both contain thezfdlx4/6lacZ transgene. C, D, Coronal sections through the telencephalon of wild-type (C) and mutant (D) E14.5 embryos. LacZ expression is virtually absent from the lateral (LGE) and medial (MGE) ganglionic eminences of the mutants, but is preserved in the rostral mantle (B, asterisk). E–P,In situ hybridization on sections adjacent to the ones shown inA–D with probes for Dlx5(E–H), Dlx6(I–L), and Dlx1(M–P). The Dlx1 probe recognizes a sequence in the 5′ end of the Dlx1 mRNA that is retained in the mutant (see Results). Theasterisk in D, H, andL denotes an area in the septal/preoptic region ofDlx1/Dlx2 mutants where Dlx5 andDlx6 expression at late embryonic stages appears not to be matched by lacZ expression. Other abbreviations as in Figure 3.
To determine whether the loss of Dlx5, Dlx6, andzfdlx4/6lacZ expression in the SVZ of theDlx1/2 mutants is attributable to a loss of those cells or to a change in gene regulation in SVZ cells, we studied the expression of Dlx1 and Dlx2 mRNAs. In theDlx1/2 mutants, the 5′ end of these genes was not deleted (Qiu et al., 1997). Thus, if the Dlx1/2 cis-acting regulatory sequences responsible for Dlx1/2expression are intact, and the truncated Dlx1 and/orDlx2 transcripts are stable, we should be able to detect the cells that normally express Dlx1/2 in these mutants. In fact, in situ hybridization demonstrates that both truncated genes are still expressed in the proliferative zones of the Dlx1/2 mutants (Fig. 6M–P;data not shown). This strongly supports the model that cells expressingDlx1/2 are maintained in the mutants and that there is molecular dysregulation within these cells leading to the loss of Dlx5, Dlx6, andzfdlx4/6lacZ expression.
Dlx proteins can upregulate transcription from conserved intergenic sequences
Dlx proteins bind DNA and can regulate transcription (Liu et al., 1997; Zhang et al., 1997). Because analysis of Dlx1/2 mutant mice suggests that Dlx1 and/or Dlx2 function is necessary for proper expression of Dlx5/Dlx6, one possibility is that this interaction is directly mediated by transcriptional activation of the intergenic enhancer(s) by Dlx1 or Dlx2. To test this model, we performed transient cotransfection assays in cultured cells. Reporter plasmids were constructed to contain either the zebrafish 1.4 kb I4/6 fragment, which contains both zI46i and zI46ii, or to contain only one of these elements. Effector plasmids were constructed to express full-length zebrafish Dlx1, Dlx2, Dlx3, Dlx4, or Dlx6 proteins under the control of the SV40 early promoter, or full-length mouse Dlx1, Dlx2, or Dlx5 proteins under the control of the cytomegalovirus promoter.
Cotransfection, into mouse P19 murine embryonic carcinoma cells, of a construct expressing the zebrafish Dlx2 protein resulted in a 20-fold increase in the activity of the CAT reporter gene placed under the control of the 1.4 kb dlx4/dlx6 intergenic fragment (Fig.7A). All of the zebrafishDlx expression vectors were able to activate expression of the reporter construct to a similar extent (data not shown). The mouse Dlx1, Dlx2, and Dlx5 expression vectors were also able to activate transcription of the same reporter construct in a neuroepithelial cell line (MNS-71; G. Yu, T. Zerucha, M. Ekker, and J. L. R. Rubenstein, unpublished observations). This indicates that Dlx proteins from either zebrafish or mouse are able to recognize similarly the zebrafish intergenic enhancer sequences in at least two different cell types. Not all homeodomain proteins could activate transcription from the 1.4 kbdlx4/dlx6 intergenic fragment; the products of severalsine oculis-related genes (six genes) were unable to activate transcription from this sequence (data not shown).
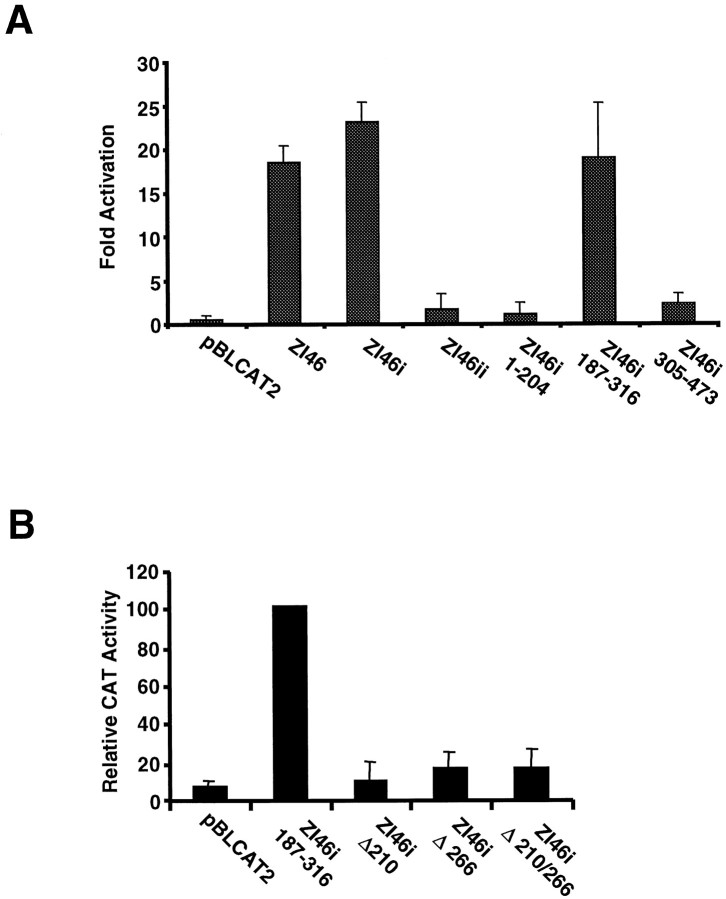
Fig. 7.
A, The zebrafish Dlx2 protein can activate transcription through intergenic regulatory sequences in transient transfection assays. Cotransfected Dlx2 activates transcription through the 1.4 kbXhoI/EcoRI fragment from the zebrafishdlx4/dlx6 intergenic region (I4/6), and specifically through zI46i, but not zI46ii. The 187–316 fragment from zI46i, but neither the 1–204 nor the 305–473 fragments (Fig.2A), is a _target for Dlx2. All values shown represent fold activation in the presence of Dlx2 relative to the same construct in the absence of cotransfected Dlx2. B,Mutagenesis of either one of the two putative binding sites for Dlx2 in zI46i 187–316 impairs activation in transient cotransfection assays. Values shown represent the percentage of the CAT activity obtained with the wild-type zI46i 187–316 fragment. All values represent three independent experiments ± SEM.
To determine if both zI46i and zI46ii contain _targets for Dlx proteins, the same Dlx expression constructs were cotransfected with reporter constructs containing either zI46i or zI46ii. All Dlx proteins examined activated expression through the zI46i element (Fig.7A; data not shown) but none activated transcription from a reporter containing zI46ii (Fig. 7A; data not shown), except perhaps for a weak (less than twofold activation) by the mouse Dlx5 protein. Furthermore, the degrees of activation produced by Dlx proteins on zI46i reporter constructs were comparable to those obtained with the 1.4 kb I4/6 fragment.
In an attempt to narrow down the region of zI46i required for activation by Dlx proteins, a series of deletion fragments of zI46i were prepared and subcloned into the reporter plasmid. The orientation of each of the deletion fragments was maintained relative to the orientation of the full-length zI46i. In transient cotransfection experiments, Dlx2 activated transcription of constructs containing the 187–316 deletion fragment to an extent similar to that observed with the full-length zI46i (Fig. 7A). However, Dlx2 did not activate transcription of constructs containing either the 1–204 or 305–473 fragments (Fig. 7A).
Recently a consensus DNA-binding site was identified for theXenopus Dlx3 ortholog, Xdll2 using a binding site selection procedure from a random oligonucleotide pool (Feledy et al., 1999). The consensus site identified is (A/C/G)TAATT(G/A)(C/G). Because of the similarity of the homeodomains of the Dlx family, it is likely that Dlx proteins other than those of the Dlx3 paralogous group will recognize a similar sequence. The 187–316 fragment of zI46i that is activated by Dlx2 contains two sites consistent with this consensus sequence. These two sites correspond to positions 207–214 and 263–270 of the zI46i sequence (Fig. 2). We mutagenized these two sites, individually or in combination. Mutagenesis of either site or of both sites almost entirely abolishes the activation of transcription by Dlx2 (Fig.7B). When tested in transgenic mice, a construct that contained mutations in both putative binding sites had little if any activity in the forebrain. Of seven primary transgenic mouse embryos, three had no detectable lacZ expression in the forebrain, and two had a few weakly positive cells at the anterior end of domain II (data not shown). These positive cells represented only a very small fraction of the endogenous pattern. Combined with our observation that activity of the forebrain enhancer is dramatically decreased in mice lacking Dlx1 and Dx2 function (Fig. 6), these results indicate that activation by Dlx proteins, presumably by Dlx1 and/or Dlx2, is essential for the activity of the I46 enhancer in the forebrain.
To determine if the Dlx proteins are able to directly interact with the zI46i element, electrophoretic mobility shift assays were performed. Nuclear extracts from SF7 SCID fibroblasts expressing a fusion of a c-myc fragment with full-length zebrafish Dlx2 (MTG-Dlx2) produced a lower mobility complex with the 187–316 deletion fragment of zI46i (Fig.8A). Migration of the lower mobility complex obtained with MTG-Dlx2 was further retarded in the presence of the 9E-10 anti-c-myc antibody, indicating the lower mobility complex contains MTG-Dlx2. No complexes of lower mobility were obtained with a control SF7 nuclear extract (Fig.8A). Furthermore, neither of the other two deletion fragments of zI46i (1–204 and 305–473) produced a complex of lower mobility in the presence of MTG-Dlx2-containing SF7 extract (Fig.8A), a result consistent with the absence of activation, by Dlx2, of reporter constructs containing these intergenic fragments in cotransfection experiments.
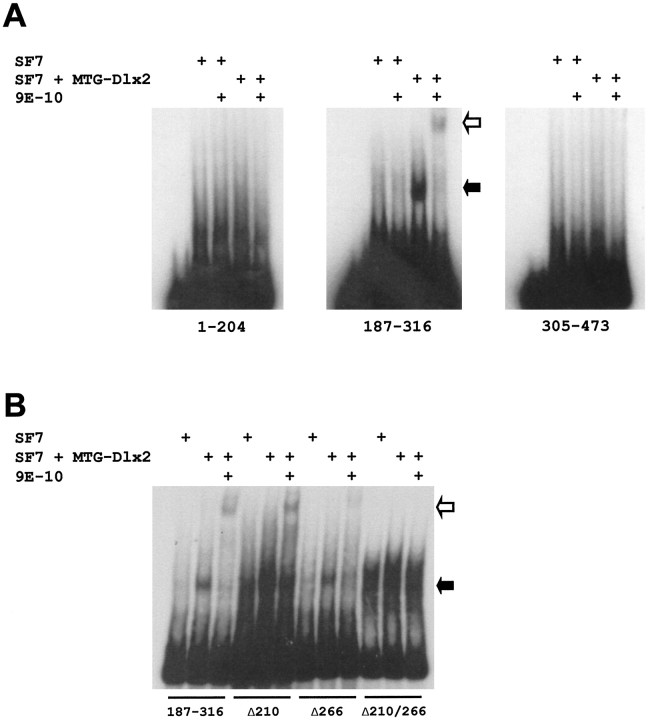
Fig. 8.
A, The zebrafish Dlx2 protein binds the 187–316 fragment of zI46i in a gel mobility shift assay. Three fragments from zI46i: 1–204, 187–316, and 305–473 were radiolabeled and incubated with a nuclear extract from an SF7-derived cell line that expresses MTG-Dlx2 or with a control SF7 nuclear extract. A lower mobility complex is indicated by the solid arrow. In the presence of the 9E-10 antibody directed against the MTG epitope of MTG-Dlx2, this mobility complex is supershifted (open arrow). B, Mutagenesis of the two putative binding sites in the zI46i 187–316 fragment impairs binding by the Dlx2 protein. Only those lower mobility complexes obtained after incubation of MTG-Dlx2 with the wild-type 187–316 or with fragments containing one mutagenized site (Δ210 or Δ266) can be supershifted by the 9E-10 antibody. A smear around the same mobility as this retarded complex can be obtained with the fragment containing the two mutations (Δ210/266), but is also seen with the control SF7 extract and is not supershifted by the 9E-10 antibody.
We next examined the effects of mutagenesis of the putative binding sites. As seen in Figure 8B, the Δ210 and Δ266 mutagenized fragments both produced a shift of lower mobility that migrated to the same place as that obtained with the wild-type 187–316 fragment. These shifts could be supershifted with the 9E-10 antibody. In contrast, we observed no supershift when the double mutant fragment was incubated with the MTG-Dlx2 and 9E-10 (Fig. 8B). Mutation at the 210 site seems to increase background binding, which is also observed in the control Sf7 extract and overlaps with the expected position for the Dlx2–DNA complex. This is especially evident for the double mutant but can also be seen in the Δ210 lanes.
The similar migrations of the lower mobility complexes obtained with the wild-type and single mutant fragments suggest that we can only observe the wild-type fragment bound by one Dlx2 molecule. This is consistent with the observation that only a small proportion of the labeled fragment is bound and suggests that Dlx2 binds to the two sites independently instead of cooperatively, which is not surprising considering the relatively large distance that separates the two binding sites (∼56 bp). The fragment bound by two Dlx2 molecules would be proportionally too weak to be observed in this assay.
Taken together with the results of our transient expression assays (Fig. 7), our observations suggest that two Dlx2 protein molecules bind to the zI46i enhancer independently but that optimal function of the enhancer requires occupancy of the two binding sites.
DISCUSSION
One intergenic enhancer is sufficient to recapitulate forebrainDlx expression
A zebrafish sequence from the intergenic region between thedlx4 and dlx6 genes is sufficient, once combined to a minimal promoter, to direct expression in cells of the telencephalon and diencephalon, of either zebrafish or mice, that normally express Dlx genes (Fig. 3). This strongly suggests that the regulatory mechanisms controlling Dlx expression in the forebrain have been conserved during vertebrate evolution, and it lends support to the idea that Dlx function during forebrain development has also been conserved. Additional evidence for conserved function of Dlx genes in forebrain development comes from the differential expression of Dlx genes in the telencephalon and diencephalon where more immature cells expressDlx1 and Dlx2 compared to Dlx5 andDlx6, as seen both for the mouse genes (Liu et al., 1997) and for their zebrafish orthologs (Fig. 4; Akimenko et al., 1994).
Functional conservation of enhancer sequences between mammals and teleost fish has been previously observed for the Otx2(Kimura et al., 1997), hoxb1 (Marshall et al., 1994), and the Hoxd-11 genes (Beckers et al., 1996; Gerard et al., 1997), although in the latter case, temporal, spatial, and mechanistic differences could be observed between the fish enhancer and its mammalian counterpart.
Comparisons of the enhancer activities of zI46i (mI56i) and zI46ii suggest the former plays a more important role in forebrain expression. zI46ii may still be necessary for optimal Dlx expression in the ventral forebrain, but this enhancer may require, to function efficiently, the presence of other regulatory sites, either from zI46i or from the promoters of one or both flanking genes.
Detailed analysis of reporter transgene activity in the mouse forebrain suggests that both the full dlx4/dlx6 intergenic construct and the zI46i (mI56i) sequences reproduce the endogenousDlx5 expression pattern more faithfully than theDlx6 expression pattern. This was observed principally by comparison of β-galactosidase expression and endogenous transcript levels in the LGE and MGE of the telencephalon (Fig. 3). Identical results were obtained with constructs from either zebrafish or mouse origin. Therefore, the observed differences cannot be attributed solely to the inability of a zebrafish enhancer to precisely recapitulateDlx expression in a mouse embryo. One possible explanation for this result is that sequences, necessary for maximal expression in cells of the mantle, are absent from our constructs, and, therefore, located outside the intergenic region. It is also possible that, although the intergenic enhancer is sufficient to direct expression to the ventral forebrain, its activity is modulated by specific interactions with other cis-acting regulatory elements, such as the promoters of each of the two flanking genes, Dlx5 andDlx6. An overall distinct set of transcriptional activators binding to upstream and intergenic regulatory sequences would be responsible for the differences in Dlx5 and Dlx6expression patterns. Experiments in zebrafish designed to examine the interactions between the intergenic forebrain enhancer and thedlx4 and dlx6 promoters are presently under way to address this issue. In summary, although it is possible that the intergenic forebrain enhancer is shared between Dlx5 andDlx6, which would explain their partially overlapping patterns of expression, additional mechanisms must account for the overall distinct expression of the two genes in the forebrain.
Transgenic animals carrying constructs containing both zI46i and zI46ii always exhibit expression of the reporter gene in the ventral forebrain. Reporter expression is often seen in the olfactory placodes, but never in regions of the embryo, such as the branchial arches, the inner ear, and the AER of the limb buds where Dlx5 andDlx6 or their zebrafish orthologs are also expressed. These results suggest that elements necessary for proper expression in the latter areas are located outside the conserved Dlx5/Dlx6intergenic region. On the other hand, several transgenic animals with the zI46i or mI56i constructs showed expression in the ectomesenchyme of the branchial arches reminiscent of endogenous Dlxexpression. The mechanisms that underlie such results are, at present, unclear but may involve integration effects. It is also possible that intergenic sequences, outside zI46i (mI56i) are necessary to restrict the activity of this enhancer to the ventral forebrain.
Dlx proteins interact with the forebrain-specific regulatory sequences
Expression of Dlx5 and Dlx6 is affected in the ventral forebrain of Dlx1/2 null mutants (Anderson et al., 1997b). Thus, Dlx5 and Dlx6transcripts are not detectable in the SVZ of the LGE at E12.5 and E14.5. Like endogenous Dlx5 and Dlx6, the activity of the zebrafish dlx4/dlx6 intergenic transgene (I4/6) is drastically reduced in the Dlx1/2 null mutants (Fig. 6). The mostly normal expression of the truncated Dlx1and Dlx2 transcripts in forebrain cells of the mutant (Fig.6; data not shown) suggests that the SVZ cells that normally expressDlx5 and Dlx6 are still present. Therefore, the reductions in Dlx5 and Dlx6 expression and in transgene activity strongly suggest that Dlx1 and/orDlx2 are required for the induction and/or maintenance ofDlx5 and Dlx6 expression and that this is mediated by the intergenic enhancer sequences. This might be achieved by direct regulation of Dlx5/Dlx6 expression by the Dlx1 or Dlx2 proteins which can function as transcriptional activators (Liu et al., 1997; Zhang et al., 1997). Such cross-regulatory interactions involving homeobox genes have been described, for example, for members of the Hox clusters (Gould et al., 1997; Nonchev et al., 1997; Studer et al., 1998) and for the zebrafish dlx genes (Zerucha et al., 1997). Alternatively, Dlx1 or Dlx2 may activate a yet unknown factor that is an essential regulator of Dlx5 andDlx6 expression. The above two mechanisms are not mutually exclusive. Our finding that the Dlx2 (Fig. 7A) and Dlx1 (data not shown) proteins, of either zebrafish or mouse origin are able to upregulate transcription of reporter constructs containing the conserved I4/6 intergenic sequences in cotransfection experiments supports the view that these sequences are the site of cross-regulatory interactions in vivo. Upregulation by Dlx2 was almost completely abolished when the putative binding sites were mutagenized (Fig. 7B). Furthermore, we were able to demonstrate binding of Dlx2 to the 187–316 fragment of zI46i in gel mobility shift assays (Fig. 8A). Mutagenesis of both sites in 187–316 abolished binding by the Dlx2 protein (Fig. 8B). The 187–316 fragment of zI46i is responsible for most if not all of the activation by Dlx proteins in transfection experiments and is able to _target transgene expression to the ventral forebrain in mice and zebrafish.
The loss of Dlx1 and Dlx2 expression only eliminates Dlx5, Dlx6, andzfdlx4/6lacZ transgene expression in the SVZ, whereas their expression is maintained in the early-born mantle cells of the rostral telencephalon (Fig. 6). This observation indicates that other transcription factors regulate Dlx5, Dlx6,and I4/6lacZ expression in a subset of early forebrain cells. Furthermore, the loss of Dlx5 and Dlx6expression in the SVZ of the Dlx1/Dlx2 null mutants also raises the possibility that the mutant phenotype may be attributable to the loss of function of all four genes, implying some functional redundancy between them. This is also supported by the recent observation that loss of Dlx5 function alone does not produce any obvious forebrain phenotype (Acampora et al., 1999; Depew et al., 1999). A better understanding of any differences in biochemical activities of Dlx proteins, such as involvement in specific protein–protein interactions, would help elucidate the functional consequences of the partially overlapping expression of these genes during development.
Intergenic region and Dlx gene evolution
The high degree of sequence similarity that we observed in the intergenic region between a pair of Dlx genes of mouse, human, and teleost fish (Fig. 2) is remarkable, considering that these sequences are outside the coding regions of either genes. High degrees of sequence similarity outside gene coding regions have been observed previously between human and mouse sequences (for example, see Becker et al., 1996; Williams et al., 1998), and some sequence conservation has also been found with sequences of distantly related vertebrates such as the pufferfish, Fugu rubripes, and zebrafish (Marshall et al., 1994; Morrison et al., 1995; Beckers et al., 1996;Kimura et al., 1997). However, none of the above examples compare in length and/or in percentage identity with the elements we report in the present study.
The convergently transcribed configuration of pairs ofdistal-less-related genes is ancient because it has been reported for the ascidian Ciona intestinalis (Di Gregorio et al., 1995). The distance that separates the two genes is relatively small (2–10 kb) for all cases reported thus far. It is likely that the paired organization arose after the divergence of arthropods from the lineage that would give rise to vertebrates because insects are thought to have only one distal-less gene. It is possible that one or a few regulatory sequences found downstream of thedistal-less gene in the common ancestor to modern day invertebrates and vertebrates were preserved after the first duplication and inversion event that produced the first pair ofdistal-less/Dlx genes. Enhancer sequences have been described downstream of the Drosophila distal-less gene (Vachon et al., 1992; O'Hara et al., 1993) and it will be interesting to determine if there is any degree of functional conservation in these enhancers and those described in the current study. A potential evolutionary advantage of enhancer-sharing by the two linked genes would be consistent with the conservation of the paired, convergently transcribed configuration and in particular the maintenance of a relatively short intergenic distance. Enhancer sharing has been previously demonstrated for some of the clustered Hox genes (van der Hoeven et al., 1996; Gould et al., 1997; Sharpe et al., 1998).
Vertebrates have at least three pairs of linked Dlx genes (Simeone et al., 1994; McGuinness et al., 1996; Ellies et al., 1997;Liu et al., 1997), the Dlx1/Dlx2, Dlx5/Dlx6(dlx4/dlx6 in zebrafish), and Dlx3/Dlx7 pairs. The presence of conserved regulatory sequences may not be unique to theDlx5/Dlx6 gene pair because the Dlx1/Dlx2intergenic region also contains highly conserved sequences (T. Zerucha, M. Qiu, J. K. Liu, J. L. R. Rubenstein, and M. Ekker, unpublished observations), although the roles of such sequences in Dlxgene regulation are, at present, unclear. The function and evolution of intergenic enhancer sequences, combined with studies of the functional specificity of Dlx genes, will enable us to understand the mechanistic basis for the concerted action of Dlx proteins in embryonic cells and the position of the Dlx genes in regulatory cascades during development.
Footnotes
This work was supported by grants from the Medical Research Council of Canada, the Natural Sciences and Engineering Research Council of Canada, and in part by Grant 1-FY99–572 from the March of Dimes Birth Defects foundation to M.E. T.S. and B.K.P. were supported by fellowships from the Deutscher Akademischer Austauschdienst and from the Korean Science and Engineering Foundation, respectively. We thank Genny Giroux, Lucille Joly, and Wei Lin for technical assistance, Allison Lewis for help in sequencing the zebrafishdlx4/dlx6 intergenic region, Ward Giffin and Robert Haché for useful discussions, and Marie-Andrée Akimenko and Lucie Jeannotte for critical reading of this manuscript. We thank Robb Krumlauf for kindly providing the p1229 and p1230 plasmids.
T.Z. and T.S. contributed equally to the work.
Correspondence should be addressed to Marc Ekker, Loeb Health Research Institute at the Ottawa Hospital, 725 Parkdale Avenue, Ottawa, Ontario K1Y 4E9, Canada. E-mail: mekker@lri.ca.
Dr. Zerucha's present address: Department of Organismal Biology and Anatomy, University of Chicago, Chicago, IL 60637.
Dr. Yu's present address: Genentech, South San Francisco, CA 94080.
REFERENCES
- 1.Acampora D, Merlo GR, Paleari L, Zeraga B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 2.Akimenko M-A, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson SA, Eisenstat DD, Shi L, Rubenstein JLR. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 4.Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JLR. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late-born striatal neurons. Neuron. 1997b;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 5.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker D, Jiang Z, Knodler P, Deinard AS, Eid R, Kidd KK, Shashikant CS, Ruddle FH, Schughart K. Conserved regulatory element involved in the early onset of Hoxb6 gene expression. Dev Dyn. 1996;205:73–81. doi: 10.1002/(SICI)1097-0177(199601)205:1<73::AID-AJA7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Beckers J, Gérard M, Duboule D. Transgenic analysis of a potential Hoxd-11 limb regulatory element present in tetrapods and fish. Dev Biol. 1996;180:543–553. doi: 10.1006/dbio.1996.0327. [DOI] [PubMed] [Google Scholar]
- 8.Bulfone A, Kim HJ, Puelles L, Porteus MH, Grippo JF, Rubenstein JLR. The mouse Dlx-2 (Tes-1) gene is expressed in spatially restricted domains of the forebrain, face and limbs in midgestation mouse embryos. Mech Dev. 1993a;40:129–140. doi: 10.1016/0925-4773(93)90071-5. [DOI] [PubMed] [Google Scholar]
- 9.Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JLR. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993b;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 11.Depew MJ, Liu JK, Long JE, Presley R, Meneses JJ, Pedersen RA, Rubenstein JLR. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- 12.Di Gregorio A, Spagnuolo A, Ristoratore F, Pischetola M, Aniello F, Branno M, Cariello L, Di Lauro R. Cloning of ascidian homeobox genes provides evidence for a primordial chordate cluster. Gene. 1995;156:253–257. doi: 10.1016/0378-1119(95)00035-5. [DOI] [PubMed] [Google Scholar]
- 13.Dollé P, Price M, Duboule D. Expression of the murine Dlx-1 homeobox gene during facial, ocular and limb development. Differentiation. 1992;49:93–99. doi: 10.1111/j.1432-0436.1992.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 14.Ellies DL, Stock DW, Hatch G, Giroux G, Weiss KM, Ekker M. Relationship between the genomic organization and the overlapping embryonic expression patterns of the zebrafish dlx genes. Genomics. 1997;45:580–590. doi: 10.1006/geno.1997.4978. [DOI] [PubMed] [Google Scholar]
- 15.Feledy JA, Morasso MI, Jang S-I, Sargent TD. Transcriptional activation by the homeodomain protein Distal-less 3. Nucleic Acids Res. 1999;27:764–770. doi: 10.1093/nar/27.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerard M, Zakany J, Duboule D. Interspecies exchange of a Hoxd enhancer in vivo induces premature transcription and anterior shift of the sacrum. Dev Biol. 1997;190:32–40. doi: 10.1006/dbio.1997.8679. [DOI] [PubMed] [Google Scholar]
- 17.Gould A, Morrison A, Sproat G, White RAH, Krumlauf R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- 18.Hogan B, Constantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1986. [Google Scholar]
- 19.Kimura C, Takeda N, Suzuki M, Oshimura M, Aizawa S, Matsuo I. Cis-acting elements conserved between mouse and pufferfish Otx2 genes govern the expression in mesencephalic neural crest cells. Development. 1997;124:3929–3941. doi: 10.1242/dev.124.20.3929. [DOI] [PubMed] [Google Scholar]
- 20.Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JLR. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 22.Luckow B, Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall H, Studer M, Pöpperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 24.McGuinness T, Porteus MH, Smiga S, Bulfone A, Kingsley C, Qiu M, Liu JK, Long JE, Xu D, Rubenstein JLR. Sequence, organization, and transcription of the Dlx-1 and Dlx-2 locus. Genomics. 1996;35:473–485. doi: 10.1006/geno.1996.0387. [DOI] [PubMed] [Google Scholar]
- 25.Morrison A, Chaudhuri C, Ariza-McNaughton L, Muchamore I, Kuroiwa A, Krumlauf R. Comparative analysis of chicken Hoxb-4 regulation in transgenic mice. Mech Dev. 1995;53:47–59. doi: 10.1016/0925-4773(95)00423-8. [DOI] [PubMed] [Google Scholar]
- 26.Nonchev S, Maconocie M, Gould A, Morrison A, Krumlauf R. Cross-regulatory interactions between Hox genes and the control of segmental expression in the vertebrate central nervous system. Cold Spring Harb Symp Quant Biol. 1997;62:313–323. [PubMed] [Google Scholar]
- 27.O'Hara E, Cohen B, Cohen SM, McGinnis W. Distal-less is a downstream gene of Deformed required for ventral maxillary identity. Development. 1993;117:847–856. doi: 10.1242/dev.117.3.847. [DOI] [PubMed] [Google Scholar]
- 28.Porteus MH, Bulfone A, Ciaranello RD, Rubenstein JLR. Isolation and characterization of a novel cDNA clone encoding a homeodomain that is developmentally regulated in the ventral forebrain. Neuron. 1991;7:221–229. doi: 10.1016/0896-6273(91)90260-7. [DOI] [PubMed] [Google Scholar]
- 29.Prefontaine GG, Lemieux ME, Giffin W, Schild-Poulter C, Pope L, LaCasse E, Walker P, Hache RJG. Recruitment of octamer transcription factors to DNA by glucocorticoid receptor. Mol Cell Biol. 1998;18:3416–3430. doi: 10.1128/mcb.18.6.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price M, Lemaistre M, Pischetola M, Di Lauro R, Duboule D. A mouse gene related to distal-less shows a restricted expression in the developing forebrain. Nature. 1991;351:748–751. doi: 10.1038/351748a0. [DOI] [PubMed] [Google Scholar]
- 31.Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JLR. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 32.Robinson GW, Wray S, Mahon KA. Spatially restricted expression of a member of a new family of murine distal-less homeobox genes in the developing forebrain. New Biol. 1991;3:1183–1194. [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual, Ed 2. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 34.Sharpe J, Nonchev S, Gould A, Whiting J, Krumlauf R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998;17:1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimamura K, Martinez S, Puelles L, Rubenstein JLR. Patterns of gene expression in the neural plate and neural tube subdivide the embryonic forebrain into transverse and longitudinal domains. Dev Neurosci. 1997;19:88–96. doi: 10.1159/000111190. [DOI] [PubMed] [Google Scholar]
- 36.Simeone A, Acampora D, Pannese M, Desposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K, Boncinelli E. Cloning and characterization of two members of the vertebrate Dlx gene family. Proc Natl Acad Sci USA. 1994;91:2250–2254. doi: 10.1073/pnas.91.6.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stock DW, Ellies DL, Zhao Z, Ekker M, Ruddle FH, Weiss KM. The evolution of the vertebrate Dlx gene family. Proc Natl Acad Sci USA. 1996;93:10858–10863. doi: 10.1073/pnas.93.20.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- 39.Thompson EA. Information gain in joint linkage analysis. IMA J Math Appl Med Biol. 1984;1:31–49. doi: 10.1093/imammb/1.1.31. [DOI] [PubMed] [Google Scholar]
- 40.Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, Cohen SM. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the _target gene distal-less. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
- 41.van der Hoeven F, Zakany J, Duboule D. Gene transposition in the HoxD complex reveal a hierarchy of regulatory controls. Cell. 1996;85:1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- 42.Williams SC, Altmann CR, Chow RL, Hemmati-Brivanlou A, Lang RA. A highly conserved lens transcriptional control element from the Pax-6 gene. Mech Dev. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- 43.Yee S-P, Rigby PWJ. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- 44.Zerucha T, Muller J-P, Chartrand N, Ekker M. Cross-interactions between two members of the Dlx family of homeobox-containing genes during zebrafish development. Biochem Cell Biol. 1997;75:613–622. [PubMed] [Google Scholar]
- 45.Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]