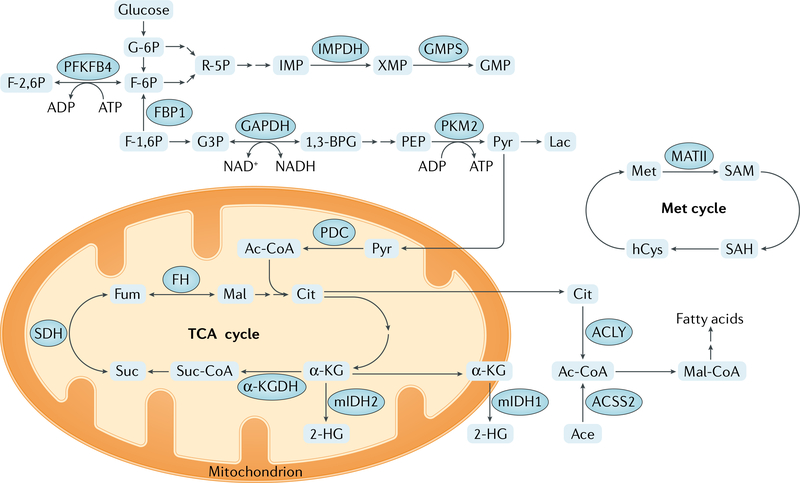
Fig. 3 |. Core metabolic functions of metabolic enzymes that also function in epigenetic modifications.
Main metabolic pathways regulated by metabolic enzymes involved in epigenetic modifications are shown. α-KG, α-ketoglutarate; α-KGDH,α-KG dehydrogenase; 1,3-BPG, 1,3-bisphosphoglycerate; 2-HG, 2-hydroxyglutarate; Ac-CoA, acetyl-CoA; Ace, acetate; ACLY, ATP citrate synthase; ACSS2, acetyl-CoA synthetase short-chain family member 2; Cit, citrate; F-1,6 P, fructose 1,6-bisphosphate; F-2,6 P, fructose 2,6-bisphosphate; F-6P, fructose 6-phosphate; FH, fumarase; Fum, fumarate; G3P, glyceraldehyde 3-phosphate; G-6P, glucose 6-phosphate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GMPS, GMP synthase; hCys, homocysteine; IMP, inosine 5′-monophosphate; IMPDH, IMP dehydrogenase; Lac, lactate; Mal, malate; Mal-CoA, malonyl-CoA; MATII, methionine adenosyltransferase II; Met, methionine; mIDH1, isocitrate dehydrogenase [NADP], cytoplasmic mutant; mIDH2, isocitrate dehydrogenase [NADP], mitochondrial mutant; PDC, pyruvate dehydrogenase complex; PEP, 2-phosphoenolpyruvate; PFKFB4, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4; PKM2, pyruvate kinase M2 isoform; Pyr, pyruvate; R-5P, ribose 5-phosphate; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SDH, succinate dehydrogenase; Suc, succinate; Suc-CoA, succinyl-CoA; TCA, tricarboxylic acid; XMP, xanthosine monophosphate.