Abstract
Development of effective vaccines against severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) is still a priority in prevention of re-emergence of SARS. Our previous studies have shown that the receptor-binding domain (RBD) of SARS-CoV spike (S) protein elicits highly potent neutralizing antibody responses in the immunized animals. But it is unknown whether RBD can also induce protective immunity in an animal model, a key aspect for vaccine development. In this study, BALB/c mice were vaccinated intramuscularly (i.m.) with 10 μg of RBD-Fc (RBD fused with human IgG1 Fc) and boosted twice at 3-week intervals and one more time at 12th month. Humoral immune responses of vaccinated mice were investigated for up to 12 months at a 1-month interval and the neutralizing titers of produced antibodies were reported at months 0, 3, 6 and 12 post-vaccination. Mice were challenged with the homologous strain of SARS-CoV 5 days after the last boost, and sacrificed 5 days after the challenge. Mouse lung tissues were collected for detection of viral load, virus replication and histopathological effects. Our results showed that RBD-Fc vaccination induced high titer of S-specific antibodies with long-term and potent SARS-CoV neutralizing activity. Four of five vaccinated mice were protected from subsequent SARS-CoV challenge because no significant virus replication, and no obvious histopathological changes were found in the lung tissues of the vaccinated mice challenged with SARS-CoV. Only one vaccinated mouse had mild alveolar damage in the lung tissues. In contrast, high copies of SARS-CoV RNA and virus replication were detected, and pathological changes were observed in the lung tissues of the control mice. In conclusion, our findings suggest that RBD, which can induce protective antibodies to SARS-CoV, may be further developed as a safe and effective SARS subunit vaccine.
Keywords: SARS-CoV, Receptor-binding domain, Protective immunity, Subunit vaccine
1. Introduction
Severe acute respiratory syndrome (SARS) is a novel infectious disease caused by SARS coronavirus (SARS-CoV), which led to several hundred deaths among thousands of cases. Although SARS has been successfully contained, re-emergence of SARS-CoV from animal reservoirs is still a potential risk for future epidemic, which is supported by continual reports of finding SARS-CoV-like coronavirus in small animals, such as civets, raccoon dogs [1], [2], [3] and bats [4], [5]. SARS outbreak may also recur in the future by the virus escaping from laboratory accidents [6], [7]. Therefore, development of safe and effective SARS vaccines for prevention of SARS-CoV infection is an important issue on current SARS research.
SARS-CoV, the causative agent of SARS, contains the genome encoding the nonstructural replicase polyprotein (rep) and structural proteins spike (S), envelope (E), membrane (M) and nucleocapsid (N) [8], [9]. Its S protein is responsible for virus binding to the receptor, angiotensin-converting enzyme 2 (ACE2), and subsequent virus entry into the host cells [10], [11]. The S protein of SARS-CoV has also been demonstrated to be the main antigen in inducing high titer of neutralizing antibodies [12], [13], [14], and in eliciting protective immunity against infection in challenged animals [15], [16], [17]. Thus, it is implied to be the main _target in development of SARS vaccines.
A number of vaccine candidates based on SARS-CoV S have been reported in terms of their abilities in inducing neutralizing antibodies and protective immunity [15], [18], [19], [20]. These candidates can be grouped into inactivated viruses-based, protein-based, DNA-based and virus-based vaccines [18], [20], [21], [22], [23]. Many viruses, including VSV, rhabdovirus, adenovirus, adeno-associated virus (AAV), modified vaccinia virus Ankara (MVA) and attenuated parainfluenza virus, have been used for expressing SARS-CoV S protein as SARS vaccine candidates [12], [15], [24], [25], [26], [27]. Bukreyev et al. [24] reported that vaccination of African green monkeys with an attenuated parainfluenza virus encoding SARS-CoV S resulted in production of SARS-CoV-specific neutralizing antibodies and protection of animals from virus challenge. These findings suggest that S protein of SARS-CoV is a good _target for development of SARS-CoV vaccines.
Since most DNA-based and virus-based vaccine candidates have still caused some safety concerns, protein-based vaccine may be an alternative candidate. Indeed, the full-length S protein or its S1 subunit could induce potent neutralizing antibody responses in the immunized animals [13], [25], [28]. However, it may also elicit antibodies that enhance virus infection [25], [29] or cause liver damage in animals challenged with SARS-CoV [23]. In this regard, the receptor-binding domain (RBD), a fragment of the S protein, which has been demonstrated to be a major neutralization determinant, has implied to be an ideal candidate for development of subunit vaccines against SARS [30], [31], [32], [33], [34]. In this study, we further evaluated its long-term immune responses and protective immunity against SARS-CoV infection in a mouse model.
2. Materials and methods
2.1. Virus, cells and recombinant proteins
The BJ01 strain of SARS-CoV (GenBank accession no. AY278488) was propagated in Vero E6 cells under the conditions of the biosafety level 3 laboratories (BSL-3) [35], [36]. Titer of this virus strain was determined by the 50% tissue-culture infectious dose (TCID50) in Vero E6 cells. A 193-aa RBD domain of SARS-CoV S protein (residues 318–510) fused with the Fc domain of human IgG1 (RBD-Fc) was produced as described previously [30], [31]. Briefly, a plasmid encoding RBD-Fc fusion protein was transfected into HEK293T cells using Fugene 6 transfection reagents (Boehringer Mannheim) according to the manufacturer's protocol. Supernatants were harvested 72 h post-transfection. Recombinant RBD-Fc protein was purified by protein A-Sepharose 4 Fast Flow (Amersham Biosciences, USA). A full-length S protein of SARS-CoV Urbani (accession no. AY278741) expressed in expresSF+® insect cells with recombinant baculovirus D3252 was purchased from the Protein Sciences Corporation (Bridgeport, CT) [13], [14].
2.2. Animal vaccination with SARS-CoV RBD-Fc
A group of five female BALB/c mice at the age of 4–6 weeks were vaccinated intramuscularly (i.m.) with 10 μg of purified RBD-Fc resuspended in phosphate-buffered solution (PBS, pH 7.2) in the presence of Freund's complete adjuvant (FCA), and boosted twice with freshly prepared emulsion of 5 μg immunogen and Freund's incomplete adjuvant (FIA) at 3-week intervals. The mice were boosted for the third time with the same amount of RBD-Fc + FIA 12 months after the first vaccination and were challenged with SARS-CoV 5 days later (Fig. 1 ). Mice injected with the same amount of PBS were used as the negative control. For each group, mouse sera were collected before immunization and at 1-month interval post-vaccination. Sera were kept at −20 °C until use.
Fig. 1.
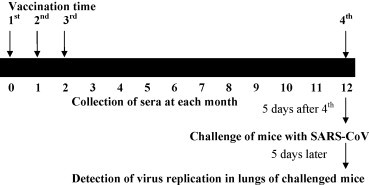
Schematics for RBD-Fc vaccination and SARS-CoV challenge. BALB/c mice were i.m. immunized with RBD-Fc fusion protein suspended in PBS plus adjuvant (vaccination group) or PBS only (control group) four times in total. Serum was collected from each mouse at a 1-month interval. The mice in both groups were intranasally challenged with SARS-CoV 5 days after the last vaccination, followed by detection of virus replication and histopathological changes in the mouse lung tissues 5 days after the challenge.
2.3. Enzyme-linked immunosorbent assay (ELISA)
Mouse sera were tested against the recombinant SARS-CoV S protein by ELISA. Briefly, 1 μg/ml of the recombinant S protein was coated to 96-well microtiter plates (Corning Costar, Acton, MA) in 0.1 M carbonate buffer (pH 9.6) at 4 °C overnight. After blocking with 2% non-fat milk, serially diluted mouse sera were added and incubated at 37 °C for 1 h, followed by three washes with PBS containing 0.1% Tween 20. Bound antibodies were detected with HRP-conjugated goat anti-mouse IgG (Zymed, South San Francisco, CA) at 37 °C for 1 h. After three washes, the reaction was visualized by addition of the substrate 3,3′,5,5′-tetramethylbenzidine (TMB) and measured for the absorbance at 450 nm (A450) by an ELISA plate reader (Victor 1420 Multilabel Counter, Perkin-Elmer, USA).
2.4. Neutralization assay for SARS-CoV infection
Titers of the neutralizing antibody in mouse sera vaccinated with the recombinant RBD-Fc protein were detected in Vero E6 cells as described by Qu et al. [37]. Briefly, Vero E6 cells were seeded at 2 × 104 cells/well in 96-well culture plates and cultured at 37 °C for 24 h to form a monolayer. Serial two-fold dilutions of serum samples were incubated with 100 TCID50 of SARS-CoV BJ01 strain at 37 °C for 1 h and subsequently added to the monolayer of Vero E6 cells at tetrad. Cells infected with 100 TCID50 SARS-CoV and without the virus were applied as positive and negative controls, respectively. Cytopathic effect (CPE) in each well was observed daily and recorded on day 3 post-infection. The neutralizing titers of mouse antisera that completely prevented CPE in 50% of the wells were calculated by Reed-Muench method.
2.5. Challenge of vaccinated mice with SARS-CoV
Five days after the last boost with RBD-Fc, mice were anaesthetized with isoflurane and intranasally (i.n.) inoculated with 50 μl of diluted SARS-CoV BJ01 strain (106 TCID50) according to the national animal care and use guidelines in a BSL-3 laboratory. Vaccinated mice were sacrificed 5 days after the challenge and lungs of the mice were removed. The lung tissues were stored at −80 °C for virological tests or were fixed with 4% paraformaldehyde and stored at 4 °C for histopathological analysis.
2.6. Quantitative reverse-transcriptase polymerase chain reaction (Q-RT-PCR)
The viral RNA copies in lung tissues were determined by Q-RT-PCR according to the protocol described by Zhai et al [38]. The limit of detection for live virus in lung samples was 1 × 103 copies/g. Briefly, total RNA was extracted from 20 mg of the lung tissues using RNeasy Mini kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. SARS-CoV RNA was quantified in a 30 μl mixture containing 10 μl RNA, 15 μl 2 × TaqMan® one-step RT-PCR Master Mix (ABI, 4309169), 0.75 μl 40 × multiscribe, 0.25 μM each forward primer Af (Taq-772F 5′-AAG CCT CGC CAA AAA CGT AC-3′), reverse primer Ar (Taq-1000R 5′-AAG TCA GCC ATG TTC CCG AA-3′) and probe (Taq-955T 5′-FAM-TCA CGC ATT GGC ATG GAA GTC ACA CT-TAMRA) (TIB Molbiol, Berlin, Germany) using a fluorometric PCR instrument (ABI 7300).
2.7. Determination of viral titers and total virus
The virus replication was determined by titration of the inoculated virus in lung tissues collected from sacrificed mice. Briefly, the lung tissues were homogenized to a final concentration of 10% (w/v) suspension in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, USA). Tissue homogenates were centrifuged, filtrated and inoculated into the monolayer of Vero E6 cells seeded in 96-well tissue-culture plates. Results were evaluated after 3 days of the culture under phase-contrast microscopy, and viral titers using a CPE-based TCID50 test were calculated by Reed-Muench method. Viral titers were expressed as Log10TCID50/g of tissues, with the lowest detection limit of 1.5 Log10TCID50/g. The total amount of virus was calculated by multiplying the weight of the lung tissues and the viral titers measured in 10% of tissue homogenates.
2.8. Histopathological assay
The lung tissues were immediately fixed in 4% buffered paraformaldehyde and embedded in paraffin wax. Sections were made at 4–6 μm thickness and mounted on slides. Histopathological changes caused by SARS-CoV infection were examined by haematoxylin and eosin (H&E) staining and viewed under light microscopy.
3. Results
3.1. Vaccination with RBD-Fc induced long-term and potent SARS-CoV S-specific antibodies with strong neutralizing activity
As shown in Fig. 2A, RBD-Fc vaccination induced a prolonged and potent humoral immune response with IgG specific to SARS-CoV S protein in the animals as tested by ELISA, reaching the highest titer (1:1.5 × 105 ± 1.6 × 104, Mean ± SE) at the 3rd and 4th month post-vaccination. Although the titer decreased slightly afterwards, high titer of S-specific IgG antibody maintained for at least 6 months until it dropped to 1:9.0 × 102 ± 1.6 × 102 at the end of the 7th month. Specific IgG antibody increased rapidly after the 3rd boost (12 months after the first vaccination) and reached the titer of 1:2.5 × 103 ± 7.7 × 102 before SARS-CoV challenge. Fig. 2B further demonstrated that the antibodies induced by vaccination of RBD-Fc exhibited strong neutralizing activity with a similar pattern of the ELISA antibodies to RBD. The neutralizing antibodies reached the highest level at the 3rd month (1:4.0 × 103 ± 3.5 × 102) after the vaccination with RBD-Fc. The neutralizing antibody level slightly decreased to 1:5.8 × 102 ± 1.0 × 102 by the end of the 6th month but dropped down to less than 1:40 at the end of the 12th month. The titer of the neutralizing antibody to SARS-CoV increased again after the third boost, reaching the titer of 1:3.2 × 102 ± 1.9 × 102 before SARS-CoV challenge. However, neither ELISA S-specific antibodies nor neutralizing antibodies were detected in the mouse sera from the control group.
Fig. 2.
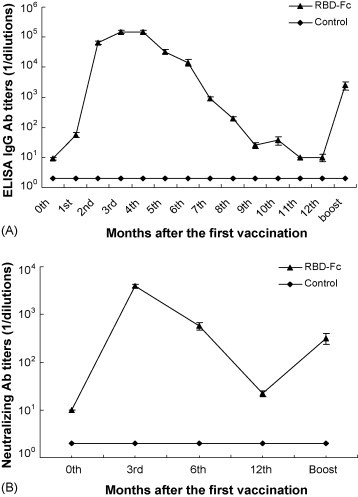
Detection of anti-S antibodies and virus neutralizing antibodies in mouse sera. (A) Titers of anti-S antibodies measured by ELISA for 12 months at monthly basis and 5 days after the last boost. Data are presented as the geometric means ± SE of five mouse sera collected at different time points. (B) Titers of the SARS-CoV neutralizing antibody in sera collected at 0, 3, 6 and 12 months post-vaccination and sera collected 5 days after the last boost. The titers of neutralizing antibodies were determined as the highest dilutions of sera that could completely prevent CPE in at least 50% of the wells and are presented as the geometric means ± SE of five mouse sera in each group.
3.2. Neutralizing antibodies elicited by RBD-Fc vaccination played an important role in inhibiting SARS-CoV infection in challenged mice
Five days after challenged with SARS-CoV through the intranasal route, mice were sacrificed and lung tissues were collected. Virus RNA copies in the lung tissues were determined by Q-RT-PCR and described as RNA copies/g of tissues. As shown in Table 1 , viral RNA copies for both the RBD-Fc group and the control group were negatively correlated with the titer of the neutralizing antibodies against SARS-CoV. The higher titer of the neutralizing antibodies was detected in the sera, the lower copy number of viral RNA was found in the lung tissues. M1 mouse in the RBD-Fc vaccination group developed the lowest titer of neutralizing antibody (1:57) and showed detectable viral RNA copies (3.13 × 103), while no viral RNA was detectable in lung tissues of the other four mice in this group as they presented higher titers of the neutralizing antibody (>1:189). Similarly, no neutralizing antibody response in the PBS control group was correlated to the higher levels of RNA copies in the lung tissues (1.39 × 105 to 3.77 × 107). These results suggested that the neutralizing antibodies induced by RBD-Fc vaccination play an important role in prevention of SARS-CoV infection in virus challenged mice.
Table 1.
SARS-CoV RNA copies in the lung tissues measured by Q-RT-PCRa
| Group | Mouse ID | Neutralizing antibody titers | RNA copies/g of lung tissues |
|---|---|---|---|
| RBD-Fc vaccination | M1 | 57 | 3.13 × 103 |
| M2 | 189 | Undetectable | |
| M3 | 505 | Undetectable | |
| M4 | 452 | Undetectable | |
| M5 | 404 | Undetectable | |
| Control | C1 | <4 | 1.39 × 107 |
| C2 | <4 | 3.77 × 107 | |
| C3 | <4 | 1.39 × 105 | |
| C4 | <4 | 1.98 × 106 | |
| C5 | <4 | 1.45 × 106 | |
Titers of neutralizing antibodies in sera collected from each mouse 5 days after the last vaccination and before the virus inoculation were determined. RNA copies in the lung tissues of mice challenged with SARS-CoV were measured by Q-RT-PCR and expressed as RNA copies/g of lung tissues. M1–M5 and C1–C5 are RBD-Fc-vaccinated mice and control mice, respectively. The limit of detection for live virus in the lung was 1 × 103 copies/g.
3.3. Vaccination with RBD-Fc suppressed SARS-CoV replication in the virus challenged mice
SARS-CoV replication was detected by titration of the inoculated virus in lung tissues to evaluate the efficacy of RBD-Fc vaccination in protecting SARS-CoV infection (Fig. 3 ). All of five mice in the control group vaccinated with PBS had high titers of virus replication in the lungs after the SARS-CoV challenge. However, the mice vaccinated with RBD-Fc were either completely (four of five mice) or partially protected (one of five mice, M1 which showed lower level of neutralizing antibody in its serum) from SARS-CoV challenge. These data indicate that vaccination with RBD-Fc is able to establish protective immunity to prevent mice from subsequent SARS-CoV challenge.
Fig. 3.
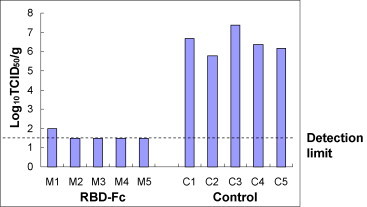
RBD-Fc protected vaccinated mice from subsequent challenge with SARS-CoV. Groups of five BALB/c mice vaccinated with RBD-Fc or PBS were intranasally challenged with 106 TCID50 of SARS-CoV 5 days after the last dose of vaccination. The titers of SARS-CoV replicated in the lung tissues of the vaccinated mice and control were detected 5 days after the challenge, and expressed as Log10TCID50/g of tissues. M1–M5 are mice vaccinated with RBD-Fc, and C1–C5 indicate the control mice. The limit of detection was 1.5 Log10TCID50/g.
3.4. Mice vaccinated with RBD-Fc did not develop histopathological changes in the lung tissues
Histopathological changes in lungs from the RBD-Fc vaccinated and control mice were observed on the H&E stained lung tissue sections. Lung tissues from the control mice (injected with PBS) revealed significant histopathological changes, including diffuse alveolar damages characterized by disruption of alveolar walls and flooding of alveolar lumina with serosanguineous exudates admixed with neutrophils and alveolar macrophages, thickened alveolar walls lined by type 2 pneumocytes, and alveolar macrophages in alveolar lumina (Fig. 4A). However, except M1 mouse with a mild level of the alveolar damage in lung tissues, other four RBD-Fc vaccinated mice exhibited no significant histopathological changes, but showed similar histological structures as the normal mouse in lung tissues (Fig. 4B). These results further confirm that RBD-Fc vaccine is able to prevent mice from subsequent SARS-CoV infection.
Fig. 4.
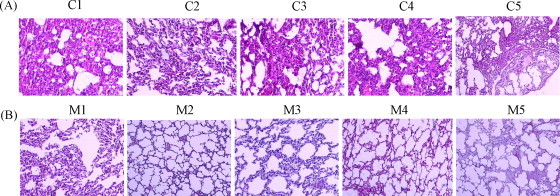
Histopathological examinations of mouse lung tissues. All sections of mouse lung tissues were stained with H&E and examined under microscopy (original magnification 200×). C1–C5 and M1–M5 are control mice and RBD-Fc vaccinated mice, respectively. (A) Histopathological changes of lung tissues from control mice administered with PBS. Obvious histopathological changes were observed in lung tissues from control mice, including diffuse alveolar damages characterized by disruption of alveolar walls and flooding of alveolar lumina with serosanguineous exudates admixed with neutrophils and alveolar macrophages, thickened alveolar walls lined by type 2 pneumocytes, and alveolar macrophages in alveolar lumina. (B) Histological characterization of lung tissues from RBD-Fc vaccinated mice. No significant histopathological changes of lung tissues were observed in four of five RBD-Fc-vaccinated mice (M2–M5). Only M1 mouse exhibited mild histopathological damages in lung tissues. But compared to the control mice, M1 mouse revealed a little damage such as thickened alveolar walls with interstitial mononuclear cells and lymphocytes infiltrates, and with congestion but no markedly hemorrhage or effusion in the alveolar spaces of lung tissues.
4. Discussion
Development of effective and safe SARS vaccines is crucial in prevention of re-occurrence of SARS epidemic. Among four structural proteins of SARS-CoV, S serves as the most important antigen for development of SARS vaccines since it plays essential roles in the host cell tropism and virus entry, and its ability to induce neutralizing antibodies and protective immunity [13], [17], [20], [39], [40]. Reports have shown that neutralizing antibodies against S protein protect vaccinated animals from SARS-CoV challenge, thus vaccines encoding the S protein and _targeting the humoral immune responses/neutralizing antibodies are indispensable in preventing SARS-CoV infection [16], [17], [20], [28], [39], [40].
Most of the currently reported SARS vaccine candidates are based on the full-length S protein of SARS-CoV. Although the full-length S protein-based vaccines can elicit neutralizing antibodies and/or protective immunity [13], [17], [20], it has been reported that this kind of vaccines may also induce harmful inflammatory and immune responses [23], [29]. This has raised concerns about the efficacy and safety of the vaccines containing or encoding the full-length S protein. We thus proposed to use RBD, a fragment of S protein, rather than the full-length S protein, for development of a safe and effective SARS subunit vaccine since it contains the major neutralizing epitopes [31], [32], [41]. Our previous studies have shown that the recombinant RBD can induce high potent neutralizing antibodies against SARS-CoV and its variants [31], [34], but we do not know whether the RBD-based vaccine can protect animals from SARS-CoV infection.
In this study, we compared the antibody responses and protective immunity in mice vaccinated with RBD-Fc and the control mice. We found that RBD-Fc elicited potent and long-term humoral immune responses in vaccinated mice. High titers of SARS-CoV S-specific ELISA antibodies and virus neutralizing antibodies were maintained for at least 6 months. Although the antibodies dropped to a low level at the end of the 12th month, their titers rebound rapidly after the mice were re-boosted with RBD-Fc. These vaccinated mice were protected from SARS-CoV challenge since no significant virus replication was detected in their lungs. Notably, presence of high titers of the neutralizing antibody was a prerequisite for protective immunity. Higher titer of neutralizing antibodies was associated with lower level of RNA copies/virus replication and stronger protective activity against SARS-CoV challenge. Among five mice vaccinated with RBD-Fc, four mice possessing high titer of neutralizing antibodies exhibited no significant virus replication and histopathological changes in their lungs while only one mouse with low sera neutralizing antibody titer showed marginal level of virus RNA copies, and mild histopathological changes. However, all of the five mice with undetectable neutralizing antibody in the control group presented high level of RNA copies/virus replication, and obvious histopathological changes in their lungs. Our results indicated that the neutralizing antibody produced in RBD-Fc vaccinated mice plays a significant role in protection of mice from the virus challenge without causing immunopathological damages.
In summary, the vaccines containing the RBD of SARS-CoV S protein may induce sufficient neutralizing antibodies and long-term protective immunity against SARS-CoV challenge in the established mouse model. This vaccine candidate may overcome the disadvantages of other vaccine candidates mentioned above, but protect the vaccinated mice from subsequent SARS-CoV challenge. Our results suggest that RBD-Fc vaccine can be further developed as an ideal subunit vaccine for prevention of SARS epidemic.
Acknowledgements
This study was supported by China 973 Grant 2005CB523000, Beijing SARS Research Grant (H030230100130) and by Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government.
References
- 1.Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- 2.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 3.Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. 2005;102(7):2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 6.Liang G., Chen Q., Xu J., Liu Y., Lim W., Peiris J.S. Laboratory diagnosis of four recent sporadic cases of community-acquired SARS, Guangdong Province, China. Emerg Infect Dis. 2004;10(10):1774–1781. doi: 10.3201/eid1010.040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Normile D. Infectious diseases. Mounting lab accidents raise SARS fears. Science. 2004;304(5671):659–661. doi: 10.1126/science.304.5671.659. [DOI] [PubMed] [Google Scholar]
- 8.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 9.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 10.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faber M., Lamirande E.W., Roberts A., Rice A.B., Koprowski H., Dietzschold B. A single immunization with a rhabdovirus-based vector expressing severe acute respiratory syndrome coronavirus (SARS-CoV) S protein results in the production of high levels of SARS-CoV neutralizing antibodies. J Gen Virol. 2005;86(Pt 5):1435–1440. doi: 10.1099/vir.0.80844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y., Li J., Heck S., Lustigman S., Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J Virol. 2006;80(12):5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Z., Post P., Chubet R., Holtz K., McPherson C., Petric M. A recombinant baculovirus-expressed S glycoprotein vaccine elicits high titers of SARS-associated coronavirus (SARS-CoV) neutralizing antibodies in mice. Vaccine. 2006;24(17):3624–3631. doi: 10.1016/j.vaccine.2006.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapadia S.U., Rose J.K., Lamirande E., Vogel L., Subbarao K., Roberts A. Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology. 2005;340(2):174–182. doi: 10.1016/j.virol.2005.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Zhang L., Qin C., Ba L., Yi C.E., Zhang F. Recombinant modified vaccinia virus ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily _targeting the receptor binding region. J Virol. 2005;79(5):2678–2688. doi: 10.1128/JVI.79.5.2678-2688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J Gen Virol. 2004;85(Pt 10):3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- 19.Qin E., Shi H., Tang L., Wang C., Chang G., Ding Z. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine. 2006;24(7):1028–1034. doi: 10.1016/j.vaccine.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong W.P., Xu L., Stadler K., Ulmer J.B., Abrignani S., Rappuoli R. Modulation of the immune response to the severe acute respiratory syndrome spike glycoprotein by gene-based and inactivated virus immunization. J Virol. 2005;79(22):13915–13923. doi: 10.1128/JVI.79.22.13915-13923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spruth M., Kistner O., Savidis-Dacho H., Hitter E., Crowe B., Gerencer M. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine. 2006;24(5):652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., St Claire M. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23(17–18):2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du L., He Y., Wang Y., Zhang H., Ma S., Wong C.K. Recombinant adeno-associated virus expressing the receptor-binding domain of severe acute respiratory syndrome coronavirus S protein elicits neutralizing antibodies: implication for developing SARS vaccines. Virology. 2006;353(1):6–16. doi: 10.1016/j.virol.2006.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhi Y., Figueredo J., Kobinger G.P., Hagan H., Calcedo R., Miller J.R. Efficacy of severe acute respiratory syndrome vaccine based on a nonhuman primate adenovirus in the presence of immunity against human adenovirus. Hum Gene Ther. 2006;17(5):500–506. doi: 10.1089/hum.2006.17.500. [DOI] [PubMed] [Google Scholar]
- 28.Bisht H., Roberts A., Vogel L., Subbarao K., Moss B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology. 2005;334(2):160–165. doi: 10.1016/j.virol.2005.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z.Y., Werner H.C., Kong W.P., Leung K., Traggiai E., Lanzavecchia A. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci USA. 2005;102(3):797–801. doi: 10.1073/pnas.0409065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of SARS coronavirus spike protein contains multiple conformational-dependant epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 32.He Y., Jiang S. Vaccine design for severe acute respiratory syndrome coronavirus. Viral Immunol. 2005;18(2):327–332. doi: 10.1089/vim.2005.18.327. [DOI] [PubMed] [Google Scholar]
- 33.He Y., Zhu Q., Liu S., Zhou Y., Yang B., Li J. Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology. 2005;334(1):74–82. doi: 10.1016/j.virol.2005.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y., Li J., Li W., Lustigman S., Farzan M., Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies _targeting the receptor-binding domain of spike protein. J Immunol. 2006;176(10):6085–6092. doi: 10.4049/jimmunol.176.10.6085. [DOI] [PubMed] [Google Scholar]
- 35.He Y., Zhou Y., Wu H., Luo B., Chen J., Li W. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J Immunol. 2004;173(6):4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 36.He Y., Zhou Y., Siddiqui P., Jiang S. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem Biophys Res Commun. 2004;325(2):445–452. doi: 10.1016/j.bbrc.2004.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qu D., Zheng B., Yao X., Guan Y., Yuan Z.H., Zhong N.S. Intranasal immunization with inactivated SARS-CoV (SARS-associated coronavirus) induced local and serum antibodies in mice. Vaccine. 2005;23(7):924–931. doi: 10.1016/j.vaccine.2004.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhai J., Briese T., Dai E., Wang X., pang X., Du Z. Real-time polymerase chain reaction for detecting SARS coronavirus, Beijing, 2003. Emerg Infect Dis. 2004;10(2):300–303. doi: 10.3201/eid1002.030799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giroglou T., Cinatl J., Jr., Rabenau H., Drosten C., Schwalbe H., Doerr H.W. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J Virol. 2004;78(17):9007–9015. doi: 10.1128/JVI.78.17.9007-9015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang S., He Y., Liu S. SARS vaccine development. Emerg Infect Dis. 2005;11(7):1016–1020. doi: 10.3201/eid1107.050219. [DOI] [PMC free article] [PubMed] [Google Scholar]


