Abstract
Trypanosoma brucei is a kinetoplastid parasite that causes African trypanosomiasis, which is fatal if left untreated. T. brucei regularly switches its major surface antigen, VSG, to evade the host immune responses. VSGs are exclusively expressed from subtelomeric expression sites (ESs) where VSG genes are flanked by upstream 70 bp repeats and downstream telomeric repeats. The telomere downstream of the active VSG is transcribed into a long-noncoding RNA (TERRA), which forms RNA:DNA hybrids (R-loops) with the telomeric DNA. At an elevated level, telomere R-loops cause more telomeric and subtelomeric Double-Strand Breaks (DSBs) and increase VSG switching rate. In addition, stabilized R-loops are observed at the 70 bp repeats and immediately downstream of ES-linked VSGs in RNase H-defective cells, which also have an increased amount of subtelomeric DSBs and more frequent VSG switching. Although subtelomere plasticity is expected to be beneficial to antigenic variation, severe defects in subtelomere integrity and stability increase cell lethality. Therefore, regulation of the telomere and 70 bp repeat R-loop levels is important for the balance between antigenic variation and cell fitness in T. brucei. Additionally, the high level of the active ES transcription favors accumulation of R-loops at the telomere and 70 bp repeats, providing an intrinsic mechanism for local DSB formation, which is a strong inducer of VSG switching.
Keywords: R-loop, TERRA, telomere, 70 bp repeats, antigenic variation, Trypanosomes
Graphical Abstract
Introduction
Transcription associated RNA:DNA hybrids or R-loops are a double-edged sword that plays important roles in certain cellular processes and causes genome instability 1. Therefore, their localization and amount need to be tightly regulated. The telomeric tandem repeats in a number of organisms are transcribed into TERRA 2, which can form telomere R-loops that often affects genome stability 3 in humans, yeast, and Trypanosoma brucei cells 4. Recent studies also showed that R-loops at the telomere and the subtelomere influence antigenic variation 5 in T. brucei 6–8. The beneficial functions and adverse effects of R-loops have been reviewed extensively previously 1, 4, 9, 10. Here we will focus on recent findings on effects of R-loops, particularly those at the telomere and the subtelomere, on antigenic variation in trypanosomes.
The R-loop structure
R-loops are three-stranded RNA-DNA structures with an RNA:DNA hybrid and a displaced single-stranded DNA 11. The replication-associated RNA:DNA hybrids are usually 11 bp long (in Okazaki fragments) and transcription-associated RNA:DNA hybrids are generally 8 bp long (within the RNA polymerase active site) 12. However, longer R-loops (> 1 kb) can also form 13. Currently the thread back hypothesis is the best working model for R-loop formation: DNA behind a transcription bubble is negatively supercoiled, which has a tendency to unwind, allowing the nascent RNA to anneal with the template strand easily 1, 14. In support of this, defective transcription elongation and termination, RNA splicing, and relaxation of supercoiled DNA all lead to elevated R-loop levels 15–18. In addition, DNA nicks on the nontemplate DNA strand downstream of the promoter and G clusters facilitate initial R-loop formation, while subsequent expansion and stabilization of the RNA:DNA hybrid are enhanced by high G density and negative supercoiling 19.
R-loops play important roles and are necessary for several cellular processes 1. First, in mammals, R-loops drive the programmed genomic rearrangement during immunoglobulin class switch in activated B-cells 20. Second, R-loops formed at the promoter regions can modulate gene expression 21–23. Third, R-loops are associated with H3 S10 phosphorylation, a heterochromatic marker, suggesting that it has an important role in chromatin structure modulation 24. Fourth, in human mitochondria, origin-specific DNA replication is initiated with a two step process: (1) transcription from an upstream promoter leads to accumulation of R- loops, and (2) processing the R-loops by RNase H1 generates 3’ ends for DNA replication by DNA polymerase γ 25. However, R-loops are also well-known to be a genome instability factor 26. The displaced single-stranded DNA can be easily mutated and contributes to transcription-associated mutagenesis 27, recombination, and DSBs 28, 29. A stable R-loop structure can also block the progression of the replication fork 19,30, although the underlying mechanism is still not fully understood 3. It is also possible that R-loops can trigger chromatin compaction 31–33, which in turn can block replication fork progression 3. Additionally, nucleotide excision repair endonucleases can process R-loops into DSBs 34.
R-loops are quite stable, as RNA:DNA interactions are thermodynamically more stable than DNA:DNA pairing 35, and many enzymes are involved in dissolution of the R-loop. Several RNA helicases including Rho 36, DHX9 37, and Senataxin 38 can unwind the RNA:DNA duplex. Ribonuclease H1 (RNase H1) can degrade the RNA strand of the RNA:DNA hybrid and requires a tract of at least four ribonucleotides in the substrate 39. Eukaryotic Ribonuclease H2 (RNase H2), usually a trimer 40, 41, can resolve the R-loop 42, 43 and remove single ribonucleotides from genomic DNA 44–46. These enzymes play important roles in maintaining an appropriate level of R-loops in cells 47.
Recent studies in a kinetoplastid parasite, Trypanosoma brucei, have shown that telomere and subtelomere R-loops can induce DNA damages, not only contributing to genome instability but also emerging as an important factor that influences antigenic variation 6–8. These results further indicate that R-loops can have both beneficial and detrimental effects, and its level needs a tight regulation.
Antigenic variation in T. brucei
Trypanosomsa brucei causes human and animal African trypanosomiasis, which are usually fatal without treatment. T. brucei is transmitted by tsetse (Glossina spp.) and threatens millions of people in sub-Saharan Africa 48. The bloodstream form (BF) T. brucei proliferates in the extracellular space of the mammalian host. Its major surface antigen, Variant Surface Glycoprotein (VSG), forms a dense protein layer 49, masks other invariant surface antigens, and can elicit strong immune responses 50. However, T. brucei regularly switches its VSG coat, effectively evading its elimination by the host 5. This antigenic variation is a critical pathogenesis mechanism and has two major aspects: VSG monoallelic expression and VSG switching.
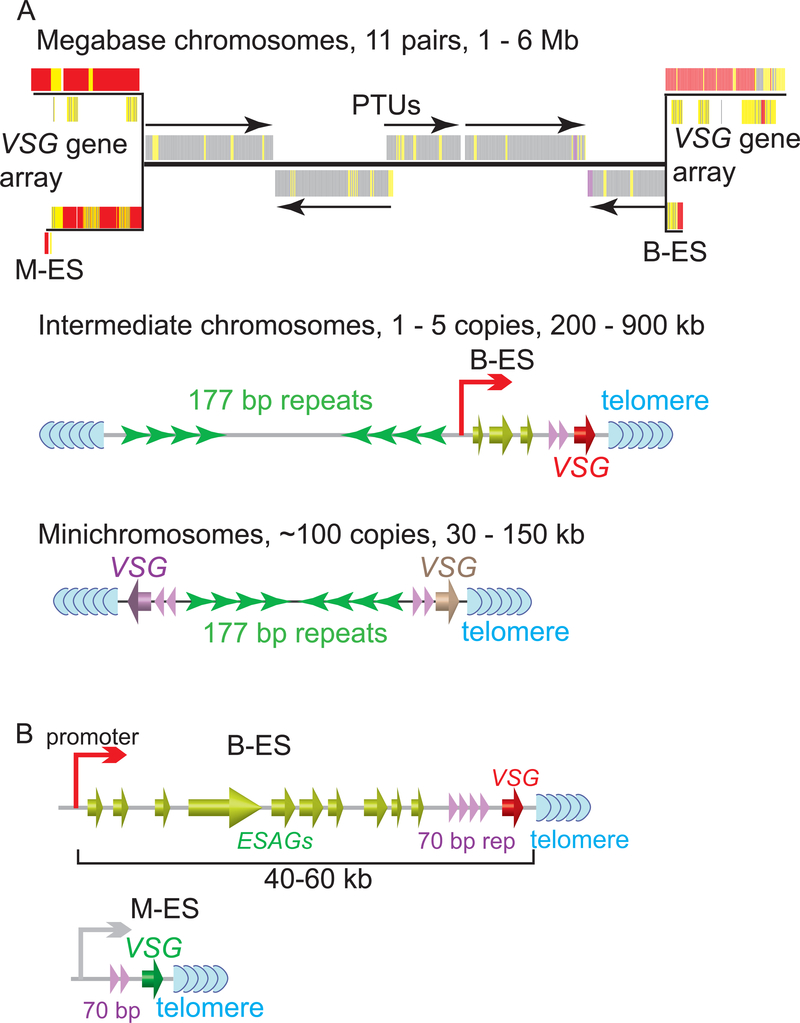
There are more than 2,500 VSG genes and pseudogenes (all are located at subtelomeric regions) in the T. brucei genome 51. Many of these are in large VSG gene arrays on megabase chromosomes (Fig. 1A, top) 52–54. Individual VSG genes are also found at two-thirds of minichromosome subtelomeres (Fig. 1A, bottom) 51, 55, 56. VSG genes located at these loci are normally not expressed. At the BF stage, VSGs are expressed exclusively from BF VSG Expression Sites (B-ESs, Fig. 1B, top), which are large polycistronic transcription units (PTUs) transcribed by RNA Pol I and are located immediately upstream of the telomere on megabase or intermediate chromosomes (Fig. 1A, top and middle) 57, 58. VSG is the last gene in any B-ES and is located within 2 kb from the telomeric TTAGGG repeats in all completely sequenced B-ESs, while the B-ES promoter is located 40 – 60 kb upstream (Fig. 1B, top) 52, 59, 60. 70 bp repeats are found immediately upstream of the VSG gene in long arrays (3 – 20 kb) in B-ESs 52, 59. T. brucei has multiple B-ESs (~ 15 in the Lister 427 strain), all with similar gene organization and ~ 90% sequence identity 59, but often having different VSG genes 59. However, at any moment, only one B-ES is fully active, presenting a single type of VSG on the cell surface 49. Monoallelic VSG expression ensures effectiveness of VSG switching, as the previously active VSG needs to be silenced for the parasite to avoid elimination by the host. Although detailed mechanisms are not fully understood, many factors have been shown to regulate VSG expression 61, such as chromatin structure 62–72, transcription elongation 73–75, inositol phosphate pathway 76, 77, nuclear lamina 78, 79, recruitment of sumoylated protein(s) to the active ES promoter 80, DNA replication initiation factors 81–84, a subtelomere and VSG-associated VEX complex 85, 86, and telomeric silencing 64, 87. The VSG coat is lost when T. brucei is ingested by its insect vector, tsetse 88. At the same time, T. brucei differentiates into the procyclic form (PF). After migrating to the salivary gland of tsetse, the infectious metacyclic form T. brucei expresses VSG again and is ready to be injected into a mammalian host 89. At this stage, VSGs are expressed from metacyclic ES (M-ESs, Fig. 1B, bottom) that are also located at subtelomeric regions and are transcribed by RNA Pol I 90, 91, except that M-ESs are monocistronic transcription units with the promoter located < 5 kb from the telomere 92–96.
Figure 1.
(A) Schematic diagram of a megabase chromosome (top), an intermediate chromosome (middle), and a minichromosome (bottom) in T. brucei, with their type, number, and size indicated. The central region of the two homologous megabase chromosomes are the same and is shown once, while their subtelomeres are different and shown separately (top). Individual genes are represented as short colored bars. Grey, functional genes; red, VSG genes; yellow, pseudogenes. PTUs and their transcription directions are marked with arrows. VSG gene arrays, a B-ES, and an M-ES are shown. A B-ES is shown at one subtelomere of the intermediate chromosome (middle). Individual VSG genes are shown at subtelomeres of the minichromosome (bottom) (B) A representative B-ES (top) and an M-ES (bottom) is shown. ESAGs: Expression Site Associated Genes.
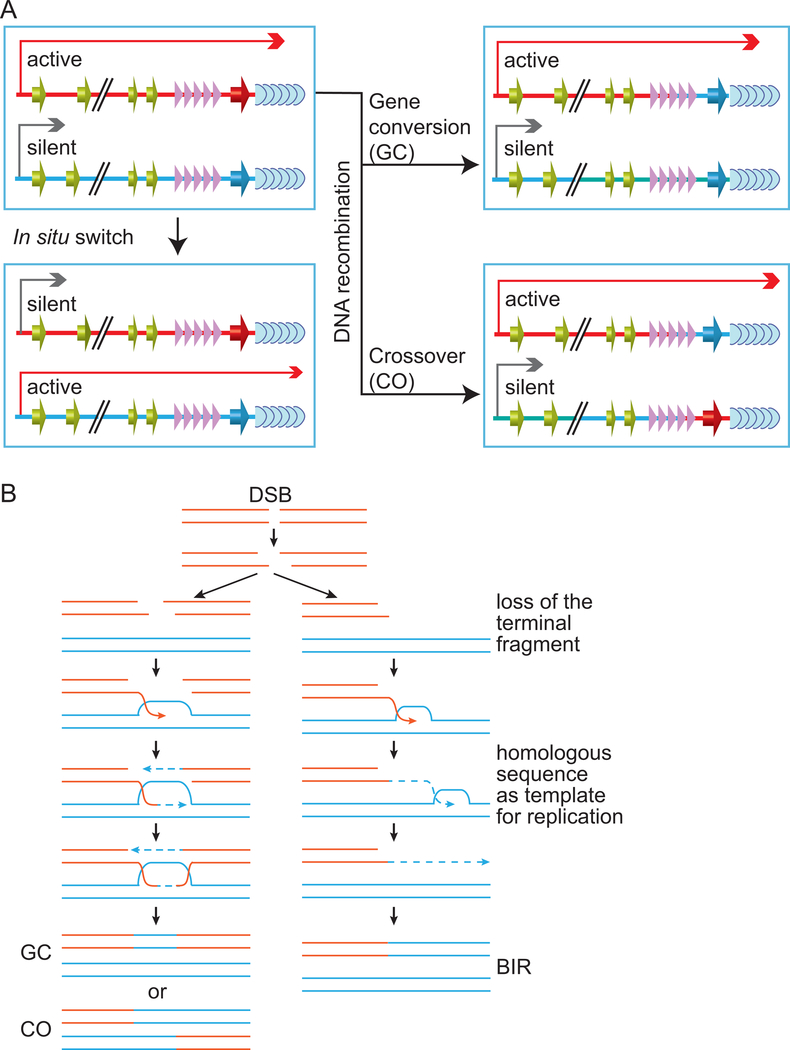
VSG switching only occurs in proliferative BF T. brucei cells 97. It has two major pathways 98, 99. In-situ switching occurs at the transcriptional level, where the originally active ES is silenced while a previously silent ES becomes fully transcribed, and no gene rearrangement is involved (Fig. 2A, bottom left) 98,99. The second and more frequent switching event is DNA recombination-mediated and includes two major types of process 100. In crossover (CO, or telomere exchange, Fig. 2A, bottom right), a silent VSG (often ES-linked or at a minichromosome subtelomere) exchange places with the active VSG reciprocally, resulting in a new VSG being expressed from the originally active ES with no loss of genetic information 101, 102. In gene conversion-mediated VSG switching (GC, Fig. 2A, top right), a previously silent VSG gene is copied into the active ES to replace the originally active VSG, which is subsequently lost 103, 104. Theoretically, any functional VSG gene in the genome can act as a donor in GC-mediated VSG switching. However, sequences of different B-ESs are 90% identical 59, GC can often occur between B-ESs as they have long homologous sequences 103, 105. Additionally, all ES-linked and minichromsome VSGs are expected to be good GC donors as they have long telomere repeats downstream and 70 bp repeats upstream, the same as the active VSG (Fig. 1). Indeed, telomere conversion-mediated VSG switching events have been observed 106. When telomere-adjacent VSGs are used as donors, it is possible that VSG switching may occur through break-induced-replication (BIR) 107 instead of a classical GC (Fig. 2B). However, whether BIR occurs in VSG switching has not been investigated. Another type of GC has also been observed in VSG switching: several VSG genes can act as donors simultaneously, each donating a piece of the gene segment, resulting in a novel mosaic VSG gene 108–111. In many published switching assays, in situ switching events are less frequent than recombination-mediated ones, while gene conversion is the most prevalent 6,112–115.
Figure 2.
(A) VSG switching mechanisms. Light blue soft arrow heads, telomere repeats; purple arrow heads, 70 bp repeats; red and dark blue 3D arrows, VSG genes; green 3D arrows, ESAGs; long red line with an arrow head, active transcription from the B-ES promoter; grey short line with an arrow head, short-distance transcription from the silent B-ES promoter. VSG switching pathways are explained in the text. (B) The classical double Holliday Junction pathway of homologous recombination to generate either GC or CO products is shown on the left. In comparison, the BIR pathway that usually occurs at the telomere is shown on the right. The BIR and GC products are indistinguishable because the telomeres downstream of the VSG genes have the same tandem repeat sequence.
Many proteins, especially those involved in DNA recombination, have been shown to play important roles in VSG switching. For example, RAD51 that mediates strand invasion in homologous recombination 116, RAD51–3 (a RAD51-related protein) 117, and BRCA2 118, all facilitate VSG switching, while Topoisomerase 3 alpha 114, the RMI1 homologue 115, replication origin binding factor TbORC1 81, and a RecQ helicase, RECQ2 119, suppress VSG switching. Additionally, telomere proteins have been shown to suppress VSG switching 6, 112, 120, 121, while cells harboring an extremely short active VSG-adjacent telomere have a ~ ten-fold higher VSG switching rate than WT cells 122. In addition, cells with defective RNase H enzymes appear to have a higher VSG switching frequency than WT cells 7, 8. However, how VSG switching is initiated and regulated is less well-understood 123, 124.
T. brucei telomere proteins and antigenic variation
VSG is expressed exclusively from subtelomeric regions 60, and the telomere complex has been shown to play important roles in antigenic variation in T. brucei 6, 87, 112, 120, 121, 125. Telomeres are nucleoprotein complexes that are essential for genome integrity and chromosome stability in eukaryotes 126, 127. Proteins that directly bind the telomere DNA or associate with the telomere chromatin through protein-protein interaction play critical roles in all aspects of telomere functions, including proper maintenance of the telomere length 128, suppression of illegitimate DNA degradation, recombination, and repair at the chromosome ends 129, assembly of a telomere heterochromatin that represses subtelomeric gene expression 130, and regulation of the telomeric transcript level 131.
In T. brucei, besides the telomerase components that are required for telomere maintenance 132–134, several other telomere proteins have been identified 87, 121, 125, 135. TbTRF directly binds the duplex TTAGGG repeats through its C-terminal Myb domain 135, and the telomere DNA binding activity is essential for cell viability and telomere/subtelomere stability: cells transiently depleted of TbTRF and the TbTRF mutants with weakened telomere-binding activities have more VSG switching events that mostly involve the loss of the active ES 112. A TbTRF-interacting factor, TbTIF2, is essential for normal cell growth and has a critical role in maintaining telomere/subtelomere integrity: depletion of TbTIF2 results in increased amount of DSBs in ESs, and a transient depletion of TbTIF2 leads to more frequent VSG switching, with most events involving the loss of the active ES 121. TelAP1 is identified in the TbTRF protein complex and in the complex that can interact with an oligonucleotide of telomeric sequence 125. It is the only non-essential telomere protein identified so far, and TelAP1 null cells exhibit a faster VSG silencing kinetics when cells are differentiated from BF to PF in vitro 125.
TbRAP1 is a TbTRF-interaction factor that associates with the telomere chromatin and is essential for cell viability 87. Depletion of TbRAP1 leads to derepression of all ES-linked silent VSGs (upto several thousand-fold), while the originally active VSG is expressed at ~ 50% of its WT level 64, 87. Because transcription from silent B-ES promoters is detected but transcription elongation attenuates within a few kb 74, 75, it is hypothesized that depletion of TbRAP1 removes a blockage of transcription elongation, resulting in basal level VSG expression from all silent ESs 87. This is consistent with the observation that promoter-less VSGs at minichromosome subelomeres are not derepressed after TbRAP1 depletion 64,87. Additionally, the TbRAP1-mediated telomeric silencing is position-dependent, exerting strongest effects on telomere-adjacent VSG genes, weaker effects on VSG pseudogenes in the middle of B-ESs, and weakest effect on reporter genes inserted immediately downstream of the B-ES promoter 87. The PF T. brucei cells proliferate in the midgut of tsetse and express procyclins as its major surface protein 136. VSGs are normally silenced at this stage 88 but derepressed upon TbRAP1 depletion 64. Additionally, telomeric and subtelomeric chromatin structure is less compact when TbRAP1 is removed in PF cells 64. TbRAP1 also plays an important role in maintaining telomere/subtelomere integrity and stability, which helps suppress VSG switching 6. Interestingly, the underlying mechanism involves the telomeric transcript (TERRA) and telomere R-loops.
TERRA in T. brucei
The UUAGGG repeat-containing telomere transcript (TERRA) was first identified in T. brucei cells and several closely related kinetoplastid parasites three decades ago 137. In recent years, TERRA has been detected in all eukaryotes tested 138–142 and has been shown to be the product of telomere repeat transcription in humans, mouse embryonic stem cells, both budding and fission yeasts, and plants 140, 141, 143–149. Often, all telomeres are not transcribed 150, 151, and transcription from intrachromosomal telomeric sequences can be abundant 149. TERRA has been shown to play important roles in telomere protection 138, 151,153, length regulation 148, 154, 157, and recombination 158 in mammalian cells and yeasts. TERRA may also play a role in gene expression regulation in mouse embryonic stem cells 152. In addition, an excessive amount of TERRA led to telomere and subtelomere instability in yeast 158, presumably due to its propensity to form telomere R-loops 159, 160, which is known to induce DSBs and cause genome instability 4,161.
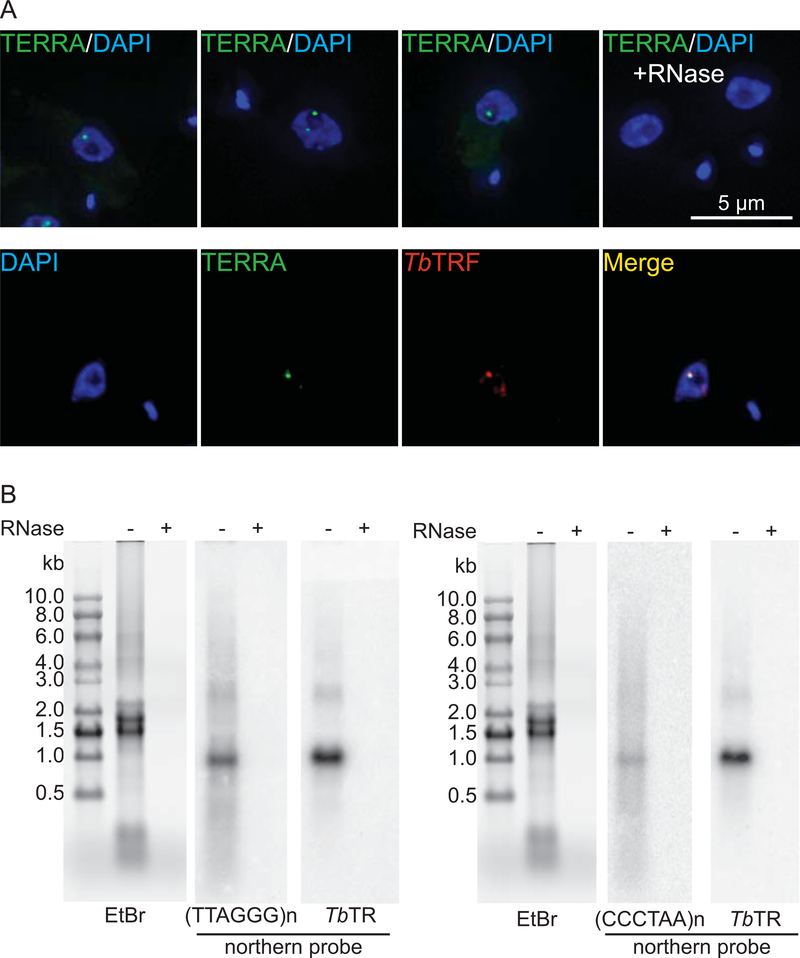
The telomere transcript was detected in both BF and PF T. brucei cells and only a fraction of this RNA is polyadenylated 137. Most strikingly, the TERRA level is resistant to 1 mg/ml alpha-amanitin, suggesting that TERRA is transcribed by RNA polymerase I 137. Knowing that the active VSG is transcribed at a very high level from a subtelomeric ES by RNA Pol I that is resistant to alpha-amanitin, it was hypothesized that TERRA is transcribed by RNA Pol I as a product of read-through into the telomere repeats downstream of the active VSG gene 137. This hypothesis was confirmed recently by Nanavaty et al. 6. All B-ESs are PTUs 52, 59, 162, and nascent polycistronic transcripts are processed through trans-splicing, where a common spliced leader (SL) sequence is added to the 5’ end of individual mRNAs 163, 164. After reverse transcription of total RNA using a CCCUAA primer, Nanavaty et al. were able to detect a PCR product using primers specific to the active VSG gene, indicating that the un-processed nascent RNA containing both the telomeric UUAGGG and the active VSG sequences exists, which confirms that TERRA indeed is transcribed from the telomere downstream of the active ES 6. No TERRA product was detected from silent ES-adjacent telomeres 6. However, T. brucei has more than 200 telomeres that do not host any ESs 52. Whether TERRA can be transcribed from ES-free telomeres is still unknown. Our lab has now performed a TERRA FISH analysis in BF T. brucei cells using the TELC-PNA probe (PNA Bio). We frequently observe only one or two TERRA foci in each T. brucei nucleus (Fig. 3A), suggesting that TERRA is transcribed from few telomeres if not only from the active ES-adjacent telomere. Additionally, we performed TERRA FISH and TbTRF IF analysis simultaneously and found that TERRA is colocalized with TbTRF at the telomere (Fig. 3A).
Figure 3.
(A) Subnuclear localization of TERRA. Top, TERRA FISH was performed in BF WT T. brucei cells with an Alexa488-conjugated PNA probe containing (CCCTAA)3 (TELC probe, PNA Bio). Hybridization was done at 37°C without denaturation. Treating cells with RNase A eliminated all nuclear punctate signals (right image in the top row), confirming that the observed signals represent RNA molecules. Bottom, TbTRF IF was performed using a rabbit antibody recognizing TbTRF 135 and an Alexa594-conjugated donkey anti-rabbit 2nd antibody. TERRA FISH was performed the same way as in (A). DNA was stained by DAPI. The large DAPI-positive circle is the T. brucei nucleus, while the smaller DAPI-positive dot is the kinetoplast. All images are of the same scale, with the scale bar shown in the top right panel. (B) Northern blot of PF T. brucei total RNA. In both left and right panels, the EtBr-stained agarose gel images are shown on the left. Hybridization using the TbTR probe was done as a loading control (shown on the right). An 800 bp (TTAGGG)n-containing DNA fragment was labeled with radioactive dGTP in the absence of dCTP to generate a probe that was used to detect the (CCCUAA)n-containing TERRA (left). The same probe was labeled with radioactive dCTP in the absence of dGTP to generate a probe that was used to detect (UUAGGG)n-containing TERRA (right). Hybridizations were done at 50°C. 10 units of RNase A and 20 units of RNase One were added in the RNase treatment control samples.
Interestingly, TERRA is detected in PF T. brucei cells where VSG is not transcribed 137, 165, suggesting that not all TERRA is transcribed as a read-through product. Our lab has also detected both UUAGGG and CCCUAA repeat-containing TERRA species in PF T. brucei cells (Fig. 3B). In addition to the TERRA species with various sizes (from 0.5 kb to 10 kb, shown in northern blotting as a smear), PF T. brucei also transcribes both G-rich and C-rich TERRAs with a more discrete size (~ 1 kb) (Fig. 3B). However, the origins of both UUAGGG and CCCUAA repeat containing TERRA species in PF T. brucei cells are currently unclear.
Although the function of T. brucei TERRA is not clear, TERRA has been shown to form telomere R-loops 6, and higher than WT levels of telomere R-loops result in an increased amount of subtelomeric and telomeric DSBs and an elevated VSG switching rate 6–8.
Telomere and subtelomere R-loops and antigenic variation
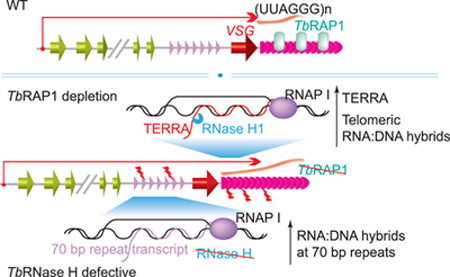
Telomere R-loops are detected in WT T. brucei cells 6 by the monoclonal antibody S9.6 that specifically recognizes the RNA:DNA hybrid 166. Depletion of TbRAP1 leads to not only a higher level of TERRA but also more telomere R-loops and an increased amount of telomeric and subtelomeric DSBs, which in turn cause more frequent VSG switching 6. The increased amount of DSBs at telomeres and subtelomeres is mainly mediated by the increased amount of telomere R-loops, as expression of an ectopic allele of TbRNase H1 in TbRAP1-depleted cells brings telomere R-loops and telomeric/subtelomeric DSBs back to WT levels, which further reduces the VSG switching rate back to its WT level 6.
Although a higher level of TERRA is frequently associated with an elevated amount of telomere R-loops and vice versa 158, 160, 167, 168, this may not always be the case in T. brucei. It has been shown that the R-loop level is influenced by RNA processing 169, 170. High transcription level and poor RNA processing can both lead to an increased level of R-loops. However, it is unknown whether TbRAP1 plays any direct roles in dissolution of the telomere R-loop or whether TbRAP1 affects premature RNA (containing both UUAGGG repeats and the upstream VSG sequences) processing. When an ectopic allele of TbRNase H1 is expressed in the TbRAP1-depleted cells, the TERRA level is still much higher than that in WT cells, even though the amount of telomere R-loop is reduced to the WT level 6, suggesting that TERRA and the telomere R-loop may be regulated independently. It would be interesting to further examine the relationship between telomere R-loop and TERRA in T. brucei.
Two recent studies on T. brucei ribonuclease H enzymes also showed that R-loops at the telomere and the subtelomere influence VSG switching frequencies 7, 8. T. brucei has two RNase H enzymes, a non-essential TbRNase H1 171 and an essential TbRNase H2 8. DRIP-seq experiments (R-loop immunoprecipitation followed by high-throughput sequencing analysis) were done to map which genomic loci have R-loops in WT and RNase H defective cells 7, 8, 172. In WT cells, R-loops are detected at the region immediately downstream of the active VSG gene, and a much lower level of R-loops is observed immediately downstream of a silent VSG gene 7, 8. More R-loops are clearly detected at both of these regions in TbRNase H1 null cells 7. Depletion of TbRNase H2A (the catalytic subunit of TbRNase H2) has a similar phenotype 8, indicating that both RNase H enzymes influence R-loop levels at the telomere and subtelomere junction.
Interestingly, in both TbRNase H1 null and TbRNase H2A-depleted cells, increased amounts of R-loops are detected across the active and silent ESs, with the most prominent increase at the 70 bp repeats 7, 8. The heterogeneous ~ 70 bp repeats are found upstream of most VSG genes (Fig. 1 & 2) 51. The repeats often contain varying numbers of tandem TAA triplets and their sizes range from 66 to 81 bp 173, 174. Two highly conserved motifs (AGTGTTGTGAGTGTG and TATAATAAGAGCAGTAAT) have been identified and 83% of the 70 bp repeats being studied contain one or both of these motifs 175. In VSG gene arrays, usually few copies of 70 bp repeats are upstream of a VSG gene 52, 162. While in B-ESs, 3 – 20 kb of 70 bp repeats are upstream of the VSG gene 59. In WT BF cells, no stable transcripts from the 70 bp repeats have been detected, even though the active ES is highly transcribed by RNA Pol I, suggesting that RNA processing is very efficient. However, in TbRNase H1 null and TbRNase H2A-depleted cells, a significant amount of R-loops are detected at the 70 bp repeat region in both active and silent ESs 7, 8. Consistent with the notion that R-loops often induce DNA damages 3, more γH2A (DNA damage associated histone H2A with phosphorylated T130 176) associates with the active ES chromatin, particularly at the telomere-proximal region, in TbRNase H1 null cells 7, suggesting telomere-proximal R-loops are more stable. When TbRNase H2A is depleted, much more γH2A associates with both active and silent ES chromatin throughout the whole ES 8. Furthermore, more switchers are observed to have shed the originally active VSG on the cell surface in the RNase H defective cells, suggesting that these cells have an increased VSG switching rate than WT cells 7, 8. Therefore, increased amounts of telomere/subtelomere R-loops in RNase H defective cells are linked with elevated DNA damage levels at ESs and more frequent VSG switching 7, 8
In TbRNase H defective cells, an increased amount of R-loops is detected at the telomere/subtelomere junction 7, 8. It is likely that telomere R-loops are also stabilized in these cells, although this has not been tested directly. It is also unknown whether TERRA levels are increased when the RNase H enzymes are deleted or depleted. In TbRNase H1 null and TbRNase H2A-depleted cells, the mRNA levels of a number of silent VSGs are increased (upto several ten-fold) 7, 8, suggesting that TbRNase H1 and H2 may be important for VSG silencing. R-loops have been shown to affect gene expression in other organisms 21–23. Therefore, it is possible that VSG expression is affected by nearby R-loops and that TERRA level is also increased in TbRNase H defective cells. On the other hand, frequent VSG switching in the RNase H defective cells 7, 8 could also lead to mildly increased VSG mRNA levels when a cell population is examined. Cells expressing both the originally active VSG and an originally silent VSG simultaneously are observed by IF in the TbRNase H defective cells 7, 8. However, these cells might be in the middle of a VSG switching process. Whether the TERRA level is affected by the RNase H enzymes needs further investigation.
It is important to note that functions of TbRNase H1 and H2 are not limited at ES regions 7, 8, 172. A recent DRIP-seq analysis detected R-loops in multiple T. brucei genome loci in WT cells 172. In TbRNase H1 null cells, increased levels of R-loops are also observed at transcription start sites of RNA Pol II transcribed PTUs, while no significant increase of DNA damage at these sites is seen 172. It is also unknown whether TbRNase H1 affects mRNA levels of genes located outside of ESs. On the other hand, depletion of TbRNase H2A leads to a dramatic increase in the amount of genomic DNA damage, particularly at the transcription initiation sites 8. Additionally, several tens of genes other than VSG, ESAG, or procyclin genes exhibit increased mRNA levels when TbRNase H2A is depleted 8.
Subtelomeric regions are often composed of various repeats and gene families 177–179. High polymorphism in the subtelomere is frequently observed among different chromosome ends and individuals in humans 180, 181, yeast 177, 182, fly 183, plant 184, and fungal pathogens 185, 186. T. brucei subtelomeres also exhibit dynamic variations: the T. brucei homologous megabase chromosome pairs often differ greatly in size (Fig. 1A) 187. Several factors contribute to this size polymorphism: subtelomeric ESs and VSG gene arrays have different sizes, telomere lengths vary at different chromosome ends, and repetitive chromosomal regions vary in size 188. Importantly, two-thirds of the size polymorphisms are due to variations in subtelomeric regions, while chromosomal core regions, containing all essential genes, are relatively stable 53.
Telomere dysfunctions are well-known to induce genome instabilities. At the telomere vicinity, unprotected telomeres lead to chromosome end-to-end fusions 189, 190, anaphase bridges 191, 192, and telomere recombination 193, 194. Telomere fusions in human cells can further induce a persistent mitotic arrest that leads to greatly increased cell lethality 195. At a global level, telomere crisis results in dicentric chromosome formation and subsequent chromothripsis and kataegis 191. Dysfunctional telomeres can also lead to subtelomere instability in yeast 196, 197. In telomerase null cells, Type I and Type II survivors use DNA recombination-dependent mechanisms to maintain their telomere length 196, where Type I survivors amplify their subtelomeric Y’ elements in a Rad51-dependent pathway 197, 198. Studies in T. brucei have shown that depletion of telomere proteins, TbTRF, TbRAP1, and TbTIF2, all result in unstable subtelomeres 6, 112, 120, 121.
Most telomere dysfunctions result in severe genome instability and cause cell growth defects 129. In T. brucei, VSG is essential 199, and damages to the active VSG gene are generally poorly tolerated: Introducing an artificial DSB (an I-SceI cut) within or near the active VSG gene leads to cell death in more than 80% of the cell population 200, which also leads to a 250-fold higher VSG switching rate 201. It is possible that the I-SceI cut is not repaired efficiently due to continued I-SceI expression. However, the location of the damage site appears to be a critical factor, as inducing the same I-SceI cut in a silent ES is much better tolerated 200. TbTIF2 and TbRAP1 are essential proteins that associate with the telomere chromatin 87, 121, and depletion of these proteins induces DNA damages mainly in the active and silent ESs 6, 121. These observations are consistent with the idea that DNA damages in the active ES are poorly tolerated. Depletion of TbRNase H2A results in global increased amounts of DNA damages at transcription initiation sites 8, suggesting that TbRNase H2A may be important for genome integrity at non-subtelomeric regions. Therefore, it is hard to interpret whether the increased amounts of DNA damages in ESs contribute significantly to the growth defect in TbRNase H2A-depleted cells. On the other hand, some telomeric dysfunction and subtelomeric damages are better tolerated, contributing to subtelomeric plasticity. Single damages within silent ESs do not severely affect cell growth 200. Telomeres downstream of the silent ESs can be as short as < 100 bp without inducing any cell growth defect 202. Additionally, cells do not experience cell cycle arrest when a single telomere (without any adjacent ES) is deleted 203. Therefore, damages that do not directly disrupt the active VSG gene seem less detrimental to cell growth than the ones that do. However, TbRNase H1 null cells represent an exception, where an increased amount of DNA damages is detected in the active ES, but the cells are viable 7. Although it is not strictly comparable among different studies, loss of TbRNase H1 does not seem to cause an as high level of DNA damage in ESs as depletion of TbTIF2 or TbRAPl 6, 7, 121. In the latter two cases, DNA damages are observed in both active and silent ESs, while loss of TbRNase H1 only causes a moderate increase in the amount of DNA damages in the active ES 6, 7, 121. Therefore, at T. brucei subtelomeres, the amount of damages probably also contributes to its effect on cell fitness. In consistence with this, some DNA breaks are detected at 70 bp repeat region in the active ES in WT cells 201, suggesting that cells can tolerate a low level of subtelomeric damages, although the exact amount of telomere and subtelomere damages in WT cells is unknown.
These observations suggest that the balance between plasticity and integrity at the T. brucei subtelomere is a key factor of keeping a balance between antigenic variation and cell fitness, both important for parasite survival. A similar balancing act is necessary for proper telomere maintenance in human ALT cells 168, where a telomerase-independent and DNA recombination-dependent telomere maintenance mechanism is critical for cell survival 204. In ALT cells, overexpression of TERRA leads to an increased level of telomere R-loops and many more telomere recombination events 168. Importantly, depletion of RNase H1 in these cells leads to an accumulation of telomere R-loops and C-circle excision-mediated rapid telomere shortening 168.
Overexpression of RNase H1 in ALT cells reduces the amount of telomere R-loops, which hinders telomere recombination potential and results in gradual telomere shortening 168. Therefore, perturbing telomere R-loop levels in ALT cells disrupts the delicate balance between recombination-mediated telomere attrition and maintenance.
Achieving a good balance between telomere/subtelomere stability and subtelomere variation may be feasible through an introduction of the right amount of telomere/subtelomere damages 205. In Kluyveromyces lactis, variation in a subtelomeric gene family encoding β-galactosidase allows yeast to better cope with different nutrition 206, a scenario not too different from VSG switching in T. brucei. Mild telomere dysfunction that does not induce global genome instability leads to increased variation of the subtelomere β-galactosidase-coding genes, while severe telomere dysfunction causes complete deletion of these genes 206. Therefore, mild telomere dysfunction can serve as a beneficial drive for subtelomere variations that allow cells to better adapt to environmental stresses 205. The TbRNase H1 null cells 7 may be a good example of having a mild telomere/subtelomere dysfunction in T. brucei, as the amount of R-loop-induced subtelomeric DNA damages appears not severe enough to trigger cell growth defect but sufficient to stimulate VSG switching, which is presumably beneficial for a long-term parasite survival inside the mammalian host. Since TbRNase H1 null cells are viable, it would be interesting to examine whether these cells have increased virulence when infecting an animal host.
Telomere/subtelomere R-loops and initiation of VSG switching
Homologous recombination (HR) is a major pathway for VSG switching 100. For ES-linked and minichromosome VSGs, the telomere sequence is found downstream of VSG51, 52, 59. Additionally, most VSG genes have a common 14 nt sequence in their 3’UTR region 51. It has been proposed that the common 14 nt VSG 3’UTR sequence and sometimes the telomere sequence can serve as the downstream homologous arm, while the 70 bp repeats can serve as the upstream homologous arm for efficient homologous recombination that mediates a VSG switch 207. However, an earlier study showed that the 70 bp repeats upstream of the VSG gene in an ES is not required for VSG switching 103. In a recent study that introduces an I-SceI cut in the active ES to initiate VSG switching, it is found that 70 bp repeats in the active ES promote selection of VSG donors from the genomic archive rather than only from silent ESs 207.
Although a DSB is not absolutely required for HR, it is a good inducer for HR and frequently repaired by HR 208. Therefore, it has been hypothesized that DSBs are an important trigger of VSG switching 123, 201. In support of this, introducing an artificial DSB immediately upstream of the active VSG leads to a more than 250-fold higher VSG switching frequency 201, and DSBs introduced near the active VSG (both upstream and downstream) result in efficient VSG switching 200. DSBs can also be detected in 70 bp repeats by Ligation mediated PCR analysis in WT T. brucei cells 201, although the amount of telomeric and subtelomeric DNA damage in WT cells has not been carefully quantified. Interestingly, the active ES-adjacent telomere experiences frequent large fragment deletions 209, which may be a consequence of TERRA transcription by RNA Pol I 6, 137 that depletes most nucleosomes in the active ES 67, 68 and may also interfere with the binding of TbTRF at the telomere. Therefore, DNA breaks in both the telomere and 70 bp repeats are possible to serve as an inducer for VSG switching. Several mechanisms are possible for DSB formation in these repetitive sequences.
First, as repetitive sequences, both telomeres and 70 bp repeats can be difficult to be replicated due to strand slippage during DNA replication 210 and secondary structure formation 211 (such as the G-quadruplex structure formed by the telomere G-rich strand DNA 212 and the non-H bonded structure formed by TTA/TAA in the 70 bp repeats 213). In mammalian and yeast cells, several telomere proteins have been shown to play important roles in ensuring proper telomere replication 214–216. Whether T. brucei telomere proteins have similar functions still requires further investigation. Nevertheless, obstacles in DNA replication often lead to stalled replication fork and eventual DSBs 217, 218. Second, it is shown that the active B-ES is replicated in early S phase 119. Hence, the transcription machinery may collide with the DNA replication machinery in the active B-ES and its adjacent telomere, which often causes fork stalling and collapsing followed by DSB formation 219. Third, the active ES and its adjacent telomere are transcribed at high levels by RNA Pol I 6, 220, which can induce more DSB formation with or without the R-loop formation. RNA Pol I transcription depletes nucleosomes from the active ES 67, 68 and can, presumably, also remove some TbTRF from the telomere. The exposed DNA in the active ES and the adjacent telomere is likely more vulnerable to nuclease attack, which can result in more DSBs. Additionally, the traverse of RNA polymerase along repetitive DNA allows formation of DNA secondary structure, which helps DSB formation 221. Furthermore, the high levels of transcription can induce transcription-associated recombination (TAR) 222, and RNA Pol I transcription in BF T. brucei cells stimulates homologous recombination > 3 fold 223. Finally, the telomere and subtelomere R-loops may further increase the chance of DSB formation in this area. We now know that TERRA forms R-loops with the telomeric DNA 6, and an increased level of telomere R-loops leads to an increased amount of DSBs in ESs and more frequent VSG switching 6. Additionally, R-loops are stabilized at the 70 bp repeats in TbRNase H1 null and TbRNase H2A-depleted cells, which is also linked with increased amounts of DNA damages in ESs and more frequent VSG switching 7, 8. Transcribing the active VSG with the flanking repetitive sequences provides a couple of intrinsic mechanisms to induce DSBs in the local region, either with or without the formation of local R-loops, which ensures a good chance of VSG switching through homologous recombination.
R-loops at the telomere and 70 bp repeats clearly contribute to more efficient antigenic variation 6–8. However, the underlying mechanisms of the formation and dissolution of these R-loops are not well-understood. Functions of T. brucei TERRA are not clear, and whether high levels of TERRA always lead to high levels of telomere R-loops is not known. Similarly, it is unclear whether the RNA transcribed from the 70 bp repeats can only exist as part of the R-loop in TbRNase H defective cells. Finally, TbRNase H1 null cells appear to have achieved a good balance between improved subtelomeric plasticity and sufficient genome stability 7. A better understanding about the detailed functions of TbRNase H1, particularly at the subtelomere, would be revealing for the positive and negative effects of telomere/subtelomere R-loops on antigenic variation and genome stability.
Telomere biology in other microbial pathogens and the potential link with pathogen virulence
Two kinetoplastid parasites that are closely related to T. brucei also cause debilitating human diseases: Trypanosoma cruzi causes Chagas disease, which can lead to serious heart and digestive problems 224. Leishmaniasis is caused by over 20 species of Leishmania. The disease severely decreases the life quality and causes heavy economic burdens 225. Telomeres in all three kinetoplastid parasites have the TTAGGG repetitive sequence 162, 226, 227. Homologues of all known T. brucei telomere proteins can be easily identified in T. cruzi and Leishmania genomes 228, 229. Additionally, TERRA has been detected in Leishmania 165. However, it is unknown whether T. cruzi transcribes its telomeric sequence. It is also unclear whether R-loops are formed in T. cruzi and Leishmania. Still, the essential telomere function in maintaining genome stability is expected to be conserved in both T. cruzi and Leishmania. In addition, subtelomere stability may be an important factor for T. cruzi virulence. T. cruzi is able to infect any host cell but mainly macrophages, fibroblasts and epithelial cells 230. Infective forms of T. cruzi express surface transsialidase proteins 231, 232, and their conserved peptide motifs are important for interacting with cytokeratin and mediate host-parasite interaction 233. Decreasing trans-sialidase expression also contributes to the loss of T. cruzi virulence 234. Interestingly, the gp85 gene family that encodes trans-sialidase is located at subtelomeric regions 235, and maintaining telomere and subtelomere stability is expected to help maintain stable gp85 gene copy numbers. Therefore, it will be interesting to investigate whether gp85 expression is affected by telomeric silencing, whether any R-loop is formed at telomere/subtelomere, and whether these R-loops affect gp85 gene family stability or their expression.
Several other microbial pathogens that undergo antigenic variation also host their variable surface antigens at subtelomeres 179, including Pneumocystis jirovecii that causes pneumonia in immunodeficient patients, in which DNA recombination appears to be the major pathway of antigenic variation 185. A better understanding of telomere functions and how telomere R-loops influence telomere and subtelomere stability will help us better understand the mechanisms of antigenic variations in these human pathogens.
Research Highlights.
In T. brucei, a protozoan parasite that causes African trypanosomiasis and undergoes antigenic variation, the major surface antigen-coding VSG gene is transcribed from telomere-adjacent expression sites, where it is flanked by the telomere and 70 bp repeats
In TbRAP1-depleted cells, an increased amount of telomere R-loops leads to an elevated amount of telomere/subtelomere DSBs and more frequent VSG switching
Removal of TbRNase H enzymes results in accumulation of R-loops at the 70 bp repeats and at the telomere-subtelomere junctions, more DSBs in VSG expression sites, and more frequent VSG switching
R-loop levels at the telomere and 70 bp repeats influence the balance between antigenic variation and cell fitness
The high level of transcription of the active ES by RNA Pol I promotes accumulation of R-loops at the telomere and 70 bp repeats. This provides an intrinsic mechanism for DSB formation in the active ES, which is a strong inducer of VSG switching.
Acknowledgements
This work is partly supported by an NIH R01 grant AI066095 to B. Li, an NSF grant MCB 1615896 to B. Li & K. Chakrabarti, and an NIH R01 grant AI127562 to H. Kim. The publication cost is partly supported by GRHD at CSU. We thank Dr. Amit Gaurav, Dr. Maiko Tonini, and Brittny Schnur for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costantino L, Koshland D. The Yin and Yang of R-loop biology. Curr Opin Cell Biol 2015;34:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diman A, Decottignies A. Genomic origin and nuclear localization of TERRA telomeric repeat-containing RNA: from Darkness to Dawn. FEBS J 2018;285:1389–1398. [DOI] [PubMed] [Google Scholar]
- 3.Rondón AG, Aguilera A. What causes an RNA-DNA hybrid to compromise genome integrity. DNA Repair (Amst) 2019102660. [DOI] [PubMed] [Google Scholar]
- 4.Toubiana S, Selig S. DNA:RNA hybrids at telomeres - when it is better to be out of the (R) loop. FEBS J 2018;285:2552–2566. [DOI] [PubMed] [Google Scholar]
- 5.Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol 2001;49:1–70. [DOI] [PubMed] [Google Scholar]
- 6.Nanavaty V, Sandhu R, Jehi SE, Pandya UM, Li B. Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids. Nucleic Acids Res 2017;45:5785–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs E, Crouch K, Lemgruber L, Lapsley C, McCulloch R. Ribonuclease H1-_targeted R-loops in surface antigen gene expression sites can direct trypanosome immune evasion. PLoS Genet 2018;14:e1007729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs E, Crouch K, Lemgruber L, Hamilton G, Lapsley C, McCulloch R. Trypanosoma brucei ribonuclease H2A is an essential R-loop processing enzyme whose loss causes DNA damage during transcription initiation and antigenic variation. Nucleic Acids Res 2019;47:9180–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crossley MP, Bocek M, Cimprich KA. R-Loops as Cellular Regulators and Genomic Threats. Mol Cell 2019;73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet 2015;16:583–597. [DOI] [PubMed] [Google Scholar]
- 11.Thomas M, White RL, Davis RW. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A 1976;73:2294–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell 2004;119:481–489. [DOI] [PubMed] [Google Scholar]
- 13.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 2003;4:442–451. [DOI] [PubMed] [Google Scholar]
- 14.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A 1987;84:7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huertas P, Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 2003;12:711–721. [DOI] [PubMed] [Google Scholar]
- 16.Gómez-González B, García-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marín A, Foiani M, Aguilera A. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J 2011;30:3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domínguez-Sánchez MS, Barroso S, Gómez-González B, Luna R, Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLoS Genet 2011;7:e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 2011;44:978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell 2012;46:115–124. [DOI] [PubMed] [Google Scholar]
- 20.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 2008;28:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 2013;340:619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 2012;45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castellano-Pozo M, Santos-Pereira JM, Rondon AG, Barroso S, Andujar E, Perez-Alegre M, Garcia-Muse T, Aguilera A. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell 2013;52:583–590. [DOI] [PubMed] [Google Scholar]
- 25.Posse V, Al-Behadili A, Uhler JP, Clausen AR, Reyes A, Zeviani M, Falkenberg M, Gustafsson CM. RNase H1 directs origin-specific initiation of DNA replication in human mitochondria. PLoS Genet 2019;15:e1007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005;122:365–378. [DOI] [PubMed] [Google Scholar]
- 27.Muers M. Mutation: the perils of transcription. Nat Rev Genet 2011;12:156. [DOI] [PubMed] [Google Scholar]
- 28.Wimberly H, Shee C, Thornton PC, Sivaramakrishnan P, Rosenberg SM, Hastings PJ. R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat Commun 2013;4:2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 2014;28:1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 2011;25:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castellano-Pozo M, Garcia-Muse T, Aguilera A. R-loops cause replication impairment and genome instability during meiosis. EMBO Rep 2012;13:923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groh M, Lufino MM, Wade-Martins R, Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet 2014;10:e1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 2014;516:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell 2014;56:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 1992;258:1463–1466. [DOI] [PubMed] [Google Scholar]
- 36.Richardson JP. Loading Rho to terminate transcription. Cell 2003;114:157–159. [DOI] [PubMed] [Google Scholar]
- 37.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 2011;10:654–665. [DOI] [PubMed] [Google Scholar]
- 38.Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, Liberi G. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 2012;151:835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J 2009;276:1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen TA, Tak YS, Lee CH, Kang YH, Cho IT, Seo YS. Analysis of subunit assembly and function of the Saccharomyces cerevisiae RNase H2 complex. FEBS J 2011;278:4927–4942. [DOI] [PubMed] [Google Scholar]
- 41.Reijns MA, Bubeck D, Gibson LC, Graham SC, Baillie GS, Jones EY, Jackson AP. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J Biol Chem 2011;286:10530–10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 2010;24:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc Natl Acad Sci U S A 2010;107:692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie 1993;75:123–126. [DOI] [PubMed] [Google Scholar]
- 45.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol 1999;19:8361–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci U S A 2002;99:16654–16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Groh M, Gromak N. Out of balance: R-loops in human disease. PLoS Genet 2014;10:e1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutherland CS, Yukich J, Goeree R, Tediosi F. A literature review of economic evaluations for a neglected tropical disease: human African trypanosomiasis (“sleeping sickness”). PLoS Negl Trop Dis 2015;9:e0003397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cross GAM. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 1975;71:393–417. [DOI] [PubMed] [Google Scholar]
- 50.Diffley P. Trypanosoma brucei: immunogenicity of the variant surface coat glycoprotein of virulent and avirulent subspecies. Exp Parasitol 1985;59:98–107. [DOI] [PubMed] [Google Scholar]
- 51.Cross GAM, Kim HS, Wickstead B. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol Biochem Parasitol 2014;195:59–73. [DOI] [PubMed] [Google Scholar]
- 52.Müller LSM, Cosentino RO, Förstner KU, Guizetti J, Wedel C, Kaplan N, Janzen CJ, Arampatzi P, Vogel J, Steinbiss S, Otto TD, Saliba AE, Sebra RP, Siegel TN. Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature 2018;563:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callejas S, Leech V, Reitter C, Melville S. Hemizygous subtelomeres of an African trypanosome chromosome may account for over 75% of chromosome length. Genome Res 2006;16:1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Sayed NM, Ghedin E, Song J, MacLeod A, Bringaud F, Larkin C, Wanless D, Peterson J, Hou L, Taylor S, Tweedie A, Biteau N, Khalak HG, Lin X, Mason T, Hannick L, Caler E, Blandin G, Bartholomeu D, Simpson AJ, Kaul S, Zhao H, Pai G, Van Aken S, Utterback T, Haas B, Koo HL, Umayam L, Suh B, Gerrard C, Leech V, Qi R, Zhou S, Schwartz D, Feldblyum T, Salzberg S, Tait A, Turner CM, Ullu E, White O, Melville S, Adams MD, Fraser CM, Donelson JE. The sequence and analysis of Trypanosoma brucei chromosome II. Nucleic Acids Res 2003;31:4856–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alsford S, Wickstead B, Ersfeld K, Gull K. Diversity and dynamics of the minichromosomal karyotype in Trypanosoma brucei. Mol Biochem Parasitol 2001;113:79–88. [DOI] [PubMed] [Google Scholar]
- 56.Williams RO, Young JR, Majiwa PAO. Genomic environment of T. brucei VSG genes: presence of a minichromosome. Nature 1982299 417–421. [DOI] [PubMed] [Google Scholar]
- 57.Crozatier M, van der Ploeg LH, Johnson PJ, Gommers-Ampt J, Borst P. Structure of a telomeric expression site for variant specific surface antigens in Trypanosoma brucei. Mol Biochem Parasitol 1990;42:1–12. [DOI] [PubMed] [Google Scholar]
- 58.Barnes DA, Mottram JC, Agabian N. Bloodstream and metacyclic variant surface glycoprotein gene expression sites of Trypanosoma brucei gambiense. MBP 1990;41:101–114. [DOI] [PubMed] [Google Scholar]
- 59.Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, Brooks K, Churcher C, Fahkro S, Goodhead I, Heath P, Kartvelishvili M, Mungall K, Harris D, Hauser H, Sanders M, Saunders D, Seeger K, Sharp S, Taylor JE, Walker D, White B, Young R, Cross GAM, Rudenko G, Barry JD, Louis EJ, Berriman M. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE 2008;3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Lange T, Borst P. Genomic environment of the expression-linked extra copies of genes for surface antigens of Trypanosoma brucei resembles the end of a chromosome. Nature 1982;299:451–453. [DOI] [PubMed] [Google Scholar]
- 61.Cestari I, Stuart K. Transcriptional Regulation of Telomeric Expression Sites and Antigenic Variation in Trypanosomes. Curr Genomics 2018;19:119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gunzl A, Kirkham JK, Nguyen TN, Badjatia N, Park SH. Mono-allelic VSG expression by RNA polymerase I in Trypanosoma brucei: Expression site control from both ends? Gene 2015;556:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Epigenetics Rudenko G. and transcriptional control in African trypanosomes. Essays Biochem 2010;48:201–219. [DOI] [PubMed] [Google Scholar]
- 64.Pandya UM, Sandhu R, Li B. Silencing subtelomeric VSGs by Trypanosoma brucei RAP1 at the insect stage involves chromatin structure changes. Nucleic Acids Res 2013;41:7673–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narayanan MS, Rudenko G. TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res 2013;41:2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Povelones ML, Gluenz E, Dembek M, Gull K, Rudenko G. Histone H1 Plays a Role in Heterochromatin Formation and VSG Expression Site Silencing in Trypanosoma brucei. PLoS Pathog 2012;8:e1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanne TM, Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot Cell 2010;9:136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Figueiredo LM, Cross GAM. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell 2010;9:148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hughes K, Wand M, Foulston L, Young R, Harley K, Terry S, Ersfeld K, Rudenko G. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J 2007;26:2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Denninger V, Rudenko G. FACT plays a major role in histone dynamics affecting VSG expression site control in Trypanosoma brucei. Mol Microbiol 2014;94:945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Denninger V, Fullbrook A, Bessat M, Ersfeld K, Rudenko G. The FACT subunit TbSpt16 is involved in cell cycle specific control of VSG expression sites in Trypanosoma brucei. Mol Microbiol 2010;78:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Navarro M, Cross GAM, Wirtz E. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J 1999;18:2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schulz D, Zaringhalam M, Papavasiliou FN, Kim HS. Base J and H3.V Regulate Transcriptional Termination in Trypanosoma brucei. PLoS Genet 2016;12:e1005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kassem A, Pays E, Vanhamme L. Transcription is initiated on silent variant surface glycoprotein expression sites despite monoallelic expression in Trypanosoma brucei. Proc Natl Acad Sci U S A 2014;111:8943–8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vanhamme L, Poelvoorde P, Pays A, Tebabi P, Van Xong H, Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol 2000;36:328–340. [DOI] [PubMed] [Google Scholar]
- 76.Cestari I, McLeland-Wieser H, Stuart K. Nuclear Phosphatidylinositol 5-Phosphatase Is Essential for Allelic Exclusion of Variant Surface Glycoprotein Genes in Trypanosomes. Mol Cell Biol 2019;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cestari I, Stuart K. Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc Natl Acad Sci U S A 2015;112:E2803–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DuBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, Bart JM, Ratushny AV, Wan Y, Bastin P, Barry JD, Navarro M, Horn D, Aitchison JD, Rout MP, Field MC. NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol 2012;10:e1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maishman L, Obado SO, Alsford S, Bart JM, Chen WM, Ratushny AV, Navarro M, Horn D, Aitchison JD, Chait BT, Rout MP, Field MC. Co-dependence between trypanosome nuclear lamina components in nuclear stability and control of gene expression. Nucleic Acids Res 2016;44:10554–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopez-Farfan D, Bart JM, Rojas-Barros DI, Navarro M. SUMOylation by the E3 Ligase TbSIZ1/PIAS1 Positively Regulates VSG Expression in Trypanosoma brucei. PLoS Pathog 2014;10:e1004545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benmerzouga I, Concepcion-Acevedo J, Kim HS, Vandoros AV, Cross GAM, Klingbeil MM, Li B. Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol Microbiol 2013;87:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tiengwe C, Marcello L, Farr H, Dickens N, Kelly S, Swiderski M, Vaughan D, Gull K, Barry JD, Bell SD, McCulloch R. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep 2012;2:185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HS, Park SH, Gunzl A, Cross GA. MCM-BP is required for repression of life-cycle specific genes transcribed by RNA polymerase I in the mammalian infectious form of Trypanosoma brucei. PLoS One 2013;8:e57001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim HS. Genome-wide function of MCM-BP in Trypanosoma brucei DNA replication and transcription. Nucleic Acids Res 2019;47:634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Faria J, Glover L, Hutchinson S, Boehm C, Field MC, Horn D. Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex. Nat Commun 2019;10:3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Glover L, Hutchinson S, Alsford S, Horn D. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc Natl Acad Sci U S A 2016;113:7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell 2009;137:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Turner CMR, Barry JD, Vickerman K. Loss of variable antigen during transformation of trypanosoma brucei rhodesiense from bloodstream to procyclic forms in the tsetse fly. Parasitol Res 1988;74:507–511. [DOI] [PubMed] [Google Scholar]
- 89.Tetley L, Turner CMR, Barry JD, Crowe JS, Vickerman,K. Onset of expression of the variant surface glycoproteins of Trypanosoma brucei in the tsetse fly studied using immunoelectron microscopy. J Cell Sci 1987;87:363–372. [DOI] [PubMed] [Google Scholar]
- 90.Kolev NG, Günzl A, Tschudi C. Metacyclic VSG expression site promoters are recognized by the same general transcription factor that is required for RNA polymerase I transcription of bloodstream expression sites. Mol Biochem Parasitol 2017;216:52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramey-Butler K, Ullu E, Kolev NG, Tschudi C. Synchronous expression of individual metacyclic variant surface glycoprotein genes in Trypanosoma brucei. Mol Biochem Parasitol 2015;200:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Graham SV, Terry S, Barry JD. A structural and transcription pattern for variant surface glycoprotein gene expression sites used in metacyclic stage Trypanosoma brucei. Molecular & Biochemical Parasitology 1999;103:141–154. [DOI] [PubMed] [Google Scholar]
- 93.Pedram M, Donelson JE. The anatomy and transcription of a monocistronic expression site for a metacyclic variant surface glycoprotein gene in Trypanosoma brucei. J Biol Chem 1999;274:16876–16883. [DOI] [PubMed] [Google Scholar]
- 94.Graham SV, Wymer B, Barry JD. Activity of a trypanosome metacyclic variant surface glycoprotein gene promoter is dependent upon life cycle stage and chromosomal context. Molecular & Cellular Biology 1998;18:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barry JD, Graham SV, Fotheringham M, Graham VS, Kobryn K, Wymer B. VSG gene control and infectivity strategy of metacyclic stage Trypanosoma brucei. Mol Biochem Parasitol 1998;91:93–105. [DOI] [PubMed] [Google Scholar]
- 96.Graham SV, Wymer B, Barry JD. A trypanosome metacyclic VSG gene promoter with two functionally distinct, life cycle stage-specific activities. Nucleic Acids Res 1998;26:1985–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Batram C, Jones NG, Janzen CJ, Markert SM, Engstler M. Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife 2014;3:e02324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Myler PJ, Allison J, Agabian N, Stuart K. Antigenic variation in African trypanosomes by gene replacement or activation of alternative telomeres. Cell 1984;39:203–211. [DOI] [PubMed] [Google Scholar]
- 99.Myler P, Nelson RG, Agabian N, Stuart K. Two mechanisms of expression of a variant antigen gene of Trypanosoma brucei. Nature 1984;309:282–284. [DOI] [PubMed] [Google Scholar]
- 100.McCulloch R, Morrison LJ, Hall JPJ. DNA Recombination Strategies During Antigenic Variation in the African Trypanosome. Microbiol Spectr 2015;3:MDNA3–0016. [DOI] [PubMed] [Google Scholar]
- 101.Pays E, Guyaux M, Aerts D, vanMeirvenne N, Steinert M. Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature 1985;316:562–564. [DOI] [PubMed] [Google Scholar]
- 102.Rudenko G, McCulloch R, Dirksmulder A, Borst P. Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol 1996;80:65–75. [DOI] [PubMed] [Google Scholar]
- 103.McCulloch R, Rudenko G, Borst P. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in variant surface glycoprotein expression sites lacking 70-base-pair repeat sequences. Molecular & Cellular Biology 1997;17:833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pays E, van Assel S, Laurent M, Darville M, Vervoort T, van Meirvenne N, Steinert M. Gene conversion as a mechanism for antigenic variation in trypanosomes. Cell 1983;34:371–381. [DOI] [PubMed] [Google Scholar]
- 105.Pays E, van Assel S, Laurent M, Dero B, Michiels F, Kronenberger P, Matthyssens G, van Meirvenne N, LeRay D, Steinert M. At least two transposed sequences are associated in the expression site of a surface antigen gene in different trypanosome clones. Cell 1983;34:359–369. [DOI] [PubMed] [Google Scholar]
- 106.de Lange T, Kooter JM, Michels PA, Borst P. Telomere conversion in trypanosomes. Nucleic Acids Res 1983;11:8149–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kramara J, Osia B, Malkova A. Break-Induced Replication: The Where, The Why, and The How. Trends Genet 2018;34:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roth C, Bringaud F, Layden RE, Baltz T, Eisen H. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc Natl Acad Sci USA 1989;86:9375–9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res 2007;17:1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dubois ME, Demick KP, Mansfield JM. Trypanosomes expressing a mosaic variant surface glycoprotein coat escape early detection by the immune system. Infect Immun 2005;73:2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mugnier MR, Cross GA, Papavasiliou FN. The in vivo dynamics of antigenic variation in Trypanosoma brucei. Science 2015;347:1470–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jehi SE, Li X, Sandhu R, Ye F, Benmerzouga I, Zhang M, Zhao Y, Li B. Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity. Nucleic Acids Res 2014;42:12899–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Morrison LJ, Marcello L, McCulloch R. Antigenic variation in the African trypanosome: molecular mechanisms and phenotypic complexity. Cell Microbiol 2009;11:1724–1734. [DOI] [PubMed] [Google Scholar]
- 114.Kim HS, Cross GAM. TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog 2010;6:e1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim HS, Cross GAM. Identification of Trypanosoma brucei RMI1/BLAP75 homologue and its roles in antigenic variation. PLoS One 2011;6:e25313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McCulloch R, Barry JD. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev 1999;13:2875–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Proudfoot C, McCulloch R. Distinct roles for two RAD51-related genes in Trypanosoma brucei antigenic variation. Nucleic Acids Res 2005;33:6906–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hartley CL, McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol Microbiol 2008;68:1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Devlin R, Marques CA, Paape D, Prorocic M, Zurita-Leal AC, Campbell SJ, Lapsley C, Dickens N, McCulloch R. Mapping replication dynamics in Trypanosoma brucei reveals a link with telomere transcription and antigenic variation. Elife 2016;5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jehi SE, Nanavaty V, Li B. Trypanosoma brucei TIF2 and TRF suppress VSG switching using overlapping and independent mechanisms. PLoS One 2016;11:e0156746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jehi SE, Wu F, Li B. Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity. Cell Res 2014;24:870–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hovel-Miner GA, Boothroyd CE, Mugnier M, Dreesen O, Cross GAM, Papavasiliou FN. Telomere length affects the frequency and mechanism of antigenic variation in Trypanosoma brucei. PLoS Pathog 2012;8:e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.da Silva MS, Hovel-Miner GA, Briggs EM, Elias MC, McCulloch R. Evaluation of mechanisms that may generate DNA lesions triggering antigenic variation in African trypanosomes. PLoS Pathog 2018;14:e1007321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McCulloch R, Cobbold CA, Figueiredo L, Jackson A, Morrison LJ, Mugnier MR, Papavasiliou N, Schnaufer A, Matthews K. Emerging challenges in understanding trypanosome antigenic variation. Emerg Top Life Sci 2017;1:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reis H, Schwebs M, Dietz S, Janzen CJ, Butter F. TelAP1 links telomere complexes with developmental expression site silencing in African trypanosomes. Nucleic Acids Res 2018;46:2820–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005;19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 127.Maciejowski J, de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 2017;18:175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Martinez P, Blasco MA. Replicating through telomeres: a means to an end. Trends Biochem Sci 2015;40:504–515. [DOI] [PubMed] [Google Scholar]
- 129.de Lange T. Shelterin-Mediated Telomere Protection. Annu Rev Genet 2018;52:223–247. [DOI] [PubMed] [Google Scholar]
- 130.Ottaviani A, Gilson E, Magdinier F. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie 2008;90:93–107. [DOI] [PubMed] [Google Scholar]
- 131.Oliva-Rico D, Herrera LA. Regulated expression of the lncRNA TERRA and its impact on telomere biology. Mech Ageing Dev 2017;167:16–23. [DOI] [PubMed] [Google Scholar]
- 132.Dreesen O, Li B, Cross GAM. Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nuc Acids Res 2005;33:4536–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sandhu R, Sanford S, Basu S, Park M, Pandya UM, Li B, Chakrabarti K. A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res 2013;23:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gupta SK, Kolet L, Doniger T, Biswas VK, Unger R, Tzfati Y, Michaeli S. The Trypanosoma brucei Telomerase RNA (TER) homologue binds core proteins of the C/D snoRNA family. FEBS Lett 2013 [DOI] [PubMed] [Google Scholar]
- 135.Li B, Espinal A, Cross GAM. Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol Cell Biol 2005;25:5011–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Roditi I, Schwarz H, Pearson TW, Beecroft RP, Liu MK, Williams RO, Overath P. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. JCB 1989;108:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rudenko G, Van der Ploeg LH. Transcription of telomere repeats in protozoa. EMBO J 1989;8:2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007;318:798–801. [DOI] [PubMed] [Google Scholar]
- 139.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 2008;10:228–236. [DOI] [PubMed] [Google Scholar]
- 140.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5’ to 3’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 2008;32:465–477. [DOI] [PubMed] [Google Scholar]
- 141.Bah A, Wischnewski H, Shchepachev V, Azzalin CM. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res 2012;40:2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Greenwood J, Cooper JP. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res 2012;40:2956–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. CpG-island promoters drive transcription of human telomeres. RNA 2009;15:2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun 2014;5:5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Feretzaki M, Lingner J. A practical qPCR approach to detect TERRA, the elusive telomeric repeat-containing RNA. Methods 2017;114:39–45. [DOI] [PubMed] [Google Scholar]
- 146.Deng Z, Wang Z, Stong N, Plasschaert R, Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei MS, Riethman H, Lieberman PM. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J 2012;31:4165–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chu HP, Froberg JE, Kesner B, Oh HJ, Ji F, Sadreyev R, Pinter SF, Lee JT. PAR-TERRA directs homologous sex chromosome pairing. Nat Struct Mol Biol 2017;24:620–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pfeiffer V, Lingner J. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS Genet 2012;8:e1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet 2010;6:e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Montero JJ, López de Silanes I, Graña O, Blasco MA. Telomeric RNAs are essential to maintain telomeres. Nat Commun 2016;7:12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lopez de Silanes I, Grana O, De Bonis ML, Dominguez O, Pisano DG, Blasco MA. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat Commun 2014;5:4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chu HP, Cifuentes-Rojas C, Kesner B, Aeby E, Lee HG, Wei C, Oh HJ, Boukhali M, Haas W, Lee JT. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 2017;170:86–101.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011;471:532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang C, Zhao L, Lu S. Role of TERRA in the Regulation of Telomere Length. Int J Biol Sci 2015;11:316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arora R, Azzalin CM. Telomere elongation chooses TERRA ALTernatives. RNA Biol 2015;12:938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Moravec M, Wischnewski H, Bah A, Hu Y, Liu N, Lafranchi L, King MC, Azzalin CM. TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe. EMBO Rep 2016;17:999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res 2010;38:5797–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yu TY, Kao YW, Lin JJ. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc Natl Acad Sci U S A 2014;111:3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet 2015;6:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hu Y, Bennett HW, Liu N, Moravec M, Williams JF, Azzalin CM, King MC. RNA-DNA Hybrids Support Recombination-Based Telomere Maintenance in Fission Yeast. Genetics 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bettin N, Oss Pegorar C, Cusanelli E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science 2005;309:416–422. [DOI] [PubMed] [Google Scholar]
- 163.Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol 2011;6:459–474. [DOI] [PubMed] [Google Scholar]
- 164.Preußer C, Jaé N, Bindereif A. mRNA splicing in trypanosomes. Int J Med Microbiol 2012;302:221–224. [DOI] [PubMed] [Google Scholar]
- 165.Damasceno JD, Silva G, Tschudi C, Tosi LR. Evidence for regulated expression of Telomeric Repeat-containing RNAs (TERRA) in parasitic trypanosomatids. Mem Inst Oswaldo Cruz 2017;112:572–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods 1986;89:123–130. [DOI] [PubMed] [Google Scholar]
- 167.Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, Luke B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 2013;20:1199–1205. [DOI] [PubMed] [Google Scholar]
- 168.Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T, Azzalin CM. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 2014;5:5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Okamoto Y, Abe M, Itaya A, Tomida J, Ishiai M, Takaori-Kondo A, Taoka M, Isobe T, Takata M. FANCD2 protects genome stability by recruiting RNA processing enzymes to resolve R-loops during mild replication stress. FEBS J 2019;286:139–150. [DOI] [PubMed] [Google Scholar]
- 170.Chakraborty P, Huang JTJ, Hiom K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat Commun 2018;9:4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kobil JH, Campbell AG. Trypanosoma brucei RNase HI requires its divergent spacer subdomain for enzymatic function and its conserved RNA binding motif for nuclear localization. Mol Biochem Parasitol 2000;107:135–142. [DOI] [PubMed] [Google Scholar]
- 172.Briggs E, Hamilton G, Crouch K, Lapsley C, McCulloch R. Genome-wide mapping reveals conserved and diverged R-loop activities in the unusual genetic landscape of the African trypanosome genome. Nucleic Acids Res 2018;46:11789–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Campbell DA, van Bree MP, Boothroyd JC. The 5’-limit of transposition and upstream barren region of a trypanosome VSG gene: tandem 76 base-pair repeats flanking (TAA)90. NAR 1984;12:2759–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Aline RF Jr., MacDonald G, Brown E, Allison J, Myler P, Rothwell V, Stuart K. (TAA)n within sequences flanking several intrachromosomal variant surface glycoprotein genes in Trypanosoma brucei. Nucleic Acids Res 1985;13:3161–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Carrington M, Miller N, Blum M, Roditi I, Wiley D, Turner MJ. Variant specific glycoprotein of Trypanosoma brucei consists of two domains each having an independently conserved pattern of cysteine residues. J Mol Biol 1991;221:823–835. [DOI] [PubMed] [Google Scholar]
- 176.Glover L, Horn D. Trypanosomal histone gammaH2A and the DNA damage response. Mol Biochem Parasitol 2012;183:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Curr Opin Genet Dev 1997;7:822–828. [DOI] [PubMed] [Google Scholar]
- 178.Mefford HC, Trask BJ. The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 2002;3:91–102. [DOI] [PubMed] [Google Scholar]
- 179.Li B. Telomere components as potential therapeutic _targets for treating microbial pathogen infections. Front Oncol 2012;2:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ambrosini A, Paul S, Hu S, Riethman H. Human subtelomeric duplicon structure and organization. Genome Biol 2007;8:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Young E, Pastor S, Rajagopalan R, McCaffrey J, Sibert J, Mak ACY, Kwok PY, Riethman H, Xiao M. High-throughput single-molecule mapping links subtelomeric variants and long-range haplotypes with specific telomeres. Nucleic Acids Res 2017;45:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Quispe X, Tapia SM, Villarroel C, Oporto C, Abarca V, García V, Martínez C, Cubillos FA. Genetic basis of mycotoxin susceptibility differences between budding yeast isolates. Sci Rep 2017;7:9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Anderson JA, Song YS, Langley CH. Molecular population genetics of Drosophila subtelomeric DNA. Genetics 2008;178:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kuo HF, Olsen KM, Richards EJ. Natural variation in a subtelomeric region of Arabidopsis: implications for the genomic dynamics of a chromosome end. Genetics 2006;173:401–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schmid-Siegert E, Richard S, Luraschi A, Muhlethaler K, Pagni M, Hauser PM. Mechanisms of Surface Antigenic Variation in the Human Pathogenic Fungus Pneumocystis jirovecii. MBio 2017;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Farman ML. Telomeres in the rice blast fungus Magnaporthe oryzae: the world of the end as we know it. FEMS Microbiol Lett 2007;273:125–132. [DOI] [PubMed] [Google Scholar]
- 187.Melville SE, Leech V, Navarro M, Cross GAM. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol Biochem Parasitol 2000;111:261–273. [DOI] [PubMed] [Google Scholar]
- 188.Melville SE, Gerrard CS, Blackwell JM. Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chromosome Research 1999;7:191–203. [DOI] [PubMed] [Google Scholar]
- 189.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell 1998;92:401–413. [DOI] [PubMed] [Google Scholar]
- 190.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 2005;7:712–718. [DOI] [PubMed] [Google Scholar]
- 191.Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015;163:1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tusell L, Pampalona J, Soler D, Frias C, Genescá A. Different outcomes of telomere-dependent anaphase bridges. Biochem Soc Trans 2010;38:1698–1703. [DOI] [PubMed] [Google Scholar]
- 193.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 2004;119:355–368. [DOI] [PubMed] [Google Scholar]
- 194.Brault ME, Autexier C. Telomeric recombination induced by dysfunctional telomeres. Mol Biol Cell 2011;22:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hayashi MT, Cesare AJ, Rivera T, Karlseder J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 2015;522:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 1993;73:347–360. [DOI] [PubMed] [Google Scholar]
- 197.Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 1999;152:143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol 2001;21:1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sheader K, Vaughan S, Minchin J, Hughes K, Gull K, Rudenko G. Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc Natl Acad Sci USA 2005;102:8716–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Glover L, Alsford S, Horn D. DNA break site at fragile subtelomeres determines probability and mechanism of antigenic variation in African trypanosomes. PLoS Pathog 2013;9:e1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Boothroyd CE, Dreesen O, Leonova T, Ly KI, Figueiredo LM, Cross GAM, Papavasiliou FN. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature 2009;459:278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dreesen O, Cross GAM. Telomerase-independent stabilization of short telomeres in Trypanosoma brucei. Mol Cell Biol 2006;26:4911–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Glover L, Alsford S, Beattie C, Horn D. Deletion of a trypanosome telomere leads to loss of silencing and progressive loss of terminal DNA in the absence of cell cycle arrest. Nuc Acids Res 2007;35:872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sobinoff AP, Pickett HA. Alternative Lengthening of Telomeres: DNA Repair Pathways Converge. Trends Genet 2017;33:921–932. [DOI] [PubMed] [Google Scholar]
- 205.Mason JMO, McEachern MJ. Chromosome ends as adaptive beginnings: the potential role of dysfunctional telomeres in subtelomeric evolvability. Curr Genet 2018;64:997–1000. [DOI] [PubMed] [Google Scholar]
- 206.Mason JMO, McEachern MJ. Mild Telomere Dysfunction as a Force for Altering the Adaptive Potential of Subtelomeric Genes. Genetics 2018;208:537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hovel-Miner G, Mugnier MR, Goldwater B, Cross GA, Papavasiliou FN. A Conserved DNA Repeat Promotes Selection of a Diverse Repertoire of Trypanosoma brucei Surface Antigens from the Genomic Archive. PLoS Genet 2016;12:e1005994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Haber JE. DNA Repair: The Search for Homology. Bioessays 2018;40:e1700229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bernards A, Michels PAM, Lincke CR, Borst P. Growth of chromosome ends in multiplying trypanosomes. Nature 1983;303:592–597. [DOI] [PubMed] [Google Scholar]
- 210.Pearson CE, Tam M, Wang YH, Montgomery SE, Dar AC, Cleary JD, Nichol K. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res 2002;30:4534–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Dere R, Napierala M, Ranum LP, Wells RD. Hairpin structure-forming propensity of the (CCTG.CAGG) tetranucleotide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J Biol Chem 2004;279:41715–41726. [DOI] [PubMed] [Google Scholar]
- 212.Williamson JR. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struct 1994;23:703–730. [DOI] [PubMed] [Google Scholar]
- 213.Ohshima K, Kang S, Larson JE, Wells RD. TTA.TAA triplet repeats in plasmids form a non-H bonded structure. Journal of Biological Chemistry 1996;271:16784–16791. [DOI] [PubMed] [Google Scholar]
- 214.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009;138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, Verhoeyen E, Britton S, Calsou P, Salles B, Bizard A, Nadal M, Salvati E, Sabatier L, Wu Y, Biroccio A, Londono-Vallejo A, Giraud-Panis MJ, Gilson E. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 2010;142:230–242. [DOI] [PubMed] [Google Scholar]
- 216.Tazumi A, Fukuura M, Nakato R, Kishimoto A, Takenaka T, Ogawa S, Song JH, Takahashi TS, Nakagawa T, Shirahige K, Masukata H. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev 2012;26:2050–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lambert S, Carr AM. Replication stress and genome rearrangements: lessons from yeast models. Curr Opin Genet Dev 2013;23:132–139. [DOI] [PubMed] [Google Scholar]
- 218.Watanabe T, Marotta M, Suzuki R, Diede SJ, Tapscott SJ, Niida A, Chen X, Mouakkad L, Kondratova A, Giuliano AE, Orsulic S, Tanaka H. Impediment of Replication Forks by Long Non-coding RNA Provokes Chromosomal Rearrangements by Error-Prone Restart. Cell Rep 2017;21:2223–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.García-Muse T, Aguilera A. Transcription-replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol 2016;17:553–563. [DOI] [PubMed] [Google Scholar]
- 220.Gunzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell 2003;2:542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wierdl M, Greene CN, Datta A, Jinks-Robertson S, Petes TD. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics 1996;143:713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gottipati P, Cassel TN, Savolainen L, Helleday T. Transcription-associated recombination is dependent on replication in Mammalian cells. Mol Cell Biol 2008;28:154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Alsford S, Horn D. RNA polymerase I transcription stimulates homologous recombination in Trypanosoma brucei. Mol Biochem Parasitol 2007;153:77–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lidani KCF, Andrade FA, Bavia L, Damasceno FS, Beltrame MH, Messias-Reason IJ, Sandri TL. Chagas Disease: From Discovery to a Worldwide Health Problem. Front Public Health 2019;7:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Souto EB, Dias-Ferreira J, Craveiro SA, Severino P, Sanchez-Lopez E, Garcia ML, Silva AM, Souto SB, Mahant S. Therapeutic Interventions for Countering Leishmaniasis and Chagas’s Disease: From Traditional Sources to Nanotechnological Systems. Pathogens 2019;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Freitas-Junior LHG, Porto RM, Pirrit LA, Schenkman S, Scherf A. Identification of the telomere in Trypanosoma cruzi reveals highly heterogeneous telomere lengths in different parasite strains. Nucleic Acids Research 1999;27:2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Müller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O’neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schäfer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ. The genome of the kinetoplastid parasite, Leishmania major. Science 2005;309:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJJ, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 2010;38:D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.da Silva MS, Perez AM, da Silveira RC, de Moraes CE, Siqueira-Neto JL, Freitas LH, Cano MI. The Leishmania amazonensis TRF (TTAGGG repeat-binding factor) homologue binds and co-localizes with telomeres. BMC Microbiol 2010;10:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lieke T, Steeg C, Graefe SE, Fleischer B, Jacobs T. Interaction of natural killer cells with Trypanosoma cruzi-infected fibroblasts. Clin Exp Immunol 2006;145:357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Alves MJM, Abuin G, Kuwajima VY, Colli W. Partial inhibition of trypomastigote entry into cultured mammalian cells by monoclonal antibodies against a surface glycoprotein of Trypanosoma cruzi. MBP 1986;21:75–83. [DOI] [PubMed] [Google Scholar]
- 232.Abuin G, Colli W, Souza WD, Alves MJM. A surface antigen of Trypanosoma cruzi involved in cell invasion (Tc-85) is heterogeneous in expression and molecular constitution. MBP 1989;35:229–237. [DOI] [PubMed] [Google Scholar]
- 233.Teixeira AA, de Vasconcelos VC, Colli W, Alves MJ, Giordano RJ. Trypanosoma cruzi Binds to Cytokeratin through Conserved Peptide Motifs Found in the Laminin-G-Like Domain of the gp85/Trans-sialidase Proteins. PLoS Negl Trop Dis 2015;9:e0004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.San Francisco J, Barría I, Gutiérrez B, Neira I, Muñoz C, Sagua H, Araya JE, Andrade JC, Zailberger A, Catalán A, Remonsellez F, Vega JL, González J. Decreased cruzipain and gp85/trans-sialidase family protein expression contributes to loss of Trypanosoma cruzi trypomastigote virulence. Microbes Infect 2017;19:55–61. [DOI] [PubMed] [Google Scholar]
- 235.Kim D, Chiurillo MA, El-Sayed N, Jones K, Santos MR, Porcile PE, Andersson B, Myler P, da Silveira JF, Ramirez JL. Telomere and subtelomere of Trypanosoma cruzi chromosomes are enriched in (pseudo)genes of retrotransposon hot spot and trans-sialidase-like gene families: the origins of T. cruzi telomeres. Gene 2005;346:153–161. [DOI] [PubMed] [Google Scholar]