Abstract
Novel coronavirus SARS-CoV-2, designated as COVID-19 by the World Health Organization (WHO) on the February 11, 2020, is one of the highly pathogenic β‐coronaviruses which infects human. Early diagnosis of COVID-19 is the most critical step to treat infection. The diagnostic tools are generally molecular methods, serology and viral culture. Recently CRISPR-based method has been investigated to diagnose and treat coronavirus infection. The emergence of 2019-nCoV during the influenza season, has led to the extensive use of antibiotics and neuraminidase enzyme inhibitors, taken orally and intravenously.
Currently, antiviral inhibitors of SARS and MERS spike proteins, neuraminidase inhibitors, anti-inflammatory drugs and EK1 peptide are the available therapeutic options for SARS-CoV-2 infected individuals. In addition, Chloroquine, which was previously used for malarial and autoimmune disease, has shown efficacy in the 2019-nCoV infection treatment. In severe hypoxaemia, a combination of antibiotics, α-interferon, lopinavir and mechanical ventilation can effectively mitigate the symptoms. Comprehensive knowledge on the innate and adaptive immune responses, will make it possible to propose potent antiviral drugs with their effective therapeutic measures for the prevention of viral infection. This therapeutic strategy will help patients worldwide to protect themselves against severe and fatal viral infections, that potentially can evolve and develop drug resistance, and to reduce mortality rates.
Keywords: Coronavirus, Immune response, Respiratory syndrome, Treatment, Pathogen
Graphical abstract
Highlights
-
•
Novel coronavirus SARS-CoV-2 which was named as COVID-19 by the WHO on the 11th of February 2020.
-
•
2019-nCoV can cause severe pulmonary infection, respiratory failure therefore early diagnosis of COVID-19 is crucial for disease control.
-
•
Due to appearance of the 2019-nCoV pneumonia neuraminidase inhibitors such as oseltamivir are has been widely used.
-
•
Nucleoside analogues, protease inhibitors, neuraminidase inhibitors, anti-inflammatory drugs, could be the treatment options for respiratory diseases.
-
•
Antiviral therapies, as well as support therapies including oxygen and mechanical ventilation have been used to the treatment of patients.
-
•
Accumulated knowledge of innate immune response system, will make it possible to propose the potent antiviral therapies for prevention of viral infection.
1. Introduction
Over the last two decades, three coronaviruses have periodically crossed animal species such as bats, transmitted to human populations, and caused an ever-increasing outbreak of a large-scale pandemic (Perlman, 2020; Wang et al., 2020a). The previously reported viral zoonotic pathogens include SARS-CoV (severe acute respiratory syndrome coronavirus) and MERS (Middle East respiratory syndrome coronavirus) (Cui et al., 2019; de Wit et al., 2016), that can cause severe respiratory disease in human (Luk et al., 2019; Phan, 2020). SARS-CoV-2, a novel coronavirus (which causes COVID-19), has fast spread like a pandemic since its outbreak in Wuhan, China, in December 2019 (Chan et al., 2020a). It causes an acute and deadly disease with a 2% mortality rate. However, this novel coronavirus is usually associated with a mild to severe respiratory disease in humans (Perlman, 2020; Zhu et al., 2019; Huang et al., 2020a). This virus has the ability of jumping between species, and causing a variety of diseases as a strange and complex pathogen (Fung and Liu, 2019). Due to the frequent interaction between humans and animals, a virus is a common source of zoonotic infection. COVID-19, due to its human-to-human transmission, has become a health emergency of global concern (Chan et al., 2020a; Organization, 2020). Currently, we have no sufficient evidence to propose that a specific wildlife animal is the virus origin. A proper study of the viral source, evolution, mode of zoonotic transmission and infectivity, would help to prevent further infections. (Cui et al., 2019; de Wit et al., 2016).
2. Structure
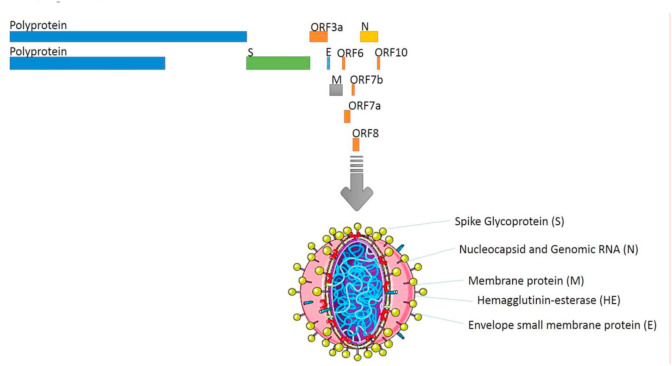
Coronaviruses (CoVs) belong to the Nidovirales order, Coronaviridae family, which comprises of two subfamilies, namely Orthocoronavirinae and Letovirinae (International Committee on Taxonomy of Viruses) (Carlos et al., 2020; Lei et al., 2018). CoVs are genotypically classified into four genera: Alpha coronaviruses (a), Beta coronaviruses (b), Gamma coronaviruses (g), and Delta coronaviruses (d), according to their phylogenetic and genomic data. Further, β‐coronavirus is subdivided into four viral lineages of A to D (Woo et al., 2012; Li, 2016). Coronavirus is an enveloped and non-segmented virus, which has a large positive‐sense single‐stranded RNA virus genome (27–32 kb), capped and polyadenylated (Song et al., 2019). Coronavirus also has crown-shape spikes projecting from its surface (80–160 nM in size), from which its name derived (Woo et al., 2010). The CoV Spike (S) glycoprotein attaches to cellular receptors on the host cell and mediates viral entry resulting in interspecies transmission and pathogenesis (Li, 2016; Zhu et al., 2018). A virion, consists of two basic components: genomic RNA and a protein capsid packaged forming a nucleocapsid. The nucleocapsid is surrounded by a phospholipid bilayer, composed of the spike glycoprotein trimmer (S) and the hemagglutinin‐esterase (HE). All viruses have Nucleocapsid (N), Spike (S), Envelope (E) and Membrane (M) structural proteins. Besides this, they also have several non-structural and accessory proteins. The membrane (M) protein and the envelope (E) protein are placed amongst the S proteins in the virus capsid (Luk et al., 2019; Li, 2016; Fahimi et al., 2018). Novel coronavirus-2019 (nCoV-2019) RNA genome, has 29891 nucleotides in length, which encode 9860 amino acids (Luk et al., 2019). nCoV-2019 genome contain following components: two flanking untranslated regions (UTRs), a single long open reading frame (ORF1ab) (7096-aa), a non-structural polyprotein (7096-aa), four structural proteins – Spike (S) (1273-aa), Envelope (E) (75-aa), Membrane (M) (222-aa), Nucleocapsid (N) (419-aa), and five accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10) (Phan, 2020; Chan et al., 2020b). (Fig. 1 ).
Fig. 1.
Coronavirus structure. Coronaviruses are enveloped, non-segmented virus with a positive‐sense single‐stranded RNA and phosphorylated nucleocapsid (N) protein.
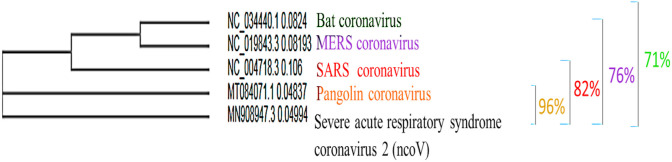
Presently, whole genome sequence of nCoV-2019 is available in GenBank (accession number MN908947) (Phan, 2020). Pair-wise sequence alignment illustrated that bat SARS-like coronavirus of bat-SL-CoVZC45 and bat-SL-CoVZXC21 (Genbank No: MG772933), sharing 88% sequence homolog were the closest strain to nCoV-2019. Recent researches on the physicochemical property of nCoV-2019 showed that it can be inactivated upon exposure to 56 °C for 30 min, ultraviolet radiation, using lipid solvents such as 75% ethanol, chlorine containing disinfectant, peroxyacetic acid and chloroform (Chen et al., 2020a). Amongst 30 known CoVs which infect humans, mammals and other animals, only α‐ and β‐CoVs are zoonotic in origin and are highly pathogenic to humans (Cui et al., 2019; Luk et al., 2019). Human α‐CoVs including NL63, 229E, OC43 and HKU1 (van der Hoek et al., 2004), generally cause mild upper respiratory infection in immunocompetent patients with common symptoms of fever, headache, and cough (Su et al., 2016; Forni et al., 2017). Human β‐CoVs include severe acute respiratory syndrome coronavirus (SARS-CoV) (Ksiazek et al., 2003; Drosten et al., 2003; Gralinski and Menachery, 2020)) and Middle East respiratory syndrome coronavirus (MERS-CoV), with no clear sign and symptom in the primary stage; then it can quickly progress to severe pneumonia, dyspnea, and even death (Drosten et al., 2003; Zaki et al., 2012), with morality rates of ~10% in immunocompetent hosts and ~35% in immunocompromised hosts (Li et al., 2020). Virus genome sequence analysis of the COVID-19 showed 96.2% identity with Bat CoV RaTG13, suggesting the bat CoV might have common ancestor with SARS-CoV-2, and predicted as origin of SARS CoV- 2 (Guo et al., 2020). It is also likely that an intermediate host exists between bats and humans. Preliminary data indicates several lineages of pangolin CoVs genetically identical to SARS-CoV-2, suggesting that pangolin may be possible intermediate host in which recombination of the bat and pangolin coronaviruses could have occurred (Guo et al., 2020; Sun et al., 2020a). Full genome phylogeny of Pangolin, SARS-CoV, MERS-CoV and Bat coronavirus showed that Pangolin possesses 96% similarity with SARS‐CoV‐2, while SARS-CoV, MERS-CoV and Bat coronavirus share 82%, 76% and 71% identity to SARS‐CoV‐2, respectively (Fig. 2 ). It seems that the high sequence identity in SARS-CoV-2 and Pangolin-CoV may be due to coincidental convergent evolution (Tang et al., 2020). Phylogenetic analysis of RBD region of the spike protein, has also led to a further presumption that the identity of SARS-CoV-2 RBD to pangolins-CoV RBD might be result of accidental mutations followed by natural selection, and/or recombination events in pangolins (Tang et al., 2020) (Fig. 2).
Fig. 2.
Phylogenetic analysis of whole genomes of SARS-CoV 2 and Pangolin-CoV, SARS-CoV, MERS-CoV and Bat-CoV).
3. Clinical symptoms and patient data
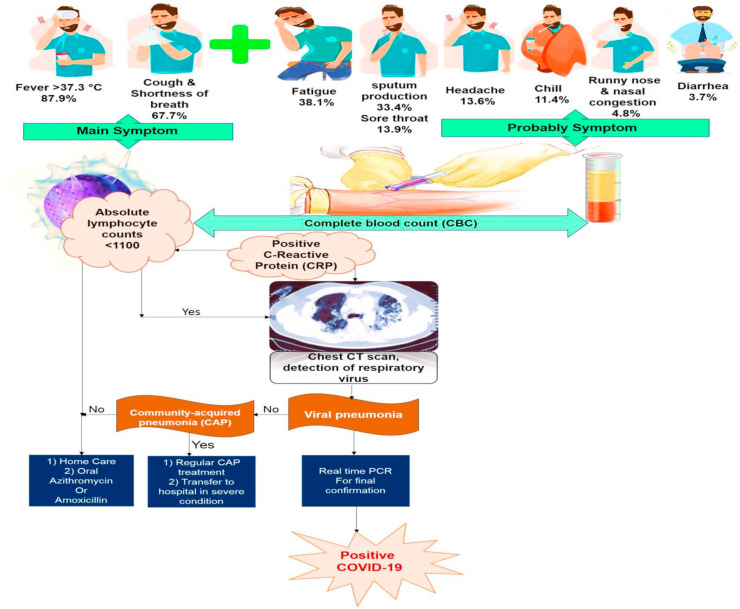
Most common clinical symptoms of COVID-19 disease are dry cough, fever and shortness of breath in the majority of patients. Some patients also experience other signs such as sore throat, headache, myalgia, fatigue and diarrhea (Chen et al., 2020b; Hui et al., 2020). In the initial phase of the disease, patients can be afebrile, only presenting with chills and respiratory symptoms. Although most cases appear to be mild, all patients have new pulmonary signs as ground-glass lung opacity on chest X-ray (Woo et al., 2010; Holshue et al., 2020). The symptoms in patients with mild pneumonia are fever, cough, sore throat, tiredness, headache or myalgia (Yang et al., 2020). They do not obviously show any of the serious symptoms or complications. Some patients were reported to have upper respiratory infection (URI), bilateral patchy opacity in lung (Chan et al., 2020a), decreased white blood cell or lymphocyte number (Zhou et al., 2020a) and increased ALT, AST, LDH, CK-MB, CRP and ESR in these stages of infection (Guan et al., 2020). Patients with severe pneumonia, suffer from acute respiratory distress syndrome (ARDS) and refractory hypoxaemia. nCoV-2019 can cause severe pulmonary infection, respiratory failure, along with organ damage and dysfunction. In case of extra-pulmonary system dysfunctions, such as derangements in hematologic and digestive system, the risk of sepsis and septic shock will be serious, resulting in considerable increase in fatality rate. The findings showed that the disease is mild in the majority of patients (81%) and only a few of them develop severe pneumonia, pulmonary edema, ARDS, or different organ damages with case morality rate of 2.3%. In children, infection generally presents with much milder clinical symptoms or even asymptomatic, compared with adult. According to previous studies, pregnant women do not seem to have a severe disease, while older patients are at a high risk of developing critical illness (Chen et al., 2020b; Yang et al., 2020). The Case Fatality Rate (CFR) increased in 50% of patients older than 80 with a history of chronic diseases, such as high blood pressure, diabetes, heart diseases, respiratory diseases, cerebrovascular diseases, endocrine system disorders, digestive system disorders and cancers. In most of cases, the cause of death is respiratory failure, septic shock or several organ failure (Chen et al., 2020b). In fact, increased C-reactive protein (CRP) is an important factor of impaired immunity, characterized by lymphopenia. So, SARS-CoV-2 is more probably to affect older people with chronic disease due to their poorer immune function (Badawi and Ryoo, 2016). Covid-19 has also been found to infect more males (average age of 55.5 years) than females (Badawi and Ryoo, 2016). The less susceptibility of females to viral infections is likely associated with the protective role of X chromosome and sex hormones, which result in stronger immune response to virus (Channappanavar et al., 2017). CT imaging findings of patients with COVID-19 revealed that most of cases had ground-glass opacities, which may manifest as crazy paving pattern, organizing pneumonia and architectural distortion. On X-rays or chest CT imaging of the examined patients, unilateral or bilateral involvement compatible with viral pneumonia was found, and bilateral multiple lobular and subsegmental regions of consolidation were noticed in cases hospitalized in the intensive care unit (Huang et al., 2020a; Moxley et al., 2002; Lucia et al., 2020) (Fig. 3 ).
Fig. 3.
Fellow diagram for COVID-19 confirmation.
4. Laboratory evaluation
Early diagnosis is the most important step to manage and treat COVID-19. The diagnostic tools are generally molecular methods, serology and viral culture. Initial laboratory investigations of hospitalized patients consist of a complete blood count, coagulation testing and serum biochemical test such as creatine kinase (CK), lactate dehydrogenase, procalcitonin, and electrolytes (Li et al., 2020; Moxley et al., 2002). Based on laboratory tests, most patients showed a significant decrease in total number of lymphocytes, suggesting that lymphocytes (particularly T lymphocytes) are likely _target of SARS-CoV-2. In the COVID infection, virus particles begin to spread through the respiratory tract and infect the surrounding uninfected cells. This leads to initiate a cytokine storm and consequently trigger a series of sever immune responses. This process results in some changes in immune cells, particularly lymphocytes, and then leads to immune system dysfunction (Li et al., 2020). Hence, the decreased number of the circulating lymphocytes could be considered as a diagnostic marker for SARS-CoV-2 infection and its severity (Chen et al., 2020b). Previous studies reported that there is a correlation between elevated level of pro-inflammatory cytokines like IL1B, IL6, IL12, IFNγ, IP10, and MCP1, and cytokines such as IFNγ, TNFα, IL15, and IL17, in SARS-CoV and MERS-CoV infection respectively, with pulmonary inflammation and lung injury. Notably, the high value of cytokines like IL1B, IFNγ, IP10, and MCP1, may activate T helper 1 cells (Th1) response. This cytokine storm is probably associated with disease severity. However, in SARS-CoV2 infection, enhanced secretion of T helper 2 (Th2) and cytokines like IL4 and IL10, reduce peripheral white blood cells and immune cells such as lymphocytes, probably leading to suppression of the inflammatory response and immune system function followed by serious lung damage, which differs from SARS-CoV infection (Moxley et al., 2002). These findings suggest that sever and uncontrolled inflammatory response have a more damaging effect on COVID-19 induced lung injury than viral pathogenicity. Therefore, in SARS-CoV-2 pneumonia, it is vital to control cytokines or chemokines to detect the impact of 2019 coronavirus on their production in the critical phase of the disease (Li et al., 2020; Moxley et al., 2002). RT-PCR (reverse‐transcription polymerase chain reaction) or Real-Time PCR and genome sequencing for respiratory or blood specimens are the next methods to confirm COVID-19 infection (Table 1 ).
Table 1.
Primers for COVID-19 detection with Real time PCR method.
| Id | Name | Sequence | Number of bases | Product size | Ref. | |
|---|---|---|---|---|---|---|
| Pair 1 | COVID-19-Forward | CCCTGTGGGTTTTACACTTAA | 21 | 199 | Lucia et al. (2020) | |
| COVID-19-Reverse | ACGATTGTGCATCAGCTGA | 19 | ||||
| Pair 2 | COVID-19- Forward | GGGGAACTTCTCCTGCTAGAAT | 22 | 99 | Jung et al. (2020) | |
| COVID-19- Reverse | CAGACATTTTGCTCTCAAGCTG | 22 | ||||
| Probe | TTGCTGCTGCTTGACAGATT | |||||
| Pair 3 | COVID-19- Forward | TTCTTGCTTTCGTGGTATTC | 20 | 89 | Chan et al. (2020a) | |
| COVID-19- Reverse | CACGTTAACAATATTGCAGC | 20 | ||||
| Pair 4 | COVID-19- Forward (ORF1b) | TGGGGYTTTACRGGTAACCT | 20 | 132 | Chu et al. (2020) | |
| COVID-19- Reverse (ORF1b) | AACRCGCTTAACAAAGCACTC | 21 | ||||
| Probe | FAM TAGTTGTGATGCWATCATGACTAG-TAMRA | 24 | ||||
| Pair 5 | COVID-19- Forward | TAATCAGACAAGGAACTGATTA | 22 | 110 | Chu et al. (2020) | |
| COVID-19- Reverse | CGAAGGTGTGACTTCCATG | 19 | ||||
| Probe | FAM-GCAAATTGTGCAATTTGCGG-TAMRA | 20 | ||||
| Pair 6 | COVID-19- Forward | ACTTCTTTTTCTTGCTTTCGTGGT | 24 | 82 | Sun et al. (2020b) | |
| COVID-19- Reverse | GCAGCAGTACGCACACAATC | 20 | ||||
| Probe | CY5-CTAGTTACACTAGCCATCCTTACTGC | 26 | ||||
| Pair 7 | COVID-19- Forward ORF1ab |
CCCTGTGGGTTTTACACTTAA | 21 | 119 | Wang et al. (2020b) | |
| COVID-19- Reverse ORF1ab |
ACGATTGTGCATCAGCTGA | 19 | ||||
| Probe | VIC-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1 | 28 | ||||
| Pair 8 | COVID-19- Forward | GGGAACTTCTCCTGCTAGAAT | 21 | 98 | Wang et al. (2020b) | |
| COVID-19- Reverse | CAGACATTTTGCTCTCAAGCTG | 22 | ||||
| Probe | FAM-TTGCTGCTGCTTGACAGATT-TAMRA | 20 | ||||
| Pair 9 | COVID-19-Forward | GACCCCAAAATCAGCGAAAT | 20 | 72 | Jung et al. (2020) | |
| COVID-19-Reverse | TCTGGTTACTGCCAGTTGAATCTG | 24 | ||||
| Probe | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | 24 | ||||
| Pair 10 | COVID-19-Forward | TTACAAACATTGGCCGCAAA | 20 | 67 | Jung et al. (2020) | |
| COVID-19-Reverse | GCGCGACATTCCGAAGAA | 18 | ||||
| Probe | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | 23 | ||||
| Pair 11 | COVID-19-Forward | GGGAGCCTTGAATACACCAAAA | 22 | 72 | Jung et al. (2020) | |
| COVID-19-Reverse | TGTAGCACGATTGCAGCATTG | 21 | ||||
| Probe | FAM-AYCACATTGGCACCCGCAATCCTG- BHQ1 | 24 | ||||
| Pair 12 | COVID-19-Forward | TCT GGT TAC TGC CAG TTG AAT CTG | 26 | 113 | Kim et al. (2020) | |
| COVID-19-Reverse | ATATTGCAGCAGTACGCACACA | 22 | ||||
| Probe | FAM- ACACTAGCCATCCTTACTGCGCTTCG- BHQ1 | 26 | ||||
| Pair 13 | COVID-19-Forward | AAATTTTGGGGACCAGGAAC | 20 | 158 | Jung et al. (2020) | |
| COVID-19-Reverse | TGGCAGCTGTGTAGGTCAAC | 20 | ||||
| Probe | FAM- ATGTCGCGCATTGGCATGGA- BHQ1 | 20 | ||||
| Pair 14 | COVID-19-Forward | CGTTTGGTGGACCCTCAGAT | 20 | 57 | Jung et al. (2020) | |
| COVID-19-Reverse | CCCCACTGCGTTCTCCATT | 19 | ||||
| Probe | FAM- CAACTGGCAGTAACCA- BHQ1 | 16 | ||||
Currently, RT‐PCR method is opted for the SARS-CoV-2 Viral RNA detection in samples collected from infected patients, which has been facilitated by availability of full genome sequence of the 2019‐nCoV in Gene Bank (Ji et al., 2020a). WHO approved real-time RT-PCR assays for detecting viral nucleic acid (RNA) of common viruses, such as SARS-CoV and MERS-CoV, influenza, avian influenza, respiratory syncytial virus, adenovirus, para-influenza virus present in respiratory samples, including oropharyngeal and nasopharyngeal swabs, sputum or bronchial aspirates, bronchoalveolar lavage fluids. Sputum is a noninvasive lower respiratory tract specimen, but only 28% of patients with 2019-nCoV in 1 case series could produce sputum for diagnostic evaluation (Huang et al., 2020a). Notably, lower respiratory tract specimens compared with upper respiratory tract samples can provide significantly more viral load and genome fraction for 2019-nCoV test (Huang et al., 2020a). In addition, stool or blood samples are also used for gene sequencing of the viral nucleic acid. Viral culture is more time-consuming method as compared to the other methods, and it was much more useful in the first stage of outbreaks, before other diagnostic methods became clinically available. Besides, viral cultures can be used in the in-vitro and in-vivo antiviral treatment and vaccine evaluation trials (Sahin et al., 2019). At present, cell culture experiment of human respiratory epithelial cells for detecting 2019-nCoV is not performable in general laboratory. Currently, serological antibody testing kit for identifying virus antigen are not available. Although RT-PCR is the common approach for detecting 2019-nCoV at current emergency condition, but shortage of RT-PCR instruments, difficulties of collection and transportation of specimens, low precision of RT-PCR, the high rates of false negative results, together with its time-consuming process and the problems associated to performance of RT-PCR kit are the main obstacles to control this epidemic (Weiss and Leibowitz, 2011). Chest CT, Compared to RT-PCR, is a fast, sensitive, easy to perform and more accurate and reliable tool for screening and diagnosis of COVID-19 (Xie et al., 2020). Chest CT can also show pulmonary abnormalities in COVID-19 patients with early negative RT-PCR results (Huang et al., 2020b; Chung et al., 2020). In the primary stage of pneumonia itself, CT images can demonstrate several small ground-glass opacity as well as some interstitial changes (Huang et al., 2020a), remarkable in the lung periphery (Chan et al., 2020a) (Sahin et al., 2019).
5. CRISPR/Cas13 a new and fast method for COVID-19 detection and treatment
Feng Zhang and his colleges from Broad Institute of MIT and Harvard, have recently developed a new and advanced CRISPR-Cas13-based SHERLOCK system to diagnose COVID-19. CRISPR-Cas13-based SHERLOCK system consists of two RNA guides, which are combined with a Cas13 protein, and form a SHERLOCK system to recognize the presence of COVID-19 viral RNA. At first the team used synthetic fragments of SARS-CoV-2 RNA as a pattern for designing two RNA guides, which are able to bind to their complementary sequences in COVID-19 RNA. In order to visual readout, they used a paper strip (as a paper strip in pregnancy test) for dipping into a prepared sample. Then, appearance of a line on the paper strip indicates the existence of virus in the sample (Zhang et al. Jonathan; Metsky et al., 2020).
Also, another research group has recently proposed a RNA-_targeting CRISPR system to _target RNA genome of SARS-CoV-2 in the laboratory, to limit its ability to reproduce (Nguyen et al., 2020). This CRISPR/Cas13d system as a novel technique is applicable for the treatment of all types of RNA virus infections. CRISPR/Cas13d system contains a guide RNAs (gRNAs) designed and specifically complementary to the virus RNA genome to bind ORF1ab and the spike (S) of SARS-CoV-2 (Nguyen et al., 2020), also a Cas13d protein to cleave the 2019-nCov RNA genome (Konermann et al., 2018). To eliminate the virus, gRNAs recognized the virus RNA sequence and through attaching its complementary sequence, recall Cas13d protein to _target site (viral RNA sequence) to cleave it. This unique system is useful, especially for _targeting different virus variants that evolve and may escape traditional drugs. Currently, a total amount of 10,333 guide RNAs have been designed to specifically _target 10 peptide-coding sites on virus RNA genome of SARS-CoV-2, which has no impact on the human transcriptome (Jordan et al., 2018). Among various virus vectors, adeno-associated virus (AAV) can be used to deliver parts of CRISPR/Cas13d system to the infected cells in SARS-CoV-2 patients, particularly to _target their lung, which is main infected organ (Jordan et al., 2018). Before therapeutic application of CRISPR/Cas13d system to patients, it is necessary to determine the safety and efficacy of this system in clearance of 2019-nCov and other viruses in animals. If researchers find this therapeutic strategy secure and beneficial, then it would be applied to kill the viruses that have the potential to evolve and also develop drug resistance (Momattin et al., 2019).
6. Recent pharmaceutical strategies to treat coronavirus
Since the emergence of 2019-nCoV, due to its rapid spread and being a serious threat to human health, researchers have made great efforts to understand the pathogenetic characteristics of this virus to develop effective drugs. Due to appearance of the 2019-nCoV during the influenza season, orally and intravenously antibiotics and neuraminidase inhibitors such as oseltamivir having been widely used as an experimental treatment for 2019-nCoV in China (Bleibtreu et al., 2018; Sheahan et al., 2020). However, there is no reliable evidence that shows oseltamivir is an effective treatment in 2019-nCoV (Sheahan et al., 2020). At present, there is no antiviral drug assumed to provide protection against COVID-19 infection; also it will take long time to develop and a vaccine and gets approval for it. (Huang et al., 2020a). Nowadays, Griffithsin, as an inhibitor of SARS and MERS spike, Remdesivir, favipiravir and ribavirin (nucleoside analogues), lopinavir/ritonavir (protease enzyme inhibitors) (Chu et al., 2004), oseltamivir (neuraminidase inhibitors), anti-inflammatory drugs and EK1 peptide (Xia et al., 2019), Umifenovir (Abidol) (Coleman et al., 2016) (RNA synthesis inhibitors (TDF, 3 TC), ShuFeng JieDu and Lianhua qingwen Capsules (chines antibacterial traditional drugs) (Ji et al., 2020b; Ding et al., 2017), are used as available drug options for new respiratory infectious diseases caused by 2019-nCoV (Chu et al., 2004; Ji et al., 2020b; Ding et al., 2017; Lu, 2020). All these drugs have been used to treat previous coronaviruses including SARS, MERS, or some other viruses such as influenza, Ebola, HIV. Among several antiviral drugs approved by FAD (including ribavirin, penciclovir, nitazoxanide, nafamostat, chloroquine) as well as two well-known antiviral agents of remdesivir and favipiravir, only two compounds (chloroquine and remdesivir) were confirmed for their highly efficiency in treatment of SARS-CoV-2 infection, in vitro. Chloroquine, which has been previously used for malarial and autoimmune diseases, has the clinical potential to be applied against the 2019-nCoV infection (Savarino et al., 2006; Yan et al., 2013). Chloroquine shows its inhibitory effects against 2019-nCoV through increasing endosomal pH required for virus cell fusion, also affecting the glycosylation of cellular receptors of SARS-CoV (Vincent et al., 2005). Chloroquine is a cheap and safe anti-viral medicine, which is orally administrated and widely distributes all over body especially in the lungs (Wang et al., 2020c). Also, remdesivir, which has a structure chemically similar to HIV reverse-transcriptase inhibitors, is currently in the phase of clinical trials for 2019-nCov (Momattin et al., 2019). The spike (S) protein is a promising _target for the development of antivirals drugs, due to its major role in the virus-receptor interaction. Griffithsin, a medicine _targeting the oligosaccharides on the surface of different spike proteins (Zumla et al., 2016), has also been tested in phase I trials for the treatment of HIV and SARS-CoV (Arabi et al., 2020). However, the efficacy of spike inhibitors for the treatment of 2019-nCoV should be re-evaluated (Zumla et al., 2016). There are multiple mechanisms of action for nucleoside analogues (adenine or guanine derivative), including lethal mutagenesis, inhibition of RNA biosynthesis, or RNA chain premature termination (Arabi et al., 2020). They inhibit synthesis of viral RNA in human coronaviruses by _targeting the RNA-dependent RNA polymerase (De Clercq, 2019). Favipiravir (T-705), a guanine analogue, is an antiviral drug _targeting the viral RNA-dependent RNA polymerase of RNA viruses such as Ebola, influenza, yellow fever, chikungunya, norovirus and enterovirus. It has been recently proposed to be effective also against coronaviruses, in vitro (De Clercq, 2019). Using fabiravir and ribavirin combined with oseltamivir, for the treatment of coronaviruses diseases such as in severe influenza, is better than oseltamivir alone (Wang et al., 2019). At present, lopinavir/ritonavir and IFN-alpha are in the initial phase of clinical trials in patients infected with 2019-nCoV (Wang et al., 2019; Bassetti et al., 2020). According to the guidelines (Wang et al., 2019), lopinavir (400 mg/100 mg bid po) used to treat HIV/AIDS infection and ritonavir as a booster, as well as IFN-alpha (5 million U bid inh) usually utilized for treatment of HBV, are recommended as antiviral therapy (Chu et al., 2004; Lu, 2020). A scholar's report from Hong Kong illustrated that use of the combination of lopinavir/ritonavir (LPV/RTV) (Anti-HIV drugs) with ribavirin, reduces the risk of acute respiratory distress syndrome (ARDS) or death in treated patients (Chu et al., 2004). Further researches are needed to understand whether these inhibitors can effectively block the 3-chymotrypsin-like and papain-like proteases of 2019-nCoV (Huang et al., 2020a). In addition, remdesivir, earlier used against the Ebola virus, has been presently determined as antiviral drug for treating MERS/SARS and related zoonotic bat CoVs in the cell cultures and animal models (Sheahan et al., 2017; Mulangu et al., 2019; Brown et al., 2019). Based on animal experiments, remdesivir effectively decreases the virus titer of mice infected with MERS-CoV and improves the lung tissue damage, in comparison to other cases treated with a compound of lopinavir/ritonavir and interferon-β (Sheahan et al., 2020). Remdesivir has also been recently suggested to inhibit 2019-nCoV (Holshue et al., 2020) by terminating the viral RNA transcription in early stage (Warren et al., 2016; Jordan et al., 2018). Although remdesivir seems be an effective drug for the treatment of 2019-nCoV (Agostini et al., 2018), more clinical researches are needed to confirm its efficacy and safety in infected patients. Since 2003, glucocorticoid has been widely used to inhibit pulmonary infection through regulating inflammatory responses in SARS-CoV patients. Among other therapeutic strategies, corticosteroid therapy has been used as a combined therapy, only when severe COVID-19 pneumonia was diagnosed by the physicians (Li and De Clercq, 2020). Systemic corticosteroids (methylprednisolone 40–120 mg per day) was applied frequently to treat severe cases of COVID-19-Induced pneumonia via reducing lung damage as a result of secreting high level of cytokines induced by SARS-CoV, MERS-CoV, and 2019-nCoV infections (Li and De Clercq, 2020; Zhou et al., 2020b; Russell et al., 2020). Although, value of lymphopenia is an important factor associated with disease severity and mortality rate in the SARS-CoV-2-related pneumonia, present evidence in SARS/MERS patients assume that receiving corticosteroids cannot prevent the coronaviruses infected patients from dying, but rather delay viral clearance. Western-type antiviral therapies with antibiotics, α-interferon and lopinavir as well as supportive therapies including oxygen therapy (eg, nasal cannula) and mechanical ventilation, have also been administered to treat COVID-19 patients, as per severity of the hypoxaemia (Russell et al., 2020).
7. Improve immune system to fight with COVID-19
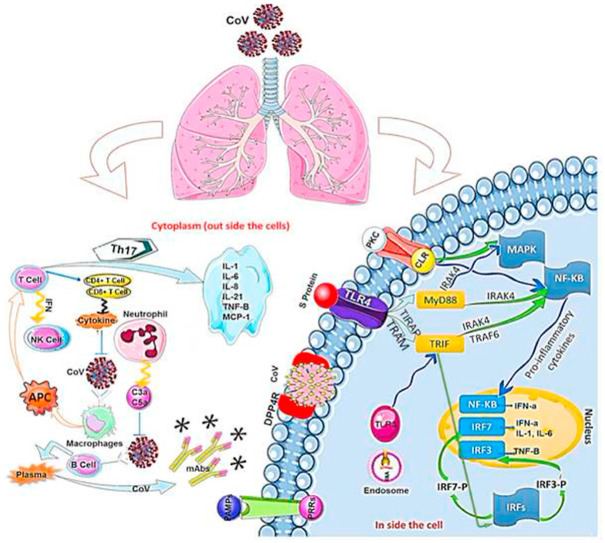
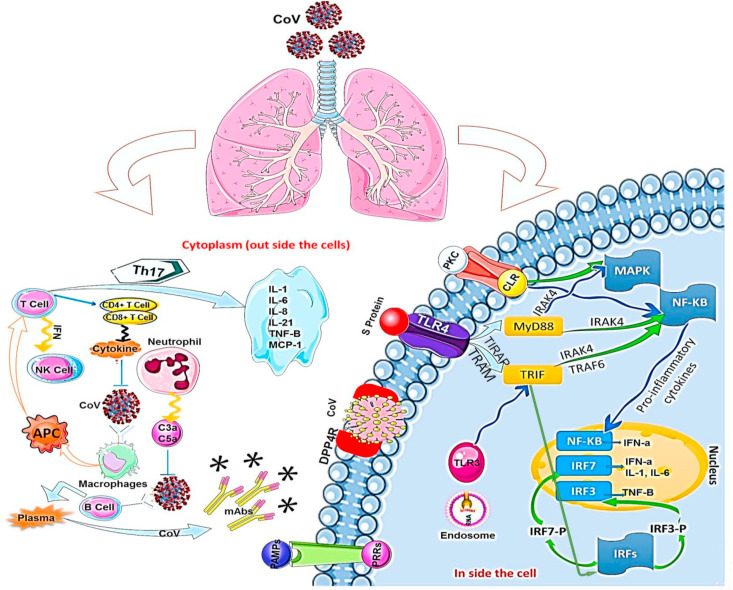
Recent advances in viral research brought a deep understanding on the immune system response to RNA viral infection. During viral infection, antiviral immune responses destroy replicating virus, leading to quick clearance of the virus. Tissue-repair response is a result of an adaptive immune response induction to clear viruses. However, uncontrolled systemic inflammatory response while recognizing coronaviruses, may leads to lung tissue injury, its dysfunction, and lung volume reduction in infected patient. The regulatory effect of chemotactic factors on positioning of leukocytes in the host lungs is critical for the function of the innate immune system against the viral infections. Therefore, change in chemotactic factors may cause sever and uncontrolled immune responses. Immune insufficiency and deficiency, may upraise viral replication which results in tissue damages (Guo et al., 2019). Conversely, exaggerated immune responses may also provoke the innate and adaptive immune responses against coronaviruses infection. Upon virus entry inside the host, it infects macrophages by attaching to DPP4 on the host cell through spike protein(s), which leads to releasing of genomic RNA in the cytoplasm. The macrophages present CoV antigens to T cells, which has a key role in anti-viral immune system. Afterward, antigen presentation stimulates T-cell activation and differentiation, and leads to generation and massive release of large quantity of cytokines for immune response reinforcement. Coronaviruses induce the continual production of the mediators that negatively affect the activation of NK, and CD8 T-cell. However, CD8 T-cells continue to produce high amount of cytokines to eliminate Coronaviruses. In fact, an immune response to dsRNA will ensue, when the viral genome begins to replicate (Wu and Zheng, 2020). TLR‐3 sensitized by dsRNA, IRFs and NF‐κB signaling pathways are activated to producing type I IFNs and pro-inflammatory cytokines. The elevated type I IFNs leads to increase in the secretion of antiviral proteins, which protect the uninfected cells. During coronaviruses replication, virus accessory proteins can interfere with TLR‐3 signaling through attaching the dsRNA to inhibit TLR‐3 activation, and then they can escape the immune response. It is likely that TLR‐4 recognizes S protein, thus allowing for activation of pro-inflammatory cytokines via the MyD88-Dependent Pathway. During coronaviruses infection, virus-host interaction induces the overproduction of immune mediators that leads to increase in secretion of chemokines and cytokines including IL‐1, IL‐6, IL‐8, IL‐21, TNF‐β, and MCP‐1 in infected cells. This high concentration of chemokines and cytokines, recruits lymphocytes and leukocytes at the _target site (viral infection site) for virus clearance. _targeted immunotherapy can be used as an appropriate alternative to some antiviral drugs with low therapeutic efficacy, or in patients showing drug resistance. A deep understanding of the innate and adaptive immune response system will make it possible to propose potent antiviral drugs with their effective therapeutic measures for detecting and preventing viral infection (Dyall et al., 2014) (Fig. 4 ).
Fig. 4.
After coronavirus infection, macrophages are activated and antigen-presenting cells (APC) display antigen to T cells. In cellular immunity, CD4 and CD8 with T cells produce cytokines to suppress the virus. T cells affect T helper cells to secrete interferons and interleukins. On the other hand, humoral immunity Th-2 activates B cells to produce memory B cells and antibodies. CoV attaches to DPP4R in the host cell through the S protein, causing genomic RNA to enter the cytoplasm. When CoV replicates, infected cell can partially start an immune response against dsRNA. The cascade of dsRNA and signaling pathways (IRF and NF-κB activation, respectively) sensitizes TLR-3 and is activated to produce type I IFN and proinflammatory cytokines. Type I IFN production plays a major role in increasing the release of antiviral proteins to protect uninfected cells.
8. Conclusion
The clinical virology of COVID-19 has yet to be developed, and there are much to learn about the behavior of the coronavirus in the human host. However, early identification of this novel coronavirus, helped us to promptly investigate antiviral compounds and to try to develop vaccines. Here, we reviewed the current information on genome sequences, protein structures and function of this novel virus, and tried to determine the evolutionary trajectories and viral-host interactions, to help virologists building a deep understanding of topic. A general knowledge on probable molecular mechanisms behind the coronavirus infection and the pathogenesis of SARS-CoV-2-related pneumonia, will help to develop both antiviral drugs and vaccines to reduce mortality in future.
Funding
The financial support for the current research was provided by Research Deputy of Kashan University of Medical Sciences, Kashan, Iran.
Availability of data and materials
Not applicable.
Authors’ contributions
ESH, HHK and NRK provided direction and guidance throughout the preparation of this manuscript. HHB, HN and HHK conducted the literature and drafted the manuscript. Other authors reviewed the manuscript and made significant revisions on the drafts. All authors read and approved the final version.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Declaration of competing interest
The authors declared that they have no competing interests.
Acknowledgment
We would like to thank all my colleagues for their assistance. The present work was part of research project (No. 99070), which was financially supported by Kashan University of Medical Sciences, Kashan, Iran.
Abbreviations:
- COVID-19
Coronavirus disease 2019
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- MERS-CoV
Middle East respiratory syndrome coronavirus
- 2019-nCoV
Novel coronavirus
- UTRs
Untranslated regions
- ORF
Open reading frame
- URI
Upper respiratory infection
- ARDS
Acute respiratory distress syndrome
- ESR
Erythrocyte sedimentation rate
- CRP
C-reactive protein
- LDH
Lactate dehydrogenase
- AST
Aspartate transaminase
- ALT
Alanine transaminase
- CK-MB
Creatine kinase myocardial band
- CFR
Case Fatality Rate
- CT
Computerized tomography
- IL
Interleukin
- MCP-1
Monocyte Chemotactic Protein-1
- IP-10
Inducible Protein 10
- IFNγ
Interferon gamma
- TNFα
Tumor necrosis factor alpha
- Th
T-helper
- RT-PCR
Reverse‐transcription polymerase chain reaction
- CRISPR
Clustered regularly interspaced short palindromic repeats\
- HIV
Human immunodeficiency viruses
- RNA
Ribonucleic acid
- LPV
Lopinavir
- RTV
Ritonavir
- DPP4
Dipeptidyl peptidase 4
- CD
Cluster of differentiation
- TLR
Toll-like receptor
- MyD88
Myeloid differentiation factor 88
- IRF
Interferon regulatory factor
- NF-κB
Nuclear factor kappa B
- AAV
Adeno-associated virus
References
- Agostini M.L. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. p. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M. Treatment of Middle East respiratory syndrome with a combination of lopinavir/ritonavir and interferon-β1b (MIRACLE trial): statistical analysis plan for a recursive two-stage group sequential randomized controlled trial. Trials. 2020;21(1):1–8. doi: 10.1186/s13063-019-3846-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawi A., Ryoo S.G. Prevalence of comorbidities in the Middle East respiratory syndrome coronavirus (MERS-CoV): a systematic review and meta-analysis. Int. J. Infect. Dis. 2016;49:129–133. doi: 10.1016/j.ijid.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti M., Vena A., Giacobbe D.R. The Novel Chinese Coronavirus (2019‐nCoV) Infections: challenges for fighting the storm. Eur. J. Clin. Invest. 2020;50(3) doi: 10.1111/eci.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleibtreu A. Clinical management of respiratory syndrome in patients hospitalized for suspected Middle East respiratory syndrome coronavirus infection in the Paris area from 2013 to 2016. BMC Infect. Dis. 2018;18(1):331. doi: 10.1186/s12879-018-3223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019;169:104541. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos W.G. Novel wuhan (2019-nCoV) coronavirus. Am. J. Respir. Crit. Care Med. 2020;201(4):P7–P8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- Chan J.F.-W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 2017;198(10):4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-M. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World Journal of Pediatrics. 2020:1–7. doi: 10.1007/s12519-020-00345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M. Radiology; 2020. CT Imaging Features of 2019 Novel Coronavirus (2019-nCoV) p. 200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.M. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and middle east respiratory syndrome coronavirus fusion. J. Virol. 2016;90(19):8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chemistry–An Asian Journal. 2019;14(22):3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. The Chinese prescription lianhuaqingwen capsule exerts anti-influenza activity through the inhibition of viral propagation and impacts immune function. BMC Compl. Alternative Med. 2017;17(1):130. doi: 10.1186/s12906-017-1585-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Dyall J. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58(8):4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahimi H. Dengue viruses and promising envelope protein domain III-based vaccines. Appl. Microbiol. Biotechnol. 2018;102(7):2977–2996. doi: 10.1007/s00253-018-8822-y. [DOI] [PubMed] [Google Scholar]
- Forni D. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-j. MedRxiv; 2020. Clinical Characteristics of 2019 Novel Coronavirus Infection in China. [Google Scholar]
- Guo L. Clinical features predicting mortality risk in patients with viral pneumonia: the MuLBSTA score. Front. Microbiol. 2019;10:2752. doi: 10.3389/fmicb.2019.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Medical Research. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295(1):22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W. Cross‐species transmission of the newly identified coronavirus 2019‐nCoV. J. Med. Virol. 2020;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S. Unique synergistic antiviral effects of Shufeng Jiedu Capsule and oseltamivir in influenza A viral-induced acute exacerbation of chronic obstructive pulmonary disease. Biomed. Pharmacother. 2020;121:109652. doi: 10.1016/j.biopha.2019.109652. [DOI] [PubMed] [Google Scholar]
- Jordan P.C. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018;14(2) doi: 10.1371/journal.ppat.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y.J. bioRxiv; 2020. Comparative Analysis of Primer-Probe Sets for the Laboratory Confirmation of SARS-CoV-2. [Google Scholar]
- Kim J.-M. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health and Research Perspectives. 2020;11(1):3. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S. Transcriptome engineering with RNA-_targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676. doi: 10.1016/j.cell.2018.02.033. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp 3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annual review of virology. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Nature Publishing Group; 2020. Therapeutic Options for the 2019 Novel Coronavirus (2019-nCoV) [DOI] [PubMed] [Google Scholar]
- Li G. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Bioscience trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- Lucia C., Federico P.-B., Alejandra G.C. bioRxiv; 2020. An Ultrasensitive, Rapid, and Portable Coronavirus SARS-CoV-2 Sequence Detection Method Based on CRISPR-Cas12. [Google Scholar]
- Luk H.K. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect. Genet. Evol. 2019 doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsky H.C. bioRxiv; 2020. CRISPR-based Surveillance for COVID-19 Using Genomically-Comprehensive Machine Learning Design. [Google Scholar]
- Momattin H., Al-Ali A.Y., Al-Tawfiq J.A. A systematic review of therapeutic agents for the treatment of the Middle East respiratory syndrome coronavirus (MERS-CoV) Trav. Med. Infect. Dis. 2019;30:9–18. doi: 10.1016/j.tmaid.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxley G. Sexual dimorphism in innate immunity. Arthritis Rheum. 2002;46(1):250–258. doi: 10.1002/1529-0131(200201)46:1<250::AID-ART10064>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Mulangu S. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.M., Zhang Y., Pandolfi P.P. Nature Publishing Group; 2020. Virus against Virus: a Potential Treatment for 2019-nCov (SARS-CoV-2) and Other RNA Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization W.H. World Health Organization (WHO); 2020. Pneumonia of Unknown Cause–China. Emergencies Preparedness, Response, Disease Outbreak News. [Google Scholar]
- Perlman S. Mass Medical Soc; 2020. Another Decade, Another Coronavirus. [Google Scholar]
- Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infect. Genet. Evol. 2020;79 doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin A.R. Novel coronavirus (COVID-19) outbreak: a review of the current literature. EJMO. 2019;4(1):1–7. 2020. [Google Scholar]
- Savarino A. New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 2006;6(2):67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396) doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1):1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 2020 doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. medRxiv; 2020. The Infection Evidence of SARS-COV-2 in Ocular Surface: a Single-Center Cross-Sectional Study. [Google Scholar]
- Tang X. On the origin and continuing evolution of SARS-CoV-2. National Science Review. 2020 doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L. Identification of a new human coronavirus. Nat. Med. 2004;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.J. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2(1):69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Comparative effectiveness of combined favipiravir and oseltamivir therapy versus oseltamivir monotherapy in critically ill patients with influenza virus infection. J. Infect. Dis. 2019 doi: 10.1093/infdis/jiz656. [DOI] [PubMed] [Google Scholar]
- Wang C. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Jama; 2020. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.R., Leibowitz J.L. Advances in Virus Research. Elsevier; 2011. Coronavirus pathogenesis; pp. 85–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Zheng M. 2020. Single-cell RNA Expression Profiling Shows that ACE2, the Putative Receptor of Wuhan 2019-nCoV, Has Significant Expression in the Nasal, Mouth, Lung and Colon Tissues, and Tends to Be Co-expressed with HLA-DRB1 in the Four Tissues. [Google Scholar]
- Xia S. A pan-coronavirus fusion inhibitor _targeting the HR1 domain of human coronavirus spike. Science advances. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. Radiology; 2020. Chest CT for Typical 2019-nCoV Pneumonia: Relationship to Negative RT-PCR Testing; p. 200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23(2):300–302. doi: 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. Potential benefits of precise corticosteroids therapy for severe 2019-nCoV pneumonia. Signal Transduction and _targeted Therapy. 2020;5(1):1–3. doi: 10.1038/s41392-020-0127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. Predicting the receptor-binding domain usage of the coronavirus based on kmer frequency on spike protein. Infect. Genet. Evol.: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2018;61:183–184. doi: 10.1016/j.meegid.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019 doi: 10.1056/NEJMoa2001017. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.