Abstract
Histone tails, representing the N- or C-terminal regions flanking the histone core, play essential roles in chromatin signaling networks. Intrinsic disorder of histone tails and their propensity for post-translational modifications allow them to serve as hubs in coordination of epigenetic processes within the nucleosomal context. Deposition of histone variants with distinct histone tail properties further enriches histone tails’ repertoire in epigenetic signaling. Given the advances in experimental techniques and in silico modelling, we review the most recent data on histone tails’ effects on nucleosome stability and dynamics, their function in regulating chromatin accessibility and folding. Finally, we discuss different molecular mechanisms to understand how histone tails are involved in nucleosome recognition by binding partners and formation of higher-order chromatin structures.
Keywords: histones, nucleosome, histone tail, chromatin
Introduction
Packing of eukaryotic DNA into chromatin engages the basic building blocks called nucleosomes. Nucleosome core particles (NCP) consist of an octamer of four types of histones (H2A, H2B, H3, H4), and ~147 DNA base pairs wrapped around them [1]. The N- and C-terminal intrinsically disordered (IDP) regions flanking histone cores represent histone tails that do not have well determined tertiary structure but rather exist in a dynamic conformational ensemble. In addition, a linker histone H1, comprising a globular domain and disordered N-and C-terminal tails, can bind to nucleosome to form so called chromatosome. The variability in histone tails can be introduced through post-translational modifications (PTM) and deposition of histone variants. The roles of histone tails in the epigenetic regulation have been elucidated through the advances in experimental approaches such as nuclear magnetic resonance (NMR) spectroscopy, cryo-electron microscopy (cryo-EM), fluorescence resonance energy transfer (FRET), chemical crosslinking and mass spectrometry (MS) [2*,3,4**,5,6,7]. A large variety of histone tail PTMs, histone modifying enzymes and reader proteins has been identified and characterized to unravel their roles in various regulatory processes [8,9]. With a rapidly increasing number of histone and nucleosome complex structures deposited in the Protein Data Bank (PDB) [10], atomic details of histone tail mediated interactions have been revealed [11*,12**,13**].
Histone tails participate in various chromatin functions. They moderate nucleosome stability and dynamics, DNA accessibility, nucleosome sliding and repositioning [14,15]. One of the most important roles of histone tails is associated with nucleosomal function to ensure the coordination in time and space of different pathways of epigenetic regulation. Indeed, nucleosomes represent hub points in chromatin signal flow and form a dense set of interactions with other macromolecules [16,17*]. This requires modularity, a crosstalk between signaling components, proofreading capability and redundancy which in turn yields high response sensitivity and robustness. In this review, we summarize the distinct biological and physico-chemical properties of histone tails which allow them to perform these above-mentioned functions in the context of the full nucleosome and chromatin.
Physico-chemical properties of histone tails are essential for chromatin signaling
IDPs are characterized by certain physico-chemical properties which are explored by histone tails. Intrinsic disorder in histone tails may speed up the search for nucleosome _targets and facilitate their interactions with partners. To achieve it, according to the fly-casting mechanism, intrinsically disordered proteins use an increased capture radius for specific binding compared to folded proteins [18]. It has been known for a long time that histone tails affect nucleosome thermodynamic stability and dynamics. Removing tails may induce a more rapid nucleosome unwrapping but the long-lived DNA detachments may lead to structural rearrangements of the H3 tails delaying of DNA re-wrapping [19,20*,21] (Figure 2c). Histone tail dynamics is correlated with the DNA unwrapping although this coupling strongly depends on the salt concentration [7,22]. The energetic barrier in nucleosome unwrapping mainly arises from the electrostatic interactions between DNA and histone core. However, according to the recent study, the unwrapping of DNA from histone core abolishes the tail-DNA interactions and results in an increased conformational entropy of histone tails. Thus, the energetic barrier arising from the electrostatic interactions can be significantly offset by such entropic contribution from histone tails (enthalpy-entropy compensation) [20*]. The same study suggested that histone modifications may modulate the stability of nucleosomes through fine-tuning of histone tails’ entropic contributions to the free energy of nucleosome unwrapping [20*].
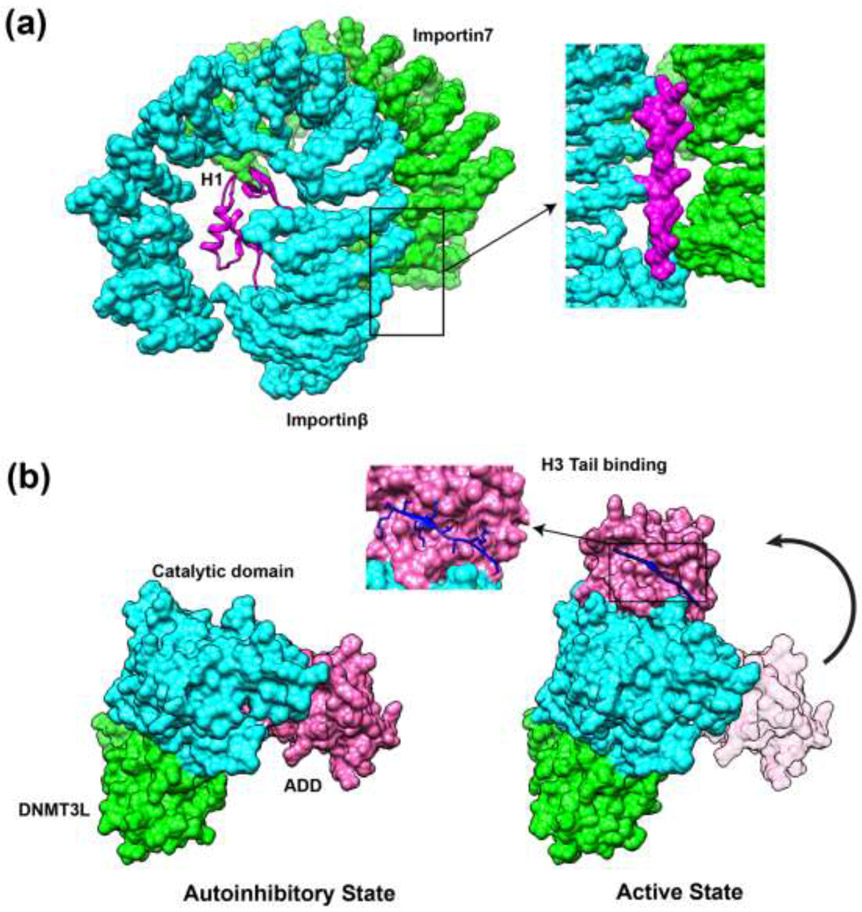
Growing evidence demonstrates the functional importance of so-called fuzzy interactions [23] mediated by histone tails when a high conformational heterogeneity of tails can be maintained in the bound state [12**,13**,24*,25]. It has been suggested that such interactions may provide high on- and off-rates, thereby enabling rapid signaling response in chromatin [24*,25]. Moreover, high conformational flexibility maintained on interaction interfaces minimizes the conformational entropy loss upon tail binding and facilitates the binding of histone modifying enzymes to multiple PTM sites [13**,24*,25]. The following example illustrates how interactions between two disordered regions of chaperone Vps75 and H3 N-terminal tail allow H3 tail to stay in close proximity to the acetyltransferase Rtt109 active site. As a consequence, different H3 acetylation sites, K9, K23 or K27, can be all dynamically exposed to the catalytic pocket with the minimal entropic penalty [13**]. Such mechanism can, in turn, promote the cooperativity between nearby epigenetic marks of different types. Finally, disordered histone tails may promote the formation of complexes, as was observed in the case of H1 C-terminal tail which stays disordered, bridges together and stabilizes the complex between importin7 and importinβ [12**] (Figure 1a).
Figure 1.
Intrinsically disordered histone tails perform their functions through different molecular mechanisms. (a) Fuzzy interactions of H1 tails stabilize the importin7-importinβ complex in histone transport (PDB: 6N88). H1 C-terminal domain can bind to acidic residues located near the importin7-importinβ binding interfaces through transient and non-specific electrostatic interactions. (b) Binding of H3 tails to ADD domain of DNMT3A allosterically induces the transition of DNMT3A from autoinhibitory state to the active state (PDB: 4UTP, 4U7T). Molecular surfaces are rendered in magenta (ADD domain of DNMT3A), cyan (catalytic domain of DNMT3A) and green (DNMT3L).
Disordered histone tails can fold upon binding undergoing so called disorder-order transitions. In such cases, the dynamic intrinsically disordered tails lose entropy, which may result in high specificity recognition even if the binding affinity is rather low. The following roles of disorder-order transitions may be directly connected to histone tails’ functions in chromatin signaling. First, disorder-order transitions may maintain the spatial clustering of residues. Recent simulation study showed that N-terminal tail of H1 was completely unstructured in the free unbound state but folded upon binding to nucleosome [26*]. Such disorder-order transition allowed to preserve clustering of basic residues (K14, K17, K20 and K21) and provided concerted positive charges on one face of the formed alpha helix promoting interactions between H1 N-terminal tail and DNA [26*]. Second, disordered tails may adopt different stable conformations upon binding to different partners. This could confer functional promiscuity enabling tails to bind to a wide range of different chromatin factors. Third, disorder-order transitions may modulate and drive the signal transduction by allosteric coupling. Multiple studies pointed to the allosteric regulation of chromatin factors by histone tails [11*,27,28]. One representative example includes an allosteric activation of de novo methyltransferase DNMT3A by H3 tails (Figure 1b). Interactions between ATRX–DNMT3–DNMT3L (ADD) domain and catalytic domain (CD) of DNMT3A cause autoinhibition of its enzymatic activity. The disordered H3 tails can interact with residues located on the ADD-CD binding interface and form an anti-parallel β sheet with the two β-strands in ADD domain, leading to a conformational change of ADD domain, thus inducing the activation of DNMT3A [11*].
The deposition of reversible PTMs and histone variants enhances the functional diversity of histone tails. The distinct patterns of covalent histone modifications form a “histone code” which offers a dynamic way to mediate the regulatory processes in chromatin on the nucleosomal scaffold. IDPs are usually enriched with modification sites and histone tails represent extreme cases with the very high density of PTMs (methylation, acetylation, ubiquitination, phosphorylation, ADP-ribosylation, crotonylation and succinylation), although recent studies point to the importance of PTMs in histone cores as well [29]. The flexibility of histone tails provides relatively easy access to their sites for chromatin factors, modifications in histone tails are recognized by various reader domains, allowing for recruitment of regulatory proteins to nucleosomes [30]. PTMs can orthosterically disrupt the interaction if the modified residue is directly located on binding interface. At the same time, post-translational modifications in tail residues can play roles of covalent allosteric effectors and modulate binding of histone tails to partners by shifting the IDP’s conformational ensembles and/or equilibrium between ordered and disordered states. For example, it was shown that charge-reducing modifications may trigger the disorder-order transitions of H1 tails in solvent and induce the formation of secondary structures [26*].
Histone tails carry multiple PTMs sites that can be modified simultaneously or in sequential time order. Previously we showed for multiple phosphorylation events that they can expand the repertoire of the recognition patterns, provide more accurate modulation of the strength of the signal and lead to cooperative binding effects [31]. All these mechanisms could be attributed to histone tail signaling. For example, it has been shown that H3S28 phosphorylation leads to the recruitment of the HP1 protein to the H3K9 methylated H3-tail since the negative charge of the phosphate group increases the H3-tail dynamics [32**]. At the same time, the enhanced conformational sampling study of H4 tail showed that progressive sequential acetylation had a cumulative effect, decreased conformational heterogeneity, and increased tail’s helical propensity [33]. Finally, it was demonstrated that H4K16ac worked cooperatively with the other three acetylated lysines on the same tail to disrupt the chromatin folding: while H4K5ac, H4K8ac and H4K12ac showed little effect on folding, the acetylation of all four lysines on the H4 tail disrupted chromatin folding significantly more than H4K16ac alone [34].
Besides the covalent modifications in histone tails, the deposition of diverse histone variants into nucleosomes brings another layer of regulatory mechanism [35]. Histone variants may have shortened or extended N- and C-terminal tails with altered physico-chemical properties compared to the canonical histones and in some cases (like H3.3 and H2A.X) may carry variant-specific PTMs [36,37]. For instance, H1 variant tails vary in sequence lengths and carry different numbers of charged amino acids and certain sequence motifs, like S/TPKK motifs [38,39]. It may fine-tune the disorder-order transitions upon DNA binding leading to different binding affinity values of H1 variants to nucleosome [26*]. Another Micrococcal nuclease digestion experiment showed that the extended C-terminal tail in H2A.W variant interacted with the linker DNA which increased the stability of nucleosome and affected the binding of linker histone H1 to nucleosome [40*]. Several “short H2A variants” including H2A.B, H2A.L, H2A.P and H2A.Q lack the portions of C-terminal tails and deposition of these variants into a nucleosome leads to unwrapping of DNA ends from the histone octamer and an increase in DNA accessibility [41,42].
Histone tail-DNA interactions modulate nucleosome recognition by chromatin factors
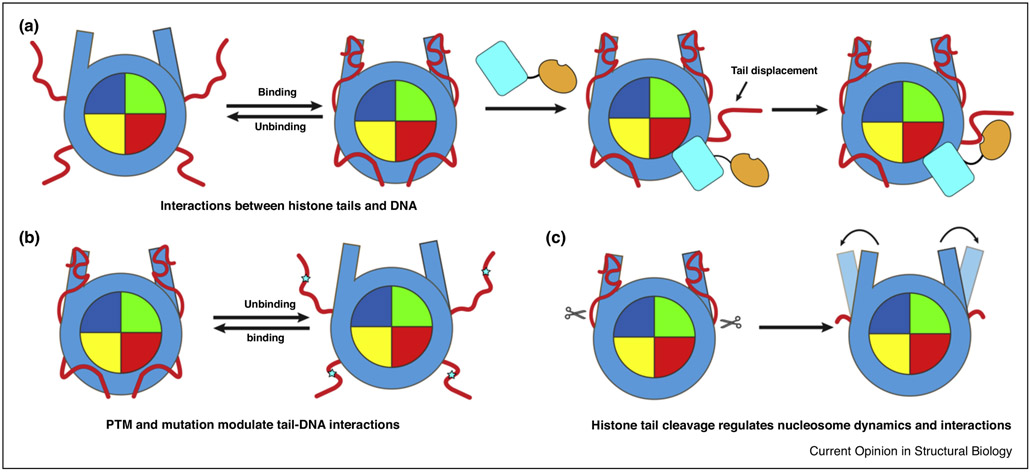
In eukaryotic cells the genetic material is tightly packed into the nucleus but at the same time, in many cases it should remain dynamically accessible during transcription and replication processes. To overcome such challenge, eukaryotic cells have different means to regulate chromatin and DNA accessibility, and intrinsic disorder of histone tails may provide a basis for such dynamic regulation. Histone tails have long been shown to have high conformational flexibility and solvent accessibility, giving an impression that they extend into the solvent and are fully accessible to binding partners. However, a growing pile of evidence points to the extensive and transient interactions of histone tails with nucleosomal and linker DNA [4**,5,7] (Figure 2a). Recent NMR studies, for example, have shown that interactions of H3 tails with DNA lead to the decreased nucleosomal and linker DNA solvent accessibility [2*,4*,15]. Moreover, the removal of histone tails causes an enhanced binding of many chromatin factors to nucleosome, most likely resulting from the increased solvent accessibility of nucleosomal and linker DNA [17*]. These findings altogether point to the fact that tail-DNA interactions within and between nucleosomes regulate the amount of surface area on histone tails and nucleosomal or linker DNA accessible for binding to other partners. The tail-DNA interactions have been suggested to compete with binding of chromatin factors to histone tails and/or to nucleosomal and linker DNA [4**,15,17*].
Figure 2.
Mechanisms of regulation of nucleosome dynamics and interactions through histone tails. (a) Extensive and transient tail-DNA interactions in nucleosome and tail displacement mechanism of nucleosome recognition. (b) PTMs and mutations in histone tails modulate tail-DNA interactions and accessibility of DNA and tails in nucleosome. (c) Histone tail cleavage regulates nucleosome dynamics and interactions.
Recently a tail displacement model has been proposed according to which the association of one protein or domain with the nucleosomal or linker DNA can potentially displace histone tails from DNA and increase tails’ accessibilities to other binding partners [2*,8] (Figure 2a). One recent example supporting this view includes the nucleosome recognition by LSD1-CoREST complex, where the interaction between the SANT2 domain and nucleosomal DNA displaces the H3 tails from DNA and facilitates tails’ interactions with the LSD1 active site [2*]. Moreover, PTMs and mutations can directly modulate tail-DNA interactions in the context of nucleosome [5,43] (Figure 2b). Charge-changing modifications, for instance, can affect the binding of histone tails to DNA by perturbating their electrostatic interactions. In concordance with this, recent NMR studies demonstrate that phosphorylation and acetylation in H3 tail can weaken the tail-DNA interactions, enhance the tail dynamics and solvent accessibility thereby facilitating the binding of reader proteins to histone tails [4**,5].
Chromatin signaling by histone tail cleavage
Besides the reversible covalent histone modifications, the structure and dynamics of chromatin can also be regulated by irreversible proteolytic processing of histone tails [44,45] Due to the critical roles of histone tails in chromatin function summarized above, not surprisingly that histone tail cleavage has profound effects on nucleosome dynamics and interactions. It is expected that compared to covalent modifications, clipping of histone tails should lead to more drastic and long lasting changes to chromatin and plays essential roles in gene expression, cell differentiation, aging and cancer development [44,45]. As shown in the previous section, histone tails may interact with the nucleosomal and linker DNA, therefore proteolytic cleavage of histone tails may enhance the DNA accessibility to nucleosome-binding proteins. Moreover, histone tail cleavage may result in irreversible deletion of critical PTM sites and disrupt the inter-nucleosomal interactions affecting chromatin compaction [3,44].
Histone tail cleavage is performed by histone proteases which clip histone tails at particular sites [44,45]. Although histone tail cleavage occurs among all core histones, clipping of H3 tail has been mostly investigated, and a comprehensive list of cleavage sites and their functional relevance has been compiled. For instance, matrix metalloproteinase 9 (MMP-9) primarily cleaves H3K18-Q19 sites in histone H3 N-terminal tail during the osteoclast differentiation [46] and Jumonji-C (JmjC) domain-containing protein 5 (JMJD5) clips the H3 N-tail at monomethyl-lysine (Kme1) sites in the DNA damage response pathways [47].
Roles of histone tails in the formation of higher order chromatin structures
Histone tails moderate the inter-nucleosomal interactions and induce chromatin compaction as they contain a myriad of basic residues which contribute to the attractive electrostatic interactions between neighboring nucleosomes [48]. Without histone tails nucleosomes cannot self-associate and compact into higher order chromatin structures due to the increased DNA-DNA electrostatic repulsion [49,50**]. It has been revealed that H3, H4 and H1 tails mostly contribute to the cooperative chromatin folding process. Recent cryo-EM structures of nucleosome core particle, biochemical and small-angle X-ray diffraction experiments suggested that the H3 tail participated in the initial contact with the neighboring nucleosomal DNA and made essential contacts between two nucleosomes [51,52]. As for the H4 tails, chemical cross-linking experiments showed that H4 tails from an adjacent nucleosome formed the inter-nucleosomal interactions by docking onto a negatively acidic patch exposed by the core H2A/H2B dimers [53]. Further evidence was provided by another cryo-EM experiments showing the involvement of the H4 and H1 tails in inter-nucleosomal interactions [54,55]. It was, for example, identified that the C-terminal tail of H1.2 was essential for interacting with H3K27me3 to trigger the chromatin compaction and gene silencing in cancer cells [55].
The roles of histone tails in chromatin folding are further aggrandized by PTMs, which can create binding sites for chromatin-associated proteins or directly modulate chromatin fiber dynamics. Of all known histone tail modifications, lysine acetylation is the most widely studied modification in relation to chromatin compaction, whose direct physico-chemical role is believed to neutralize the basic charge of lysine residue. Lysine acetylation results in a reduction of the electrostatic attraction between tails and the negatively charged DNA and thus directly impacts the nucleosome stability and chromatin folding [34]. For example, it has been observed that the acetylation of H4K16 has a negative effect on nucleosome array compaction in vitro and is associated with transcriptionally active chromatin [56]. A subsequent study demonstrated that H4K16ac lead toa more open chromatin in vivo [57]. Studies of H4 tail using molecular dynamics simulations have suggested that the H4K16ac impairs and weakens the H4 tail-acidic patch binding and reduces the inter-nucleosome interaction [3]. However, it should be noted that the outcome of histone tail acetylation depends on histone type and the locations of modified sites, and a recent study using reconstituted nucleosome arrays found that acetylation of H3K18 and H3K27, unlike H4K16ac, had no impact on higher-order chromatin structure [58*].
Conclusions and outlook
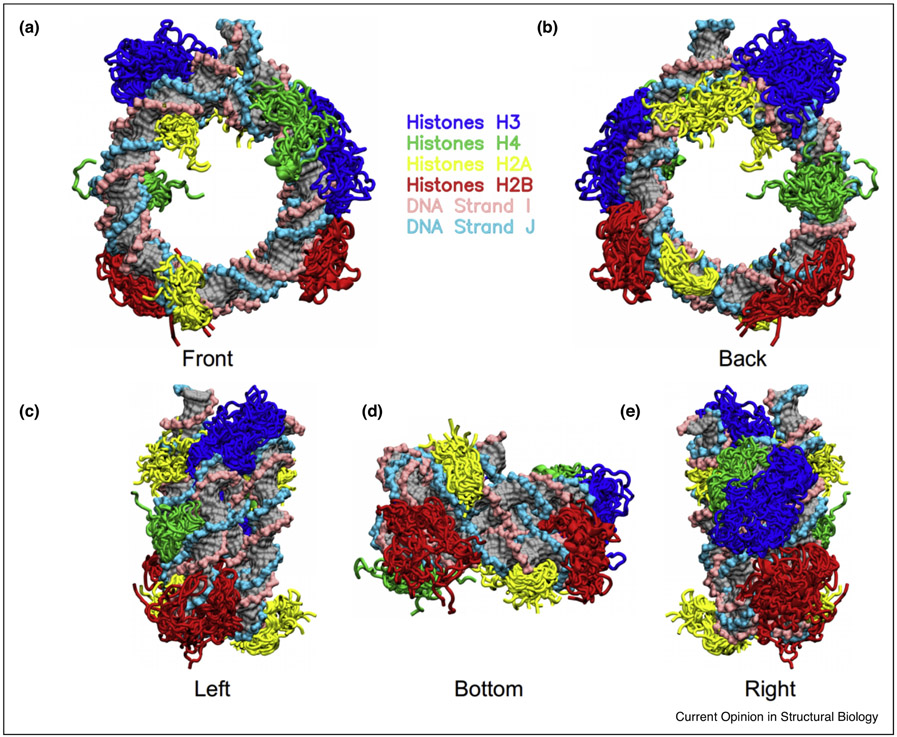
Disordered histone tails are essential components in epigenetic regulatory networks. PTMs along with histone variants dramatically enhance the functional diversity of disordered histone tails and enable them to modulate a variety of signaling processes through different molecular mechanisms. The high flexibility of histone tails is necessary for their signaling functions but poses major difficulties in characterizing their dynamics and interactions. Recent developments in experimental approaches along with the increased computational power and advances in in silico simulations using improved water models and forcefields for IDP and DNA have helped to elucidate the mechanisms of histone tails in mediating intra-/inter-nucleosome interactions (Figure 3). Notably, many hybrid methods that integrate experimental data with the molecular modelling have deliverednovel insights into how histone tails modulate nucleosome recognition by partners. However, the functional importance of histone tails merits more extensive studies focusing on their kinetics and dynamics in providing chromatin signaling with high spatio-temporal precision.
Figure 3.
Representative binding modes between disordered histone tails and nucleosomal DNA observed in molecular dynamics simulations. The cartoon representations of histone core domains are hidden for clarifications.
Highlights.
Intrinsic disorder empowers histone tails’ diverse roles in chromatin signaling.
Introducing PTMs and histone variants dramatically enhances the functional diversity of histone tails.
Interactions of histone tails with nucleosomal and linker DNA modulate the nucleosome recognition by binding partners.
Histone tail cleavage regulates chromatin structure and function.
Histone tails play critical roles in the formation of higher-order chromatin structure.
Acknowledgements
YP and DL were supported by the Intramural Research Program of the National Library of Medicine at the U.S. National Institutes of Health. SL and ARP were supported by the Department of Pathology and Molecular Medicine, Queen’s University, Canada. ARP is the recipient of a Senior Canada Research Chair in Computational Biology and Biophysics and a Senior Investigator award from the Ontario Institute of Cancer Research, Canada.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of specific interest
** of outstanding interest
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ: Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2.Pilotto S, Speranzini V, Tortorici M, Durand D, Fish A, Valente S, Forneris F, Mai A, Sixma TK, Vachette P, et al. : Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation. Proc Natl Acad Sci U S A 2015, 112:2752–2757.*Authors propose a tail displacement model for LSD1-mediated H3 demethylation where the interaction between the SANT2 domain and nucleosomal DNA displaces the H3 tails from DNA and make them avaibable for capture by the LSD1 active site.
- 3.Zhang R, Erler J, Langowski J: Histone Acetylation Regulates Chromatin Accessibility: Role of H4K16 in Inter-nucleosome Interaction. Biophys J 2017, 112:450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J, Musselman CA: The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife 2018, 7.**A study demonstrates that H3 tails extensively interact with nucleosome DNA which inhibits the binding activity of PHD finger to histone tail. It further shows that histone modifications and mutations weaken the tail-DNA interactions and increase the accessibiliy of histone tail.
- 5.Stutzer A, Liokatis S, Kiesel A, Schwarzer D, Sprangers R, Soding J, Selenko P, Fischle W: Modulations of DNA Contacts by Linker Histones and Post-translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Mol Cell 2016, 61:247–259. [DOI] [PubMed] [Google Scholar]
- 6.Kirsch R, Jensen ON, Schwammle V: Visualization of the dynamics of histone modifications and their crosstalk using PTM-CrossTalkMapper. Methods 2020, 10.1016/j.ymeth.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Shaytan AK, Armeev GA, Goncearenco A, Zhurkin VB, Landsman D, Panchenko AR: Coupling between Histone Conformations and DNA Geometry in Nucleosomes on a Microsecond Timescale: Atomistic Insights into Nucleosome Functions. J Mol Biol 2016, 428:221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver TM, Morrison EA, Musselman CA: Reading More than Histones: The Prevalence of Nucleic Acid Binding among Reader Domains. Molecules 2018, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S, Yue Y, Li Y, Li H: Identification and characterization of 'readers' for novel histone modifications. Curr Opin Chem Biol 2019, 51:57–65. [DOI] [PubMed] [Google Scholar]
- 10.ww PDBc: Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res 2019, 47:D520–D528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, et al. : Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 2015, 517:640–644.*A study reports the crystal structures of de novo DNA methyltransferases DNMT3A in autoinhibitory and active states. It shows that binding of H3 tails to the ADD domain induces a conformational change and allosterically activates DNMT3A.
- 12.Ivic N, Potocnjak M, Solis-Mezarino V, Herzog F, Bilokapic S, Halic M: Fuzzy Interactions Form and Shape the Histone Transport Complex. Mol Cell 2019, 73:1191–1203 e1196.**A study reports a cryo-EM structure of the importin7 and importinβ in complex with H1. It shows that H1 C-terminal domain stabilizes the importin7-importinβ complex through transient and non-specific electrostatic interactions.
- 13.Danilenko N, Lercher L, Kirkpatrick J, Gabel F, Codutti L, Carlomagno T: Histone chaperone exploits intrinsic disorder to switch acetylation specificity. Nat Commun 2019. 10:3435.**This paper reports a solution structural model of acetyltransferase Rtt109 in complex with histone chaperones Asf1 and Vps75 and the histone dimer H3-H4. It shows that Asf1 and Vps75 exert the fuzzy electrostatic interactions between H3 tail and the disordered C-terminal domain of Vps75 to guide lysine residues of H3 tail into the active pocket and to faciliate the acetylation.
- 14.Chakraborty K, Kang M, Loverde SM: Molecular Mechanism for the Role of the H2A and H2B Histone Tails in Nucleosome Repositioning. J Phys Chem B 2018, 122:11827–11840. [DOI] [PubMed] [Google Scholar]
- 15.Gatchalian J, Wang X, Ikebe J, Cox KL, Tencer AH, Zhang Y, Burge NL, Di L, Gibson MD, Musselman CA, et al. : Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun 2017, 8:1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kale S, Goncearenco A, Markov Y, Landsman D, Panchenko AR: Molecular recognition of nucleosomes by binding partners. Curr Opin Struct Biol 2019, 56:164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skrajna A, Goldfarb D, Kedziora KM, Cousins EM, Grant GD, Spangler CJ, Barbour EH, Yan X, Hathaway NA, Brown NG, et al. : Comprehensive nucleosome interactome screen establishes fundamental principles of nucleosome binding. Nucleic Acids Res 2020, 10.1093/nar/gkaa544.*A large-scale analysis of nucleosome interactions with binding partners in mouse. The study characterizes binding hotspots on nucleosome interfaces and reveals that removal of histone tails can enhance binding of many chromatin factors to nucleosomes.
- 18.Shoemaker BA, Portman JJ, Wolynes PG: Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci U S A 2000, 97:8868–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erler J, Zhang R, Petridis L, Cheng X, Smith JC, Langowski J: The role of histone tails in the nucleosome: a computational study. Biophys J 2014, 107:2911–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons T, Zhang B: Critical role of histone tail entropy in nucleosome unwinding. J Chem Phys 2019, 150:185103.*A simulation study elucidates the contribution of histone tail entropy to nucleosome unwrapping. It demonstrates an enthalpy-entropy compensation in nucleosome unwrapping where the energetic barrier arisen from electrostatic interactions is significantly offset by an entropic contribution of histone tails.
- 21.Chakraborty K, Loverde SM: Asymmetric breathing motions of nucleosomal DNA and the role of histone tails. J Chem Phys 2017, 147:065101. [DOI] [PubMed] [Google Scholar]
- 22.Kenzaki H, Takada S: Partial Unwrapping and Histone Tail Dynamics in Nucleosome Revealed by Coarse-Grained Molecular Simulations. PLoS Comput Biol 2015, 11:e1004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuxreiter M: Fuzziness in Protein Interactions-A Historical Perspective. J Mol Biol 2018, 430:2278–2287. [DOI] [PubMed] [Google Scholar]
- 24.Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, et al. : Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555:61–66.*The authors show that H1 tails retain high binding affinity with nuclear chaperone, but fully maintain their structural disorder in the bound state. It further deomonstrates the abundance of fuzzy interactions in eukaryotes.
- 25.Langini C, Caflisch A, Vitalis A: The ATAD2 bromodomain binds different acetylation marks on the histone H4 in similar fuzzy complexes. J Biol Chem 2017, 292:16734–16745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sridhar A, Orozco M, Collepardo-Guevara R: Protein disorder-to-order transition enhances the nucleosome-binding affinity of H1. Nucleic Acids Res 2020, 48:5318–5331.*A simulation study which shows that the disorder-order transition of H1 N-terminal tail enhances its binding affinity with the nucleosomal DNA. It further shows that helical propensity of H1 N-terminal tail depends on the H1 variant types and may correlate with the differential binding affinity of H1 variants to nucleosome.
- 27.Hwang WL, Deindl S, Harada BT, Zhuang X: Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA. Nature 2014, 512:213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longbotham JE, Chio CM, Dharmarajan V, Trnka MJ, Torres IO, Goswami D, Ruiz K, Burlingame AL, Griffin PR, Fujimori DG: Histone H3 binding to the PHD1 domain of histone demethylase KDM5A enables active site remodeling. Nat Commun 2019, 10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenley AT, Anandakrishnan R, Kidane YH, Onufriev AV: Modulation of nucleosomal DNA accessibility via charge-altering post-translational modifications in histone core. Epigenetics Chromatin 2018, 11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su Z, Denu JM: Reading the Combinatorial Histone Language. ACS Chem Biol 2016, 11:564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishi H, Demir E, Panchenko AR: Crosstalk between signaling pathways provided by single and multiple protein phosphorylation sites. J Mol Biol 2015, 427:511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aparicio Pelaz D, Yerkesh Z, Kirchgäßner S, Mahler H, Kharchenko V, Azhibek D, Jaremko M, Mootz HD, Jaremko Ł, Schwarzer D, et al. : Examining histone modification crosstalk using immobilized libraries established from ligation-ready nucleosomes. Chemical Science 2020, 11:9218–9225.** A study performs comprehensive analysis of PTMs’ effects on the recruitment of chromatin factor, HP1 to nucleosmes. It shows that H3S28 phosphorylation can enhance the binding of HP1 to H3 tail by increasing the tail dynamics.
- 33.Winogradoff D, Echeverria I, Potoyan DA, Papoian GA: The Acetylation Landscape of the H4 Histone Tail: Disentangling the Interplay between the Specific and Cumulative Effects. J Am Chem Soc 2015, 137:6245–6253. [DOI] [PubMed] [Google Scholar]
- 34.Mishra LN, Pepenella S, Rogge R, Hansen JC, Hayes JJ: Acetylation Mimics Within a Single Nucleosome Alter Local DNA Accessibility In Compacted Nucleosome Arrays. Scientific Reports 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Draizen EJ, Shaytan AK, Marino-Ramirez L, Talbert PB, Landsman D, Panchenko AR: HistoneDB 2.0: a histone database with variants—an integrated resource to explore histones and their variants. Database (Oxford) 2016, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang FT, Chan FL, JD RM, Udugama M, Mayne L, Collas P, Mann JR, Wong LH: CHK1-driven histone H3.3 serine 31 phosphorylation is important for chromatin maintenance and cell survival in human ALT cancer cells. Nucleic Acids Res 2015, 43:2603–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ismail IH, Hendzel MJ: The gamma-H2A.X: is it just a surrogate marker of double-strand breaks or much more? Environ Mol Mutagen 2008, 49:73–82. [DOI] [PubMed] [Google Scholar]
- 38.Churchill ME, Suzuki M: ‘SPKK’ motifs prefer to bind to DNA at A/T-rich sites. The EMBO Journal 1989, 8:4189–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woods DC, Wereszczynski J: Elucidating the influence of linker histone variants on chromatosome dynamics and energetics. Nucleic Acids Res 2020, 48:3591–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osakabe A, Lorkovic ZJ, Kobayashi W, Tachiwana H, Yelagandula R, Kurumizaka H, Berger F: Histone H2A variants confer specific properties to nucleosomes and impact on chromatin accessibility. Nucleic Acids Res 2018, 46:7675–7685.*This study demonstrates that H2A variant deposition affects the nucleosome stability and play essential roles in regulation of chromatin accessibility in plants. It futher characterizes the interactions of C-terminal tails of H2A variants with the linker DNA.
- 41.Jiang X, Soboleva TA, Tremethick DJ: Short Histone H2A Variants: Small in Stature but not in Function. Cells 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martire S, Banaszynski LA: The roles of histone variants in fine-tuning chromatin organization and function. Nat Rev Mol Cell Biol 2020, 21:522–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakamori M, Fujii Y, Suka N, Shirouzu M, Sakamoto K, Umehara T, Yokoyama S: Intra- and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci Rep 2015, 5:17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi SJ, Kim K: Histone tail cleavage as a novel epigenetic regulatory mechanism for gene expression. BMB Rep 2018, 51:211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azad GK, Swagatika S, Kumawat M, Kumawat R, Tomar RS: Modifying Chromatin by Histone Tail Clipping. J Mol Biol 2018, 430:3051–3067. [DOI] [PubMed] [Google Scholar]
- 46.Kim K, Punj V, Kim JM, Lee S, Ulmer TS, Lu W, Rice JC, An W: MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev 2016, 30:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen J, Xiang X, Chen L, Wang H, Wu L, Sun Y, Ma L, Gu X, Liu H, Wang L, et al. : JMJD5 cleaves monomethylated histone H3 N-tail under DNA damaging stress. EMBO Rep 2017, 18:2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allahverdi A, Chen QM, Korolev N, Nordenskiold L: Chromatin compaction under mixed salt conditions: Opposite effects of sodium and potassium ions on nucleosome array folding. Scientific Reports 2015, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Kono H: Distinct Roles of Histone H3 and H2A Tails in Nucleosome Stability. Sci Rep 2016, 6:31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwasaki W, Miya Y, Horikoshi N, Osakabe A, Taguchi H, Tachiwana H, Shibata T, Kagawa W, Kurumizaka H: Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio 2013, 3:363–369.**Using x-ray crystallography, structures of four mutant nucleosomes lacking one N-terminal tail (H2A, H2B, H3, or H4) were determined. The authors found that the deletion of the N-terminal tail of H2B or H3 had negative effects on the histone–DNA interactions and substantially decreased nucleosome stability.
- 51.Bilokapic S, Strauss M, Halic M: Cryo-EM of nucleosome core particle interactions in trans. Scientific Reports 2018, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berezhnoy NV, Liu Y, Allahverdi A, Yang RL, Su CJ, Liu CF, Korolev N, Nordenskiold L: The Influence of Ionic Environment and Histone Tails on Columnar Order of Nucleosome Core Particles. Biophysical Journal 2016, 110:1720–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen QM, Yang RL, Korolev N, Liu CF, Nordenskiold L: Regulation of Nucleosome Stacking and Chromatin Compaction by the Histone H4 N-Terminal Tail-H2A Acidic Patch Interaction. Journal of Molecular Biology 2017, 429:2075–2092. [DOI] [PubMed] [Google Scholar]
- 54.Song F, Chen P, Sun DP, Wang MZ, Dong LP, Liang D, Xu RM, Zhu P, Li GH: Cryo-EM Study of the Chromatin Fiber Reveals a Double Helix Twisted by Tetranucleosomal Units. Science 2014, 344:376–380. [DOI] [PubMed] [Google Scholar]
- 55.Kim JM, Kim K, Punj V, Liang G, Ulmer TS, Lu W, An W: Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci Rep 2015, 5:16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL: Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 2006, 311:844–847. [DOI] [PubMed] [Google Scholar]
- 57.Henikoff S, Smith MM: Histone Variants and Epigenetics. Cold Spring Harbor Perspectives in Biology 2015, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Banerjee DR, Deckard CE, Zeng Y, Sczepanski JT: Acetylation of the histone H3 tail domain regulates base excision repair on higher-order chromatin structures. Scientific Reports 2019, 9.*The acetylation of histone H3 tails was analyzed by reconstituting of the 12-mer nucleosome arrays consisting of homogeneously acetylated histone H3 (H3K18 and H3K27). It was found that H3K18ac and H3K27ac differentially influenced activities of uracil DNA glycosylase and apurinic/apyrimidinic endonuclease in chromatin. However, the simultaneous acetylation of H3K18 and H3K27 did not influence the higher-order chromatin folding.