Abstract
Aim of the study
Intrahepatic covalently closed circular DNA (cccDNA) is the main cause of hepatitis B virus (HBV) persistence. Therefore, a noninvasive serum biomarker that can reflect intrahepatic cccDNA is required for evaluation of HBV virological, biochemical activity and therapeutic response. Aim of the study was to assess serum hepatitis B pregenomic RNA in low viremia patients (HBV DNA < 2000 IU/ml) and high viremia (HBV DNA > 2000 IU/ml).
Material and methods
This study was carried out on two groups of chronic hepatitis B patients: group A – 40 patients with low viremia (HBV DNA < 2000 IU/ml); group B – 40 patients with high viremia (HBV DNA > 2000 IU/ml when diagnosed). They were assessed before treatment and after 6 months of treatment (entecavir 0.5 mg/24 h). Serum HBV pregenomic RNA was quantified using RT-PCR.
Results
Pregenomic RNA (pgRNA) was significantly lower in group A than in group B (before treatment). Moreover, it was significantly lower after 6 months of treatment than before treatment in group B. A significant positive correlation was observed between pgRNA and HBV DNA in groups A and B (before treatment); however, after 6 months of treatment of group B patients, although 35 patients had undetectable HBV DNA, they showed detectable levels of serum pgRNA and pgRNA > 4000 IU/ml was associated with virological and biochemical activity.
Conclusions
Serum HBV pregenomic RNA might be a promising marker for assessment of HBV virological, biochemical activity and evaluating therapeutic responses.
Keywords: entecavir, HBV pregenomic RNA, noninvasive, biomarker, HBV cccDNA
Introduction
Chronic hepatitis B infection is an important health problem worldwide; deaths from chronic hepatitis B related cirrhosis and hepatocellular carcinoma were 310,000 and 340,000 annually respectively [1].
Hepatitis B virus (HBV) is a DNA hepatotropic virus which belongs to the Hepadnaviridae family [2]. HBV is known as a DNA virus with reverse transcription during its replication. HBV replicates by reverse transcription of an RNA intermediate known as pregenomic RNA (pgRNA). Pregenomic RNA is a unique template between parental and new HBV DNA. The pgRNA has multiple roles during replication. Serum HBV RNA level is an important early predictor of chronic hepatitis B reactivation after stopping the antiviral therapy.
Once the liver cell is infected, the nucleocapsid is released into the cytoplasm and the relaxed circular DNA (rcDNA) is transported to the nucleus where it is converted into covalently closed circular DNA (cccDNA). After transcription of the viral RNAs, pgRNA is reverse transcribed by HBV polymerase. Through Golgi and endoplasmic reticulum (ER) the core particles acquire an envelope and are released. The cccDNA pool can be amplified through viral entry and retransport of the newly produced HBV DNA into the nucleus [3].
cccDNA uses the cellular transcriptional tools to produce all viral RNAs including HBV messenger RNAs (mRNAs) and pgRNAs that are important for protein production and viral replication [3].
pgRNA is required for synthesis of the HBV rcDNA found in the mature virus. Within the cytoplasm, HBV capsid protein (Core) assembles around HBV polymerase (Pol) and a single pgRNA copy to form the nascent virion. Within the capsid, HBV DNA is reverse transcribed by Pol (with concomitant pgRNA degradation) [4].
HBV infection cannot be totally cured due to presence of cccDNA in the infected hepatocytes [5]. Liver biopsy shows an invasive nature, so it is impractical to quantify intrahepatic cccDNA. Recently, serum HBV pregenomic RNA is known as a biomarker for estimation of intrahepatic cccDNA [6].
The aim of this work was to assess serum hepatitis B pregenomic RNA in patients with: low viremia (HBV DNA less than 2000 IU/ml) and high viremia (HBV DNA more than 2000 IU/ml).
Material and methods
Materials
Serum samples were taken from 80 patients attending the Tropical Medicine clinic and diagnosed with chronic hepatitis B by having the HBsAg (hepatitis B surface antigen) positive test for more than 6 months. Informed consent was obtained from all patients. None of the patients suffered from other causes of chronic hepatitis such as chronic hepatitis C virus (HCV), steatohepatitis, metabolic liver diseases or autoimmune liver disease. Patients in this study were assigned to two groups as follows: group A: 40 patients with low viremia (HBV DNA less than 2000 IU/ml) (not fulfilling the criteria of treatment according to EASL guidelines) [7] and group B: 40 patients with high viremia (HBV DNA more than 2000 IU/ml at time of diagnosis) (fulfilling the criteria of treatment according to EASL Guidelines) [7]. Group B patients were assessed before treatment and after 6 months of receiving entecavir (0.5 mg/24 h). Serum samples were properly kept in aliquots at –80°C to ensure stability of viral RNA.
All patients in this study were subjected to complete history taking and detailed clinical examination. Also routine laboratory investigations and liver function tests [complete blood count (CBC), prothrombin time (PT), international normalized ratio (INR), serum creatinine, urea, total bilirubin, direct bilirubin, serum albumin, alanine aminotransferase (ALT), aspartate aminotransferase (AST), ALP] and hepatitis B specific investigations (HBsAg, HBeAg, anti-HBc IgG, HBV DNA PCR) were performed. Noninvasive liver fibrosis activity markers [8] – FIB-4 score (age × AST/platelets × √ALT), AST to Platelet Ratio Index (APRI) [(AST/(ULN) AST × 100]/platelet count – were measured and calculated at the time of diagnosis in patients of group A, before the beginning of treatment and after 6 months of entecavir treatment in patients of group B. Ultrasound of the abdomen and pelvis was done to exclude cirrhosis and portal hypertension (not the only measure used to exclude cirrhosis but also noninvasive liver fibrosis activity markers were used).
Measurement of serum HBV DNA level
Serum HBV DNA level was quantified by TaqMan 48 automatic fluorescence quantitative polymerase chain reaction (q-PCR) kits using a Roche COBAS AmpliPrep/COBAS TaqMan 48 Analyzer (Roche Diagnostics, Mannheim, Germany); the detection limit was 12 IU/ml [9].
Measurement of serum level of HBV pregenomic RNA
All serum samples of both groups were primarily subjected to RNA extraction using a Qiagen RNA extraction kit QIAamp Viral RNA Mini kit spin column, Thermo Fisher Scientific Inc. Extracted RNA was estimated by nanodrop to ensure availability of sufficient RNA yield [10]. A one-step procedure of reverse transcription and amplification was performed using a HERA RT-qPCR kit (Willowfort, UK). Primers and probes of pgRNA were added to the reaction mixture: Forward primer: GGTCCCCTAGAAGAAGAACTCCCT, reverse primer: CATTGAGATTCCCGAGATTGAGAT, and probe sequence: TCTCAATCGCCGCGTCGCAGA. 18S RNA was used as the reference for normalization of RNA levels (QuantumRNA 18S Invitrogen Internal Standards, Thermo Fisher). Amplification was performed in the Step One Real-Time PCR Thermo Fisher Scientific thermal cycler apparatus using the following protocol: 15 min at 50-55°C of reverse transcription followed by 5 min at 95°C for enzyme activation and then 40 cycles of denaturation at 95°C for 10 s and annealing/extension for 30 s at 60oC.
Statistical analysis
Computerized analysis of data was done using IBM SPSS software package version 20.0 (Armonk, NY: IBM Corp). The Kolmogorov-Smirnov test was used to verify variables’ normality of distribution. Comparisons of both groups for categorical variables were made using the χ2 test. McNemar’s test was used to analyze the significance between the different stages. Student’s t-test was used to compare the groups for quantitative variables that were normally distributed. The Mann-Whitney test was used to compare quantitative variables between two groups that were not normally distributed. The paired t-test was assessed to compare different periods for normally distributed quantitative variables. The Wilcoxon signed ranks test was assessed to compare different periods for quantitative variables that were abnormally distributed. Spearman’s coefficient was used to correlate between quantitative variables. A receiver operating characteristic curve (ROC) was developed by plotting sensitivity (TP) on the Y axis versus 1-specificity (FP) on the X axis at different cut-off values. The area under the ROC curve denotes the diagnostic performance of the test. Area more than 50% gives acceptable performance and area about 100% is the best performance for the test. The ROC curve also allows a comparison of performance between two tests. Sensitivity – the capability of the test to correctly identify individuals who are diseased in a population – “true positives”. The more the sensitivity, the smaller is the number of unidentified cases – “false negatives” Specificity – the capability of the test to correctly exclude individuals who are free of the disease – “true negatives”. The more the specificity, the fewer “false positives” will be included. Positive predictive value (PPV) – the probability of the presence of the disease, among those with positive diagnostic test results. Negative predictive value (NPV) – the probability of absence of the disease, among those whose diagnostic test results were negative. Accuracy rate of agreement = (true positives + true negatives)/total tested × 100. Significance of the obtained results was judged at the 5% level.
Ethical approval
All patients who participated in the present study signed an informed consent form. The study protocol was approved by the ethics committee of the Faculty of Medicine, Alexandria University, Egypt and is in agreement with the Helsinki Declaration of 1975.
Results
Basic features of the investigation participants
All cases of both groups were HBeAg –ve, anti HBc IgG +ve, HBsAg +ve.
The age range in group A patients was 18-43 years, with a mean of 24.8 ±4.5 years; 60% of them were male (24 patients) and 40% of them were female (16 patients).
The age range in group B patients was 20-44 years with a mean of 32.7 ±5.2 years; 62.5% of them were male (25 patients) and 37.5% of them were females (15 patients).
ALT and AST values were significantly higher in group B than in group A (p < 0.001). There was no statistically significant difference between the groups regarding the values of CBC, alkaline phosphatase, serum albumin, PT%, INR, total, direct bilirubin, serum urea and creatinine (Table 1).
Table 1.
Comparison between the two studied groups according to the different biochemical parameters (CBC, ALT, AST, ALP, serum albumin, PT%, INR, total, direct bilirubin, urea and serum creatinine)
| Parameter | Group A n = 40 | Group B before treatment n = 40 | t-test p-value | |
|---|---|---|---|---|
| HGB (gm/dl) | ||||
| Range | 12-17.5 | 11-17.5 | 0.321 | |
| Mean ±SD | 14 ±1.54 | 13.99 ±1.67 | 0.50 NS | |
| RBCs (106/mm3) | ||||
| Range | 3.7-5.5 | 3.1-5.9 | 0.984 | |
| Mean ±SD | 4.62 ±0.44 | 4.46 ±0.75 | 0.127 NS | |
| Platelets (103/mm3) | ||||
| Range | 146-450 | 198-440 | 1.880 | |
| Mean ±SD | 302.7 ±79.6 | 331.2 ±53.8 | 0.064 | |
| WBCs (103/mm3) | ||||
| Range | 3.6-14 | 3.7-12 | 1.89 | |
| Mean ±SD | 7.8 ±3 | 6.9 ±2 | 0.0656 NS | |
| ALT (IU/ml) | ||||
| Range | 13-32 | 88-220 | 18.738* | |
| Mean ±SD | 22.6 ±4.3 | 135.5 ±37.9 | < 0.001* | |
| AST (IU/ml) | ||||
| Range | 14-29 | 69-150 | 22.456* | |
| Mean ±SD | 19.2 ±3.6 | 95 ±21 | < 0.001* | |
| ALP (IU/ml) | ||||
| Range | 67-145 | 38-150 | 1.71 | |
| Mean ±SD | 99.13 ±20.28 | 108.20 ±30.65 | 0.0612 NS | |
| Serum albumin (gm/dl) | ||||
| Range | 3.3-5 | 3.6-4.4 | 0.96 | |
| Mean ±SD | 3.98 ±0.32 | 3.98 ±0.16 | 0.4471 NS | |
| PT% | ||||
| Range | 88-100 | 90-100 | 0.485 | |
| Mean ±SD | 98.35 ±3.17 | 98.35 ±2.18 | 0.500 NS | |
| INR | ||||
| Range | 1.0-1.2 | 1.0-1.0 | 1.98 | |
| Mean ±SD | 1.02 ±0.05 | 1.00 ±0.00 | 0.068 | |
| Direct bilirubin (mg/dl) | ||||
| Range | 0.1-0.3 | 0.1-0.4 | U = 726.0 | |
| Median (IQR) | 0.2 (0.10-0.20) | 0.2 (0.10-0.20) | 0.434 | |
| Total bilirubin (mg/dl) | ||||
| Range | 0.7-1.2 | 0.8-1.2 | 0.111 | |
| Mean ±SD | 1.0 ±0.1 | 1.0 ±0.1 | 0.912 | |
| Urea (mg/dl) | ||||
| Range | 16-40 | 19-42 | 0.1415 NS | |
| Mean ±SD | 31.98 ±9.37 | 34.00 ±7.25 | ||
| Creatinine (mg/dl) | ||||
| Range | 0.3-1.3 | 0.4-1.4 | 0.072 NS | |
| Mean ±SD | 0.7 ±0.25 | 0.86 ±0.29 | ||
t – Student’s t-test, p was significant if ≤ 0.05, significant at level 0.05, NS – not significant
HBV DNA, HBV pregenomic RNA, FIB-4 and APRI in both studied groups at time 0
HBV DNA and HBV pregenomic RNA levels were significantly higher in group B before treatment than in group A (p < 0.001). Moreover, APRI and FIB-4 score were significantly higher in group B before treatment than in group A (p < 0.001) (Table 2).
Table 2.
Comparison between the two studied groups according to different parameters at time 0
| Group A (n = 40) | Group B (n = 40) | U | p | ||
|---|---|---|---|---|---|
| HBV DNA (IU/ml) | |||||
| Min.-Max. | 110-680 | 12,000-70,000 | 0.0* | < 0.001* | |
| Median (IQR) | 255 (160-320) | 26,000 (18,000-40,000) | |||
| Hepatitis B pregenomic RNA (IU/ml) | |||||
| Min.-Max. | 100-4,300 | 4,000-70,000 | 1.0* | < 0.001* | |
| Median (IQR) | 450 (364.5-865) | 35,000 (22,000-44,500) | |||
| FIB-4 | |||||
| Min.-Max. | 0.2-0.6 | 0.4-1.8 | 34.0* | < 0.001* | |
| Median (IQR) | 0.3 (0.3-0.4) | 0.8 (0.6-1.1) | |||
| APRI | |||||
| Min.-Max. | 0.1-0.3 | 0.5-1.0 | 0.0* | < 0.001* | |
| Median (IQR) | 0.2 (0.1-0.2) | 0.6 (0.6-0.8) | |||
U – Mann-Whitney test, p – p value for comparing between the studied groups, *statistically significant at p ≤ 0.05
Comparison between different parameters in group B before treatment and after 6 months of treatment
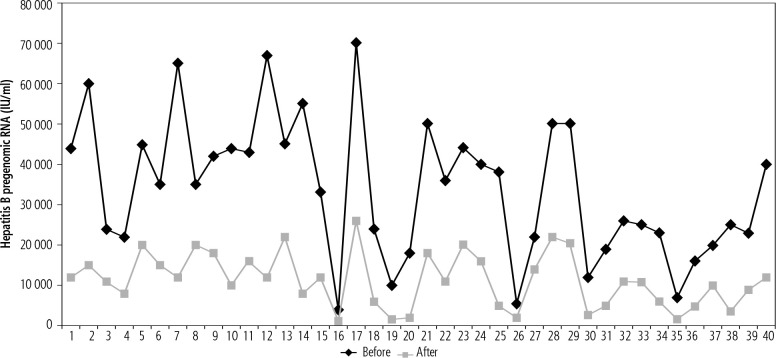
HBV pregenomic RNA values were significantly lower after 6 months of treatment than pretreatment values (p < 0.001) (Fig. 1). Furthermore, ALT, AST, FIB-4 score and APRI were significantly lower after 6 months of treatment than pretreatment values (p < 0.001). As regards HBV DNA, after 6 months of follow-up, 35 patients had undetectable HBV DNA while 5 patients had detectable values (Tables 3 and 4).
Fig. 1.
Comparison between pregenomic RNA in group B before and after 6 months of treatment (n = 40)
Table 3.
Comparison between pregenomic RNA in group B before and after 6 months of treatment (n = 40)
| Hepatitis B pregenomic RNA (IU/ml) | Before treatment | After 6 months of treatment | Z | p |
|---|---|---|---|---|
| Min.-Max. | 4000-70,000 | 1,250-26,000 | 5.512* | < 0.001* |
| Median (IQR) | 35,000 (22,000-44,500) | 11,000 (55,00-16,000) |
Z – Wilcoxon signed ranks test, p – p value for comparing between the studied periods, *statistically significant at p ≤ 0.05
Table 4.
Comparison between the different parameters in group B before and after 6 months of treatment (n = 40)
| Before treatment | After 6 months of treatment | Test of sig. | p | ||
|---|---|---|---|---|---|
| AST (IU/ml) | |||||
| Min.-Max. | 69-150 | 10-25 | t = 23.02* | < 0.001* | |
| Mean ±SD | 95 ±21 | 15.6 ±4 | |||
| ALT (IU/ml) | |||||
| Min.-Max. | 88-220 | 11-30 | t = 19.797* | < 0.001* | |
| Mean ±SD | 135.5 ±37.9 | 19.5 ±6.4 | |||
| HBV DNA (IU/ml) | |||||
| Negative | 0 (0%) | 35 (87.5%) | χ2 = 62.222* | McNp < 0.001* | |
| Positive | 40 (100%) | 5 (12.5%) | |||
| FIB-4 | |||||
| Min.-Max. | 0.4-1.8 | 0.2-1.0 | Z = 5.512* | < 0.001* | |
| Median (IQR) | 0.8 (0.6-1.1) | 0.3 (0.3-0.4) | |||
| APRI | |||||
| Min.-Max. | 0.5-1.0 | 0.1-0.3 | Z = 5.512* | < 0.001* | |
| Median (IQR) | 0.6 (0.6-0.8) | 0.1 (0.1-0.1) | |||
χ2 – chi square test, McN – McNemar test, t – paired t-test, Z – Wilcoxon signed ranks test, p – p value for comparing between the studied periods, *statistically significant at p ≤ 0.05
Relation between HBV DNA and pregenomic RNA in group B after 6 months of treatment (n = 40)
In group B, after 6 months of treatment, 35 patients showed undetectable levels of HBV DNA, with median pgRNA = 11,000 IU/ml, whereas 5 patients showed detectable levels of HBV DNA; the median was 20,000 IU/ml. Although the median of HBV pregenomic RNA was lower in HBV DNA negative patients than that of HBV DNA positive patients, it was statistically insignificant (p = 0.183) (Table 5).
Table 5.
Relation between HBV DNA and pregenomic RNA in group B after 6 months of treatment (n = 40)
| Hepatitis B pregenomic RNA at time 6 (IU/ml) | HBV DNA after 6 months of treatment | U | p | |
|---|---|---|---|---|
| Negative (n = 35) | Positive (n = 5) | |||
| Min.-Max. | 1,250-26,000 | 8,000-22,000 | 54.500 | 0.183 |
| Median (IQR) | 11,000 (5,000-15,000) | 20,000 (8,000-20,000) | ||
U – Mann-Whitney test, p – p value for association between different categories
Correlations between HBV-pregenomic RNA and different parameters
In group A, there was a statistically significant positive correlation between HBV pregenomic RNA and HBV DNA (p = 0.001) (Fig. 2). Meanwhile there was an insignificant relation between hepatitis B pregenomic RNA and other noninvasive markers; FIB-4, APRI (p = 0.156, 0.075 respectively).
Fig. 2.
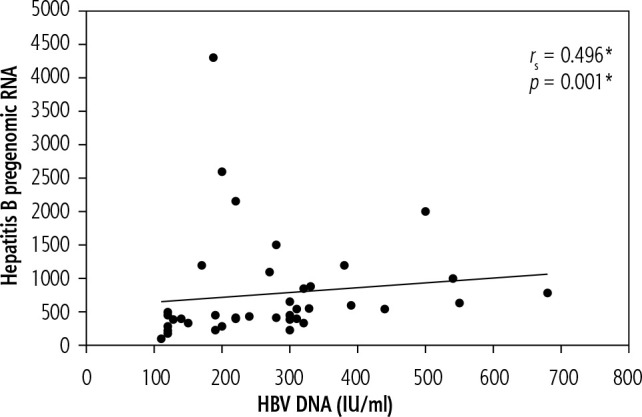
Correlation between HBV pregenomic RNA and HBV DNA in group A
In group B, before the start of treatment, there was a statistically significant positive correlation between HBV pregenomic RNA and HBV DNA (p < 0.001) (Fig. 3). Meanwhile there was an insignificant relation between hepatitis B pregenomic RNA and the other noninvasive markers: FIB-4, APRI (p = 0.810, 0.732 respectively).
Fig. 3.
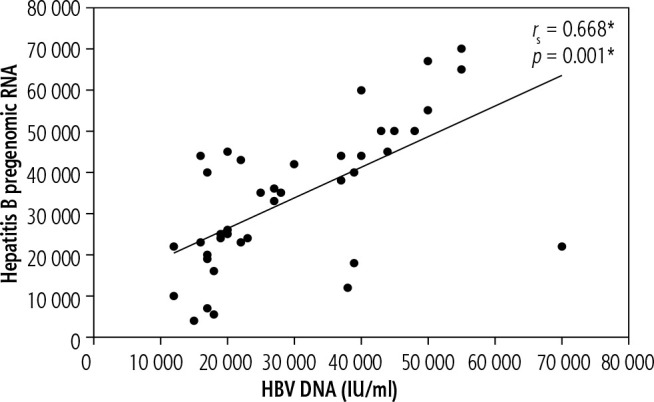
Correlation between HBV pregenomic RNA and HBV DNA in group B before the beginning of treatment
The sensitivity and specificity of hepatitis B pregenomic RNA to predict the disease activity (virological activity) – Figure 4 shows that at a cut-off value of 4000 of hepatitis B pregenomic RNA (IU/ml), the area under the curve was 0.999; the sensitivity, specificity, positive predictive values, negative predictive values and accuracy were 97.50 (Table 6).
Fig. 4.
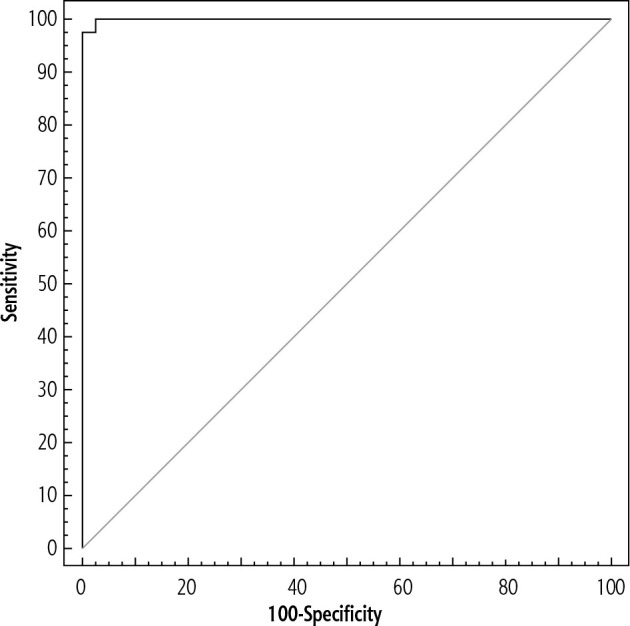
Agreement (sensitivity, specificity) for chronic active cases (group B) from chronic inactive cases (group A) (40/40)
Table 6.
Agreement (sensitivity, specificity) for chronic active cases (group B) from chronic inactive cases (group A) (40/40)
| AUC | p | 95% CI | Cut-off# | Sensitivity | Specificity | PPV | NPV | Accuracy | |
|---|---|---|---|---|---|---|---|---|---|
| Hepatitis B pregenomic RNA | 0.999 | < 0.001* | 0.997-1.001 | > 4,000# | 97.50 | 97.50 | 97.5 | 97.5 | 97.5 |
AUC – area under curve, p value – probability value, CI – confidence intervals, PPV – positive predictive value, NPV – negative predictive value, *statistically significant at p ≤ 0.05, #cut-off was choose according to Youden index
Discussion
Chronic HBV infection remains a global problem affecting millions of people worldwide. Entecavir (ETV) can inhibit replication of HBV and reduce serum HBV DNA to an undetectable level, but it is still difficult to achieve complete elimination of HBV due to the existence of cccDNA in the affected hepatocytes [11].
cccDNA is the template for the HBV mRNA transcripts; and the 3.5-kb pgRNA, template for reverse transcription and synthesis of the viral genome. Transcription of pgRNA from cccDNA is a key step in the HBV life cycle and the process is under the control of the HBV core promoter (CP) [12] which directs the production of two overlapping 3.5-kb viral transcripts [13, 14]. Translation of the longer precore (preC) mRNA leads to synthesis of the precore protein, precursor of the HBe antigen (HBeAg) [15]. The shorter pgRNA serves as mRNA for the core, polymerase genes and after encapsidation it serves as template for reverse transcription to produce viral DNA. Intracellular cycling of DNA containing nucleocapsids to the nucleus is important for cccDNA pool preservation [16].
Serum pregenomic RNA (pgRNA) is a new biomarker of chronic hepatitis B infection, where it is considered the main primer for viral particles and DNAs. Here, we focus on the role of HBV pregenomic RNA in guiding antiviral treatment including nucleot(s)ide analogues and as a marker of hepatitis activity [17].
Our study aim was to assess serum hepatitis B pregenomic RNA in patients with low viremia (HBV PCR less than 2000 IU/ml) to identify new criteria for treatment of those patients and high viremia (HBV PCR more than 2000 IU/ml) to determine new treatment endpoints.
Our results revealed that HBV pregenomic RNA was significantly higher in group B than in group A. Furthermore, there was a statistically significant positive correlation between serum HBV pregenomic RNA and HBV DNA in both groups.
In a study conducted by Halgand et al. [18], a highly positive significant correlation between high viremia and level of hepatitis B pregenomic RNA was found and it was consistent with our results.
In another study carried out by Malmstrom et al. [19], they found a highly positive significant correlation between pgRNA and viral DNA, revealing that reduction of viral load explains most of the reduction of pgRNA levels. These findings were consistent with the results of our research.
Ledesma et al. [20], confirmed the relation between viral load and hepatic damage in HBeAg negative patients, whereas Alam et al. [21], reported that a low level of viremia in some patients indicates progressive disease and does not always indicate improvement of the condition, which was consistent with the results of our research.
Therefore, measurement of serum HBV DNA and its correlation with liver function tests may provide a guide for the beginning and termination of treatment, but this needs to be augmented by a new biomarker for better assessment.
In another study conducted by Prakash et al. [22] serum HBV RNA from patients with chronic HBV was analyzed. A strong positive significant correlation between viral RNA and DNA was detected and these results were consistent with the results of our research.
In our study, serum HBV-pregenomic RNA was significantly lower after 6 months of therapy in group B patients. Surprisingly, although 35 patients became HBV-DNA negative they had detectable levels of HBV-pgRNA in their sera.
Lin et al. [23] investigated whether serum pgRNA is a strong marker that reflects HBV cccDNA values and analyzed the relation between serum pgRNA, serum HBV DNA, and HBsAg. They concluded that HBV pgRNA levels in serum can be a marker that reflects intrahepatic HBV cccDNA compared with serum HBV DNA and HBsAg. This is important for selection of proper timing for stoppage of antiviral medications, especially when HBV DNA levels are below the detection levels, and this was consistent with the results of our research.
In another study conducted by Shen et al. [24], it was found that HBV pgRNA levels increased after using nucleos(t)ide analogues (NA) treatment, which was not consistent with the final outcome of our research.
In another study conducted by Jansen et al. [25], during long-term NA treatment of chronic hepatitis B (CHB) patients, HBV RNA levels remained higher than HBV DNA levels. HBV RNA levels differed significantly from those of established viral markers during treatment, which highlights its importance as an independent marker in the assessment of patients with CHB, and that was consistent with the final outcome of our research.
In another study conducted by Li et al. [26], noninvasive fibrosis activity markers were used for assessment of hepatic fibrosis in chronic hepatitis B individuals. They aimed to determine the effect of antiviral therapy on APRI, FIB-4 and FibroScan, in patients with significant liver histological changes (SLHC). APRI, FIB-4 and FibroScan values decreased significantly after 36 months of ETV treatment, which indicates that these activity markers might be useful for following regression of hepatic fibrosis and determining treatment efficacy during long-term treatment, and that was consistent with the results of our research.
In an attempt to find a beneficial marker to be included in guidelines of chronic HBV treatment, we performed ROC curve analysis in this study; we observed that serum PgRNA at cutoff values more than 4000 IU/ml is associated with virological and biochemical disease activity; therefore future studies on a large scale might be of great value.
However, the limitations of our study are the short follow-up period, and a larger study group is recommended. Thus our results may provide a baseline for understanding the importance of serum pgRNA levels in predicting the course and outcome of chronic HBV infection.
Conclusions
From the results we can conclude that serum pgRNA might be used as a promising marker for assessment of HBV virological and biochemical activity. Moreover, persistence of serum pgRNA in those with undetectable HBV DNA after treatment highlights its advantage over HBV DNA as a future promising marker of monitoring therapeutic response as well as treatment endpoint.
Acknowledgements
The authors appreciate the consent of our patients to sharing their data.
Disclosure
The authors declare no conflict of interest.
References
- 1.Spinall EJ, Hawkins G, Fraser A, et al. Hepatitis B prevention, diagnosis, treatment and care. Occup Med (Lond) 2011; 61: 531-540. [DOI] [PubMed] [Google Scholar]
- 2.Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol 2007; 13: 14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levrero M, Pollicino T, Petersen J, et al. Control of cccDNA function in hepatitis B virus infection. J Hepatol 2009; 51: 581-592. [DOI] [PubMed] [Google Scholar]
- 4.van Campenhout MJ, van Bömmel F, Pfefferkorn M, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 2018; 68: 839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rehermann B, Ferrari C, Pasquinelli C, et al. The hepatitis B virus persists for decades after patients’ recovery from acute viral hepatitis despite active maintenance of a cytotoxic T-lymphocyte response. Nat Med 1996; 2: 1104-1108. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Zhou B, Valdes JD, et al. Serum hepatitis B virus RNA: A new potential Biomarker for chronic hepatitis B virus infection. Hepatology 2019; 69: 1816-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Association for the Study of the Liver . EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol 2017; 67: 370-398. [DOI] [PubMed] [Google Scholar]
- 8.European Association for Study of Liver Asociacion Latinoamericana para el Estudio Del Higado . EASL-ALEH Clinical Practice Guidelines: Noninvasive tests for evaluation of liver disease severity and prognosis. J Hepatol 2015; 63: 237-264. [DOI] [PubMed] [Google Scholar]
- 9.Gordillo RM, Gutiérrez J, Casal M. Evaluation of the COBAS TaqMan 48 real-time PCR system for quantitation of hepatitis B virus DNA. J Clin Microbiol 2005; 43: 3504-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlberg H, Bragstad K, Lindegren G, et al. Rapid bedside inactivation of ebola virus for safe nucleic acid tests. J Clin Microbiol 2016; 54: 2521-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devin R, Ivane J, Mindie H, et al. Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol 2018; 3: 383-403. [DOI] [PubMed] [Google Scholar]
- 12.Kramvis A, Kew MC. The core promoter of hepatitis B virus. J Viral Hepat 1999; 6: 415-427. [DOI] [PubMed] [Google Scholar]
- 13.Yaginuma K, Shirakata Y, Kobayashi M, et al. Hepatitis B virus (HBV)particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA 1987; 84: 2678-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Will H, Reiser W, Weimer T, et al. Replication strategy of human hepatitis B virus. J Virol 1987; 61: 904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Kishimoto S, Ohori K, et al. Molecular heterogeneity of e antigen polypeptides in sera fromcarriers of hepatitis B virus. J Immunol 1991; 147: 3156-3160. [PubMed] [Google Scholar]
- 16.Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 1986;47:451-460. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization . Global Hepatitis Report, 2017. Available from: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/ (accessed Aug, 2020).
- 18.Halgand B, Desterke C, Riviere L, et al. Hepatitis B virus pregenomic RNA in hepatocellular carcinoma: a nosological and prognostic determinant. Hepatol 2018; 67: 86-96. [DOI] [PubMed] [Google Scholar]
- 19.Malmström S, Larsson SB, Hannoun C, et al. Hepatitis B viral DNA decline at loss of HBeAg is mainly explained by reduced cccDNA load–down-regulated transcription of PgRNA has limited impact. PLoS One 2012; 7: e36349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ledesma MM, Galdame O, Bouzas B, et al. Characterization of the basal core promoter and precore regions in anti-HBe-positive inactive carriers of hepatitis B virus. Int J Infect Dis 2011; 15: e314-e320. [DOI] [PubMed] [Google Scholar]
- 21.Alam S, Ahmad N, Alam K, et al. Correlation between hepatitis B viral DNA load and extent of liver pathology in patients with chronic hepatitis B. Hepat Mon 2008; 8: 185-189. [Google Scholar]
- 22.Prakash K, Rydell GE, Larsson SB, et al. High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles. Virol J 2018; 15: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin N, Ye A, Wu S, et al. Diagnostic value of detection of pregenomic RNA in sera of hepatitis B virus-infected patients with different clinical outcomes. J Clin Microbiol 2020; 58: e01275-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen T, Wang J, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol 2016; 65: 700-710. [DOI] [PubMed] [Google Scholar]
- 25.Jansen L, Kootstra NA, Van Dort KA, et al. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon alfa-2a and nucleos(t)ide analogues. J Infect Dis 2016; 213: 224-332. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Chen L, Zhou Y. Changes of FibroScan, APRI, and FIB4 in chronic hepatitis B patients with significant liver histological changes receiving 3 year entecavir therapy. Clin Exp Med 2018; 18: 273-282. [DOI] [PubMed] [Google Scholar]